Abstract
Purpose
Although the prevalence of community-acquired respiratory bacterial coinfection upon hospital admission in patients with coronavirus disease 2019 (COVID-19) has been reported to be < 5%, almost three-quarters of patients received antibiotics. We aim to investigate whether procalcitonin (PCT) or C-reactive protein (CRP) upon admission could be helpful biomarkers to identify bacterial coinfection among patients with COVID-19 pneumonia.
Methods
We carried out a multicentre, observational cohort study including consecutive COVID-19 patients admitted to 55 Spanish intensive care units (ICUs). The primary outcome was to explore whether PCT or CRP serum levels upon hospital admission could predict bacterial coinfection among patients with COVID-19 pneumonia. The secondary outcome was the evaluation of their association with mortality. We also conducted subgroups analyses in higher risk profile populations.
Results
Between 5 February 2020 and 21 December 2021, 4076 patients were included, 133 (3%) of whom presented bacterial coinfection. PCT and CRP had low area under curve (AUC) scores at the receiver operating characteristic (ROC) curve analysis [0.57 (95% confidence interval (CI) 0.51–0.61) and 0.6 (95% CI, 0.55–0.64), respectively], but high negative predictive values (NPV) [97.5% (95% CI 96.5–98.5) and 98.2% (95% CI 97.5–98.9) for PCT and CRP, respectively]. CRP alone was associated with bacterial coinfection (OR 2, 95% CI 1.25–3.19; p = 0.004). The overall 15, 30 and 90 days mortality had a higher trend in the bacterial coinfection group, but without significant difference. PCT ≥ 0.12 ng/mL was associated with higher 90 days mortality.
Conclusion
Our study suggests that measurements of PCT and CRP, alone and at a single time point, are not useful for ruling in or out bacterial coinfection in viral pneumonia by COVID-19.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
In critically ill patients with pneumonia associated to coronavirus disease 2019 a single determination of either procalcitonin or C-reactive protein, at a single time point, is not enough to rule out a bacterial coinfection. |
Introduction
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS‑CoV‑2), had a strong impact on healthcare systems worldwide [1]. Patients with severe pneumonia present high mortality due to the large number of complications they face [2]. Particularly, bacterial respiratory coinfection is a complication extensively studied in the field of virus pandemics such as severe influenza H1N1 pneumonia and is reported to be associated with the severity of illness, worse outcomes, and even with an increased risk of mortality [3].
In COVID-19 the prevalence of community-acquired bacterial respiratory coinfections (from now, bacterial coinfection) is reported to be inferior to 5% upon hospital admission [4,5,6]. Although the impact on mortality of this complication is still widely debated, almost three-quarters of patients with COVID-19 receive antibiotics, so prescribing is significantly higher than the estimated prevalence of bacterial coinfection [7]. Rule out bacterial coinfection upon admission to the hospital in these patients could potentially limit antibiotics overuse and so reduce hospitalization costs, potential side-effects and the development of multi-drug resistant infections [8].
Procalcitonin (PCT) is a serum biomarker that has shown promise in discriminating between viral and bacterial infections [9, 10], being routinely used in combination with clinical evidences as a guide to antibiotic de-escalation/discontinuation in critically ill patients [11, 12]. The role of PCT in community-acquired pneumonia (CAP) has been evaluated in multiple studies, with no scientific consensus on its usefulness for diagnosis or antibiotic initiation [13, 14]. On the other hand, PCT showed promising results in the early recognition of bacterial coinfection in influenza pneumonia [15].
C-reactive protein (CRP), another well-known systemic marker of inflammation and tissue damage, is extensively used by clinicians to monitor infections [16]. In patients with COVID-19, admission CRP correlated with disease severity and tended to be a good predictor of adverse outcome [17]. Furthermore, in patients with CAP PCT is reported to be superior when assessing disease severity in the first 48 h since symptoms onset whereas CRP is a better choice in patients with symptoms for 3 or more days [18].
We therefore aimed to determine the role of PCT and CRP in the identification of bacterial coinfection among patients with COVID-19 pneumonia.
Methods
Study design and patients
We retrospectively analysed patients from the CIBERESUCICOVID study (NCT04457505), which had prospectively included consecutive patients aged ≥ 18 years with laboratory-confirmed SARS-CoV-2 infection from across 55 Spanish intensive care units (ICUs) between 5 February 2020 and 21 December 2021 (Supplementary Table 1). All consecutive patients admitted to the intensive care unit (ICU) were enrolled if reason for admission was COVID-19. Exclusion criteria for patients included: (1) unconfirmed SARS-CoV-2 infection; (2) lack of data at baseline or hospital discharge; (3) lack of data on testing for bacterial infections and on the presence of bacterial pneumonia during hospital stay; (4) lack of data on both PCT and CRP at hospital and ICU admission; (5) patient transfer from another ICU or nursing-home; (6) antibiotic administration previous of hospital admission was also an exclusion criterion for patients not presenting a bacterial coinfection, in order to rule out possible false negative.
The study received approval by the Institution’s Internal Review Board (Comité Ètic d’Investigació Clínica, registry number HCB/2020/0370). Local researchers maintained contact with a study team member, and participating hospitals obtained local ethics committee approval. We reported results in accordance with the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) guidelines [19].
Data collection and definitions
We collected data on demographics, comorbidities, clinical symptoms, and treatment. Standard laboratory and clinical data were collected at hospital and ICU admission, particularly PCT and CRP levels. The pharmacological treatments administered and interventions performed during hospital admission until either discharge from hospital or death were also collected, as well as outcomes [20].
Bacterial coinfection was defined as any respiratory bacterial infection diagnosed by the treating physicians within 48 h since hospital admission, taking into account clinical, laboratory and imaging evidences, with the isolation of at least one respiratory pathogen in the blood or in a good quality respiratory sample (sputum, tracheal aspirate, or bronchoalveolar lavage) and/or positive urinary antigens for Streptococcus pneumoniae or Legionella pneumophila. Infections occurring later than 48 h since hospital admission were considered as nosocomial infections and not included in the present analysis [21]. Patients were then classified according to whether or not the diagnosis of bacterial coinfection was met.
Primary and secondary outcomes
The primary outcome was to investigate whether PCT or CRP serum levels upon admission could predict bacterial coinfection among patients with COVID-19 pneumonia. The secondary outcome was to evaluate the association of PCT and CRP with 30 days and 90 days mortality.
Biomarkers in higher risk profile subpopulations and according to symptoms onset
In order to evaluate the clinical usefulness of PCT and CRP in patients with an higher risk profile for bacterial coinfection, we conducted subgroups analyses considering the presence of septic shock and non-respiratory Sequential Organ Failure Assessment (SOFA) score at hospital admission.
Furthermore, as PCT and CRP present different kinetics, we explored the potential correlation between biomarkers values and time since symptoms onset, by dividing the population in ≤ 48 h and > 48 h from symptoms onset to hospital admission.
Statistical analysis
We reported the number and percentage of patients as categorical variables, and the median [first quartile (Q1); third quartile (Q3)] as continuous variables. Categorical variables were compared using the chi-squared test or Fisher’s exact test, whereas continuous variables were compared using the nonparametric Mann–Whitney U test.
To determine the predictive capacity of PCT and CRP at hospital admission for bacterial coinfection, we determined sensitivity, specificity, positive and negative predictive value (NPV), and positive and negative likelihood ratio (LR) [22,23,24], along with the 95% confidence intervals (CIs). We performed these analyses using different PCT and CRP cut-off values based on different clinical significance thresholds.
We analysed the association between PCT and CRP at hospital admission (at 48 h) and bacterial coinfection by means of a mixed-effects multivariable model [25, 26], defined by a binomial probability distribution and a logit link function, with centres as a random effect. The multivariable model included the following variables based on clinical relevance only: age, sex, body mass index, comorbidities, SOFA (without respiratory system component), PaO2/FiO2 ratio at hospital admission, leukocytosis (≥ 11 × 109/L) at hospital admission, PCT at hospital admission, CRP at hospital admission and COVID-19 wave. Odds ratios (ORs) and their 95% CIs were calculated. Discrimination was assessed by the area under the receiver operating characteristic curve (AUC).
To describe 30 days mortality, we utilized a Cox regression model stratified on the centre variable [27]. A list of candidate predictors was established a priori based on previous findings and clinical constraints: age, sex, body mass index, comorbidities, SOFA (without respiratory system component), PaO2/FiO2 ratio at hospital admission PCT at hospital admission, CRP at hospital admission, leukocytosis (≥ 11 × 109/L) at hospital admission, respiratory support at hospital admission, septic shock at hospital admission, bacterial coinfection and COVID-19 wave. Hazard ratios (HRs) and their 95% CIs were calculated. Proportional hazards assumptions were tested with log minus log plots. Patients who were transferred to another hospital were censored in survival analyses. The same survival analysis was performed for 30 days mortality.
Single collinearity in multivariable analyses was evaluated using the Pearson correlation (r) and multicollinearity was examined by means of the variance inflation factor (VIF).
To account for missing data we used the multiple imputation method [28]; further details regarding the employed imputation methodology are presented in Supplementary Table 2.
In addition, subgroup analysis were carried out taking into account the presence of septic shock, non-respiratory SOFA score (i.e., < 2 and ≥ 2) and time from first symptoms to hospital admission [i.e., early admission (≤ 48 h) and late admission (> 48 h)].
The level of significance was set at 0.05 (two-tailed). No adjustments were made for multiple comparisons. All analyses were performed using IBM SPSS version 26.0 (IBM Corp., Armonk, NY, USA) and R version 4.1.1 (R Foundation for Statistical Computing, Vienna, Austria).
Results
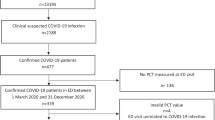
Between 5 February 2020 and 21 December 2021, 6225 patients with COVID-19 were admitted to 55 ICUs. We included 4076 patients in this analysis, 133 (3%) of whom presented a community-acquired respiratory coinfection (Supplementary Fig. 1).
Demographics and characteristics
Patient characteristics at hospital admission are reported in Table 1. Patients without bacterial coinfection presented less comorbidities than patient with coinfection (45 vs. 55%; p = 0.019). Time from symptoms onset to ICU admission and from hospital to ICU admission were both shorter for the bacterial coinfection group. At hospital admission, patients with bacterial coinfection had lower PaO2/FiO2 ratio [200 (105; 277) vs. 165 (100; 237; 0.017)] and they were more likely to undergo invasive mechanical ventilation (12 vs. 44%; p < 0.001); they also had higher leukocyte, neutrophil and platelet counts.
Only 10 patients out of the 133 with bacterial coinfection received an empirical antibiotic treatment before hospital admission.
Supplementary Table 3 reports data on antibiotic prescription, with 74% of patients without bacterial coinfection receiving antibiotics within the first 48 h from hospital admission.
Data on microorganisms isolated in the coinfection group are displayed in Supplementary Table 4. The most commonly found bacteria were Staphylococcus aureus, Streptococcus pneumoniae and Haemophilus influenzae.
Supplementary Table 5 reports data on the sampling techniques used to isolate the microorganisms responsible for the bacterial coinfections.
Primary outcome
In our population, median values of PCT [0.19 (0.1; 0.44) vs. 0.26 (0.12; 0.94) ng/mL; p = 0.01] and CRP [131 (70; 213) vs. 163 (109; 242) mg/L; p < 0.001] were significantly higher in the bacterial coinfection group (Table 1).
In order to determine the best PCT and CRP values to identify bacterial coinfection, we analysed different cut-off points from the receiver operating characteristic (ROC) curve, resulting in PCT ≥ 0.12 ng/mL and CRP ≥ 97 mg/L being the best thresholds to identify bacterial coinfection with a sensitivity higher than 80% (Table 2); both biomarkers had low AUC scores at the ROC curve analysis [0.57 (95% CI 0.51–0.61) and 0.6 (95% CI, 0.55–0.64) for PCT and CRP, respectively] (Supplementary Fig. 2), but high NPV [97.5% (95% CI 96.5–98.5) and 98.2% (95% CI 97.5–98.9) for PCT and CRP, respectively] (Table 2).
In the multivariable models we analysed the association between single biomarkers levels or their combination and bacterial coinfection. In Model 1 PCT ≥ 0.12 was not associated with coinfection (OR 1.11, 95% CI 0.69–1.79; p = 0.659), while a significance was detected both for CRP alone and, in model 2, for the combination of the two biomarkers (OR 2.0, 95% CI 1.25–3.19; p = 0.004 and OR 1.72, 95% CI 1.15–2.56; p = 0.008 for CRP in Model 1 and for the combination of PCT and CRP in Model 2) (Table 3).
Secondary outcome
The overall 15, 30 and 90 days mortality had a higher trend in the bacterial coinfection group, but without significant difference, as shown in Supplementary Table 6. In the multivariable models assessing predictors of 30 days mortality neither PCT ≥ 0.12 ng/mL nor CRP ≥ 97 mg/L showed a significant association with higher mortality (OR 1.17, 95% CI 0.97–1.40; p = 0.094 and OR 1.02, 95% CI 0.87–1.19; p = 0.839 for PCT and CRP respectively) (Supplementary Table 7), while for the 90-day mortality a positive signal was only detected for PCT (OR 1.2, 95% CI 1.03–1.4; p = 0.022)(Supplementary Table 8).
Biomarkers in higher risk profile subpopulations and according to symptoms onset
In order to evaluate the performance of PCT and CRP in patients with a higher risk profile, we conducted subgroup analysis considering non-respiratory SOFA score and the presence of septic shock.
The analysis of the cut-off points from the ROC curves was repeated, in order to identify the biomarkers thresholds with an 80% sensitivity for bacterial coinfection.
In patients presenting septic shock at hospital admission, levels of PCT ≥ 0.11 ng/mL where associated with bacterial coinfection, but with a very wide confidence interval (OR 81.1, 95% CI 1.71–3850.77; p = 0.026) (Table 4 and Supplementary Table 9). At the ROC curve analysis for identification of bacterial coinfection, PCT had an AUC of 0.82 (95% CI 0.67–0.98) in patients with septic shock (Fig. 1 and Supplementary Fig. 3).
Conversely, in patients with a non-respiratory SOFA ≥ 2, only CRP > 107 mg/L was associated with bacterial coinfection (OR 2.27, 95% CI 1.08–4.75; p = 0.03) (Table 4 and Supplementary Table 10), but with an AUC of 0.6 (95% CI 0.53–0.67) at the ROC curve analysis (Fig. 1 and Supplementary Fig. 4). CRP was also associated with bacterial coinfection in patients without septic shock (OR 2.08, 95% CI 1.24–3.51; p = 0.006) and with a non-respiratory SOFA < 2 (OR 3.18, 95% CI 1.32–7.62; p = 0.010).
Finally, acknowledging differences in PCT and CRP kinetics, we explored the potential correlation between time since symptoms onset and biomarkers values.
Supplementary Fig. 5 displays differences in PCT and CRP values at hospital admission, stratified according to time from symptoms onset. In patients with bacteria coinfection presenting symptoms from < 3 days, PCT had higher values (p = 0.006). When comparing the two study populations, coinfected patients had higher PCT and CRP in the ≤ 48 h group (PCT p = 0.001, CRP p = 0.027), while in the > 48 h group an increase from the baseline was shown only for CRP values (p = 0.001).
In the multivariable models for association with bacterial coinfection we found no significant association between biomarkers thresholds and bacterial coinfection, but with a positive trend for PCT ≥ 0.44 ng/mL in the early symptoms group (OR 14.97; 95% CI 0.96–234.03; p = 0.054)(Table 4 and Supplementary Table 11). At the ROC curve analysis for identification of bacterial coinfection, PCT had an AUC of 0.76 (95% CI 0.61–0.92) in the group with ≤ 48 h since symptoms onset (Fig. 1 and Supplementary Fig. 6).
Discussion
In this large cohort of critically ill patient with COVID-19 we found that the combined values of PCT and CRP upon admission, as well as CRP values alone, were associated with bacterial respiratory coinfection.
However, our results suggest that a single determination of these biomarkers is not enough to identify a bacterial coinfection, as their significance should always be contextualized with a thorough clinical evaluation [29].
In our population, the incidence of bacterial coinfection was 3% and the more commonly isolated bacteria were S. aureus and S. pneumoniae, consistently with the previous studies [4,5,6].
Although PCT and CRP alone had low accuracy in predicting bacterial coinfection based on the ROC curve analysis (AUC of 0.57 and 0.6 for PCT and CRP, respectively), likely due to low specificity, our study found that CRP alone and the combination of PCT and CRP were still associated with bacterial coinfection in the multivariable model. Moreover, the identified biomarkers thresholds demonstrated a sensitivity greater than 80% and high negative predictive values, suggesting that values of PCT < 0.12 ng/mL and CRP < 97 mg/L may still be useful to rule out bacterial coinfection, similarly to what was found by Ingram et al. in patients with severe 2009 H1N1 influenza infection [30]. Notably, in order to address the possible bias created by the derivation of thresholds values and evaluation of diagnostic test accuracy in the same population, we reported commonly used biomarkers cut-offs in Table 2, showing the consistency of our thresholds [31,32,33].
When interpreting these results it is important to consider that serum CRP is an acute-phase protein and it rises whenever an inflammatory process is present [34]. In a recent study it was found that high CRP levels were a good predictor of CAP in patient with clinical diagnosis and false-negative chest X-ray, confirming that very high levels of this biomarker are more specific for bacterial infection [35, 36]. Nevertheless, its serum concentration depends on the intensity of the stimulus and, at the early stage of COVID-19, CRP levels were found to be positively correlated with lung lesions [37]. Procalcitonin conversely has higher sensitivity and specificity as compared to CRP for differentiating bacterial from viral infections [38].
Clearly a single biomarker could never capture the complex and variable interactions of an infection and its values should be interpreted in light of the overall clinical context [29]; in order to evaluate the clinical usefulness of PCT and CRP, we therefore assessed the association between biomarkers and bacterial coinfection in patients with an higher risk profile for coinfection. We found an association between values of PCT ≥ 0.11 ng/mL and bacterial coinfection in patients presenting septic shock, with a very wide confidence interval (OR 81.1, 95% CI 1.71–3850.77; p = 0.026) but with an AUC of 0.82 at the ROC curve analysis. Conversely, CRP was associated with bacterial coinfection in various subgroups throughout the models (no septic shock, non-respiratory SOFA both under and over 2), but always with low AUC scores. These results are in favour of a stronger connection between PCT levels and bacterial coinfection, while CRP seems to have a lower discriminatory ability [31, 38, 39]. However, given the high number of variables we adjusted for and the small sample size of the subgroups, these analysis have low statistical power and must be discussed with caution.
When considering symptoms timing, in particular the first 48 h since symptoms onset, we found that PCT had an AUC of 0.76 (95% CI 0.61–0.92), with no significant association but with a positive trend between bacterial coinfection and values of PCT ≥ 0.44 ng/mL, suggesting improved discrimination abilities in the first hours since symptoms onset and the possible application of an higher cut off as already proposed by Bouadma et al. [35] in the PRORATA trial.
The importance of taking into account the kinetics of PCT and CRP has already been stated by Méndez et al. [18], who showed that the inflammatory response at the time of CAP diagnosis was influenced by the time since symptoms onset, with a greater expression of PCT in the first 2 days and slower kinetics for CRP. Our results may agree with their conclusions, suggesting how PCT could be a more reliable indicator in the initial phase of infection.
According to the meta-analysis of Langford et al. [7] during the COVID-19 pandemic there has been a reckless use of antibiotics: they were prescribed in three-quarters of patients and, given the low rate of coinfections, most of them were unnecessarily administered. In our study the number of patients treated with antibiotics was accordingly high, with 74% rate of administration in patients without bacterial coinfection within the first 48 h from hospital admission. In light of these results, finding a way to predict the presence of bacterial coinfection at hospital admission becomes even more crucial, to improve antibiotic stewardship and picture the possible outcomes.
In our population patients with a bacterial coinfection were admitted earlier to the ICU and had longer ICU stay, reflecting the increased severity of the disease. Mortality had a higher trend in coinfected patients (15 days mortality 11 vs 15%, 30 days mortality 22 vs 26%, 90 days mortality 31 vs 37%, respectively in not coinfected and coinfected patients), but without significant differences. In our opinion these result may be linked to several factors: first of all the high rate of antibiotic treatment at hospital admission in the coinfected group, therefore the high number of patients appropriately treated; secondly, treated bacterial coinfections may not have had a more significant impact on mortality than other complications, as age, comorbidities, immunosuppression, lack of knowledge and resources reportedly had, especially during the first two waves on the pandemic. In the multivariable models neither PCT nor CRP were associated with 30 days mortality, but PCT ≥ 0.12 ng/mL was associated with 90 days mortality, showing an overall positive trend, accordingly to what already found by Carbonell et al. [40].
Major strengths of this study include its multicentre nature, the consecutive inclusion of all patients from each intensive care unit, the high number of patients analysed and long-term follow-up. We also considered antibiotics administered previously of hospital admission: we excluded from the no bacterial coinfection group patients that had been taking antibiotics, in order to avoid the inclusion in this group of those with an appropriately treated bacterial coinfection.
We acknowledge the presence of some limitations that should be taken into account. First of all, the poor sensitivity and incomplete use of diagnostic tests, that represent a critical barrier to coinfection detection and antibiotic stewardship [41]. It has been reported that identification of the pathogen responsible for community-acquired pneumonia may be elusive, with less than 50% of cases having a confirmed aetiology [42, 43]. In our population bacterial coinfection was more common in patients intubated at hospital admission, which could be due to illness severity SOFA < 2 but also to a more invasive sampling strategy. By limiting the inclusion in the bacterial coinfection group to patient with a microbiologically confirmed aetiology, we might have misclassified some no-coinfected patient and created a bias. However, this limitation did not affect haemocultures and urinary antigens testing and the prevalence of bacterial coinfection observed in our population is similar to that reported in the previous studies [4, 5, 30]. Second, we didn’t include radiological evidence and data on the microbiological work-up in our analysis, as the CIBERESUCICOVID database was not designed to register these information. However, radiological evidences were taken into account by clinicians at the time of diagnosis of the bacterial coinfection.
Third, in the subgroup analysis we considered the variable “time since symptoms onset”, which is necessarily self-reported, dependent on the subjective perception of symptoms and therefore susceptible to bias; however, the exact onset of symptoms is not otherwise objectifiable for a community-acquired infection [18].
Fourth, we acknowledge limitations associated to the retrospective design of the study and the plausible influence on our results of selecting patients from different waves of the pandemic, as well as those connected to the observational nature of the study and missing data. Particularly, in our population the rate of missing values was higher for PCT when compared with CRP, as the former is not routinely measured. The impact of PCT-based algorithms on antibiotic sparing, and therefore their cost-effectiveness, is still debated [11]; the decision to measure PCT is normally based on the clinician request, introducing a possible bias in observational studies involving this biomarker. However, we have adjusted all of our analyses for these confounders.
Finally, our results bring new interesting aspects to the interpretation of biomarkers in the context of bacterial coinfection, but they refer to COVID-19 patients and may not be applicable in other clinical settings.
Conclusions
Our study suggests that measurements of PCT and CRP, alone and at a single time point, are unfortunately not useful for ruling in or out bacterial coinfection in viral pneumonia by COVID-19.
Considering the combined values of these biomarkers, as well as symptoms timing and the broader clinical context, appears to be relevant in their interpretation and therefore in the design of further studies.
Data Availability
Access to each dataset for research purposes may be available upon reasonable request.
Abbreviations
- AUC:
-
Area under the curve
- CAP:
-
Community-acquired pneumonia
- CI:
-
Confidence interval
- CRP:
-
C-reactive protein
- HR:
-
Hazard ratio
- ICU:
-
Intensive care unit
- LR:
-
Likelihood ratio
- NPV:
-
Negative predictive value
- OR:
-
Odds ratio
- PCT:
-
Procalcitonin
- ROC:
-
Receiving-operating characteristic
- VIF:
-
Variance inflation factor
References
Arsenault C, Gage A, Kim MK, Kapoor NR, Akweongo P, Amponsah F, Aryal A, Asai D, Awoonor-Williams JK, Ayele W, Bedregal P, Doubova SV, Dulal M, Gadeka DD, Gordon-Strachan G, Mariam DH, Hensman D, Joseph JP, Kaewkamjornchai P, Eshetu MK, Gelaw SK, Kubota S, Leerapan B, Margozzini P, Mebratie AD, Mehata S, Moshabela M, Mthethwa L, Nega A, Oh J, Park S, Passi-Solar A, Perez-Cuevas R, Phengsavanh A, Reddy T, Rittiphairoj T, Sapag JC, Thermidor R, Tlou B, Valenzuela Guinez F, Bauhoff S, Kruk ME (2022) COVID-19 and resilience of healthcare systems in ten countries. Nat Med 28:1314–1324
Mahendra M, Nuchin A, Kumar R, Shreedhar S, Mahesh PA (2021) Predictors of mortality in patients with severe COVID-19 pneumonia—a retrospective study. Adv Respir Med 89:135–144
MacIntyre CR, Chughtai AA, Barnes M, Ridda I, Seale H, Toms R, Heywood A (2018) The role of pneumonia and secondary bacterial infection in fatal and serious outcomes of pandemic influenza a(H1N1)pdm09. BMC Infect Dis 18:637
Westblade LF, Simon MS, Satlin MJ (2021) Bacterial coinfections in coronavirus disease 2019. Trends Microbiol 29:930–941
Langford BJ, So M, Raybardhan S, Leung V, Westwood D, MacFadden DR, Soucy JR, Daneman N (2020) Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin Microbiol Infect 26:1622–1629
Langford BJ, So M, Simeonova M, Leung V, Lo J, Kan T, Raybardhan S, Sapin ME, Mponponsuo K, Farrell A, Leung E, Soucy JR, Cassini A, MacFadden D, Daneman N, Bertagnolio S (2023) Antimicrobial resistance in patients with COVID-19: a systematic review and meta-analysis. Lancet Microbe 4:e179–e191
Langford BJ, So M, Raybardhan S, Leung V, Soucy JR, Westwood D, Daneman N, MacFadden DR (2021) Antibiotic prescribing in patients with COVID-19: rapid review and meta-analysis. Clin Microbiol Infect 27:520–531
Rothe K, Feihl S, Schneider J, Wallnofer F, Wurst M, Lukas M, Treiber M, Lahmer T, Heim M, Dommasch M, Waschulzik B, Zink A, Querbach C, Busch DH, Schmid RM, Schneider G, Spinner CD (2021) Rates of bacterial co-infections and antimicrobial use in COVID-19 patients: a retrospective cohort study in light of antibiotic stewardship. Eur J Clin Microbiol Infect Dis 40:859–869
Assicot M, Gendrel D, Carsin H, Raymond J, Guilbaud J, Bohuon C (1993) High serum procalcitonin concentrations in patients with sepsis and infection. Lancet 341:515–518
Gilbert DN (2011) Procalcitonin as a biomarker in respiratory tract infection. Clin Infect Dis 52(Suppl 4):S346-350
Pepper DJ, Sun J, Rhee C, Welsh J, Powers JH 3rd, Danner RL, Kadri SS (2019) Procalcitonin-guided antibiotic discontinuation and mortality in critically Ill adults: a systematic review and meta-analysis. Chest 155:1109–1118
Evans L, Rhodes A, Alhazzani W, Antonelli M, Coopersmith CM, French C, Machado FR, McIntyre L, Ostermann M, Prescott HC, Schorr C, Simpson S, Wiersinga WJ, Alshamsi F, Angus DC, Arabi Y, Azevedo L, Beale R, Beilman G, Belley-Cote E, Burry L, Cecconi M, Centofanti J, Coz Yataco A, De Waele J, Dellinger RP, Doi K, Du B, Estenssoro E, Ferrer R, Gomersall C, Hodgson C, Moller MH, Iwashyna T, Jacob S, Kleinpell R, Klompas M, Koh Y, Kumar A, Kwizera A, Lobo S, Masur H, McGloughlin S, Mehta S, Mehta Y, Mer M, Nunnally M, Oczkowski S, Osborn T, Papathanassoglou E, Perner A, Puskarich M, Roberts J, Schweickert W, Seckel M, Sevransky J, Sprung CL, Welte T, Zimmerman J, Levy M (2021) Surviving sepsis campaign: international guidelines for management of sepsis and septic shock 2021. Intensive Care Med 47:1181–1247
Metlay JP, Waterer GW, Long AC, Anzueto A, Brozek J, Crothers K, Cooley LA, Dean NC, Fine MJ, Flanders SA, Griffin MR, Metersky ML, Musher DM, Restrepo MI, Whitney CG (2019) Diagnosis and treatment of adults with community-acquired pneumonia. An official clinical practice guideline of the American Thoracic Society and Infectious Diseases Society of America. Am J Respir Crit Care Med 200:e45–e67
Self WH, Balk RA, Grijalva CG, Williams DJ, Zhu Y, Anderson EJ, Waterer GW, Courtney DM, Bramley AM, Trabue C, Fakhran S, Blaschke AJ, Jain S, Edwards KM, Wunderink RG (2017) Procalcitonin as a marker of etiology in adults hospitalized with community-acquired pneumonia. Clin Infect Dis 65:183–190
Rodriguez AH, Aviles-Jurado FX, Diaz E, Schuetz P, Trefler SI, Sole-Violan J, Cordero L, Vidaur L, Estella A, Pozo Laderas JC, Socias L, Vergara JC, Zaragoza R, Bonastre J, Guerrero JE, Suberviola B, Cilloniz C, Restrepo MI, Martin-Loeches I, Group SGW (2016) Procalcitonin (PCT) levels for ruling-out bacterial coinfection in ICU patients with influenza: a CHAID decision-tree analysis. J Infect 72:143–151
Clyne B, Olshaker JS (1999) The C-reactive protein. J Emerg Med 17:1019–1025
Luo X, Zhou W, Yan X, Guo T, Wang B, Xia H, Ye L, Xiong J, Jiang Z, Liu Y, Zhang B, Yang W (2020) Prognostic value of C-reactive protein in patients with coronavirus 2019. Clin Infect Dis 71:2174–2179
Mendez R, Menendez R, Cilloniz C, Amara-Elori I, Amaro R, Gonzalez P, Posadas T, Gimeno A, Espana PP, Almirall J, Torres A (2018) Initial inflammatory profile in community-acquired pneumonia depends on time since onset of symptoms. Am J Respir Crit Care Med 198:370–378
Cuschieri S (2019) The STROBE guidelines. Saudi J Anaesth 13:S31–S34
Torres A, Motos A, Ceccato A, Bermejo-Martin J, de Gonzalo-Calvo D, Perez R, Barroso M, Pascual IZ, Gonzalez J, Fernandez-Barat L, Ferrer R, Riera J, Garcia-Gasulla D, Penuelas O, Lorente JA, Almansa R, Menendez R, Kiarostami K, Canseco J, Villar RA, Anon JM, Marino AB, Barbera C, Barberan J, Ortiz AB, Boado MV, Bustamante-Munguira E, Caballero J, Canton-Bulnes ML, Perez CC, Carbonell N, Catalan-Gonzalez M, de Frutos R, Franco N, Galban C, Gumucio-Sanguino VD, Torre MDC, Diaz E, Estella A, Gallego E, Garmendia JLG, Gomez JM, Huerta A, Garcia RNJ, Loza-Vazquez A, Marin-Corral J, Delgado MCM, Gandara AM, Varela IM, Messa JL, Albaiceta GM, Nieto M, Novo MA, Penasco Y, Perez-Garcia F, Pozo-Laderas JC, Ricart P, Sagredo V, Sanchez-Miralles A, Chinesta SS, Serra-Fortuny M, Socias L, Sole-Violan J, Suarez-Sipmann F, Lomas LT, Trenado J, Ubeda A, Valdivia LJ, Vidal P, Barbe F, Project C (2022) Methodology of a large multicenter observational study of patients with COVID-19 in Spanish intensive care units. Arch Bronconeumol 58(Suppl 1):22–31
Torres A, Niederman MS, Chastre J, Ewig S, Fernandez-Vandellos P, Hanberger H, Kollef M, Li Bassi G, Luna CM, Martin-Loeches I, Paiva JA, Read RC, Rigau D, Timsit JF, Welte T, Wunderink R (2017) International ERS/ESICM/ESCMID/ALAT guidelines for the management of hospital-acquired pneumonia and ventilator-associated pneumonia: guidelines for the management of hospital-acquired pneumonia (HAP)/ventilator-associated pneumonia (VAP) of the European Respiratory Society (ERS), European Society of Intensive Care Medicine (ESICM), European Society of Clinical Microbiology and Infectious Diseases (ESCMID) and Asociacion Latinoamericana del Torax (ALAT). Eur Respir J. https://doi.org/10.1183/13993003.00582-2017
Deeks JJ, Altman DG (2004) Diagnostic tests 4: likelihood ratios. BMJ 329:168–169
Altman DG, Bland JM (1994) Diagnostic tests. 1: sensitivity and specificity. BMJ 308:1552
Altman DG, Bland JM (1994) Diagnostic tests 2: predictive values. BMJ 309:102
Andrew Gelman JH (2007) Data analysis using regression and multilevel/Hierarchical models. Cambridge University Press, New York
James W, Hardin JMH (2007) Generalized linear models and extensions, 2nd edn. Stata Press
Collett D (2003) Modelling survival data in medical research. Chapman and Hall/CRC
Sterne JA, White IR, Carlin JB, Spratt M, Royston P, Kenward MG, Wood AM, Carpenter JR (2009) Multiple imputation for missing data in epidemiological and clinical research: potential and pitfalls. BMJ 338:b2393
Povoa P, Coelho L, Dal-Pizzol F, Ferrer R, Huttner A, Conway Morris A, Nobre V, Ramirez P, Rouze A, Salluh J, Singer M, Sweeney DA, Torres A, Waterer G, Kalil AC (2023) How to use biomarkers of infection or sepsis at the bedside: guide to clinicians. Intensive Care Med 49:142–153
Ingram PR, Inglis T, Moxon D, Speers D (2010) Procalcitonin and C-reactive protein in severe 2009 H1N1 influenza infection. Intensive Care Med 36:528–532
Bouadma L, Luyt CE, Tubach F, Cracco C, Alvarez A, Schwebel C, Schortgen F, Lasocki S, Veber B, Dehoux M, Bernard M, Pasquet B, Regnier B, Brun-Buisson C, Chastre J, Wolff M, Group Pt (2010) Use of procalcitonin to reduce patients’ exposure to antibiotics in intensive care units (PRORATA trial): a multicentre randomised controlled trial. Lancet 375:463–474
Petel D, Winters N, Gore GC, Papenburg J, Beltempo M, Lacroix J, Fontela PS (2018) Use of C-reactive protein to tailor antibiotic use: a systematic review and meta-analysis. BMJ Open 8:e022133
Linnet K, Bossuyt PM, Moons KG, Reitsma JB (2012) Quantifying the accuracy of a diagnostic test or marker. Clin Chem 58:1292–1301
Povoa P (2002) C-reactive protein: a valuable marker of sepsis. Intensive Care Med 28:235–243
Claessens YE, Debray MP, Tubach F, Brun AL, Rammaert B, Hausfater P, Naccache JM, Ray P, Choquet C, Carette MF, Mayaud C, Leport C, Duval X (2015) Early chest computed tomography scan to assist diagnosis and guide treatment decision for suspected community-acquired pneumonia. Am J Respir Crit Care Med 192:974–982
Haran JP, Beaudoin FL, Suner S, Lu S (2013) C-reactive protein as predictor of bacterial infection among patients with an influenza-like illness. Am J Emerg Med 31:137–144
Wang L (2020) C-reactive protein levels in the early stage of COVID-19. Med Mal Infect 50:332–334
Simon L, Gauvin F, Amre DK, Saint-Louis P, Lacroix J (2004) Serum procalcitonin and C-reactive protein levels as markers of bacterial infection: a systematic review and meta-analysis. Clin Infect Dis 39:206–217
Becker KL, Snider R, Nylen ES (2008) Procalcitonin assay in systemic inflammation, infection, and sepsis: clinical utility and limitations. Crit Care Med 36:941–952
Carbonell R, Urgeles S, Salgado M, Rodriguez A, Reyes LF, Fuentes YV, Serrano CC, Caceres EL, Bodi M, Martin-Loeches I, Sole-Violan J, Diaz E, Gomez J, Trefler S, Vallverdu M, Murcia J, Albaya A, Loza A, Socias L, Ballesteros JC, Papiol E, Vina L, Sancho S, Nieto M, Del M, Lorente C, Badallo O, Fraile V, Armestar F, Estella A, Abanses P, Sancho I, Guasch N, Moreno G, Group C-SW, the L-C-I (2022) Negative predictive value of procalcitonin to rule out bacterial respiratory co-infection in critical covid-19 patients. J Infect. https://doi.org/10.1016/j.jinf.2022.06.024
Vaughn VM, Gandhi TN, Petty LA, Patel PK, Prescott HC, Malani AN, Ratz D, McLaughlin E, Chopra V, Flanders SA (2021) Empiric antibacterial therapy and community-onset bacterial coinfection in patients hospitalized with coronavirus disease 2019 (COVID-19): a multi-hospital cohort study. Clin Infect Dis 72:e533–e541
Jain S, Self WH, Wunderink RG, Fakhran S, Balk R, Bramley AM, Reed C, Grijalva CG, Anderson EJ, Courtney DM, Chappell JD, Qi C, Hart EM, Carroll F, Trabue C, Donnelly HK, Williams DJ, Zhu Y, Arnold SR, Ampofo K, Waterer GW, Levine M, Lindstrom S, Winchell JM, Katz JM, Erdman D, Schneider E, Hicks LA, McCullers JA, Pavia AT, Edwards KM, Finelli L, Team CES (2015) Community-acquired pneumonia requiring hospitalization among U.S. adults. N Engl J Med 373:415–427
Shoar S, Musher DM (2020) Etiology of community-acquired pneumonia in adults: a systematic review. Pneumonia (Nathan) 12:11
Acknowledgements
CIBERESUCICOVID Project Investigators (COV20/00110, ISCIII): Rafael Mañez, Felipe Rodríguez de Castro, María Mora Aznar, Mateu Torres, María Martinez, Cynthia Alegre, Sofía Contreras, Javier Trujillano, Montse Vallverdú, Miguel León, Mariona Badía, Begoña Balsera, Lluís Servià, Judit Vilanova, Silvia Rodríguez, Neus Montserrat, Silvia Iglesias, Javier Prados, Sula Carvalho, Mar Miralbés, Josman Monclou, Gabriel Jiménez, Jordi Codina, Estela Val, Pablo Pagliarani, Jorge Rubio, Dulce Morales, Andrés Pujol, Àngels Furro, Beatriz García, Gerard Torres, Javier Vengoechea, Silvia Gomez, Lorena Forcelledo Espina, Emilio García Prieto, Paula Martín Vicente, Cecilia Del Busto Martínez, María Aguilar Cabello, Carmen Eulalia Martínez Fernández, María Luisa Blasco Cortés, Ainhoa Serrano Lázaro, Mar Juan Díaz, María Teresa Bouza Vieiro, Inés Esmorís Arijón, David Campi Hermoso, Rafaela Nogueras Salinas, Teresa Farre Monjo, Ramon Nogue Bou, Gregorio Marco Naya, Núria Ramon Coll, Juan Carlos Montejo-González, Gloria Renedo Sanchez-Giron, Juan Bustamante-Munguira, Ramon Cicuendez Avila, Nuria Mamolar Herrera, Alexander Agrifoglio, Lucia Cachafeiro, Emilio Maseda, Albert Figueras, Maria Teresa Janer, Laura Soliva, Marta Ocón, Luisa Clar, J Ignacio Ayestarán, Sandra Campos Fernández, Eva Forcadell-Ferreres, Immaculada Salvador-Adell, Neus Bofill, Berta Adell-Serrano, Josep Pedregosa Díaz, Núria Casacuberta-Barberà, Luis Urrelo-Cerrón, Àngels Piñol-Tena, Pablo Ryan Murúa, Covadonga Rodríguez Ruíz, Laura Carrión García, Juan Ilazo Álvarez, Desire Macias Guerrero, Daniel Tognetti, Carlos García Redruello, David Mosquera Rodríguez, Eva María Menor Fernández, Sabela Vara Adrio, Vanesa Gómez Casal, Marta Segura Pensado, María Digna Rivas Vilas, Amaia García Sagastume, Raul de Pablo Sánchez, David Pestaña Laguna, Tommaso Bardi, Carmen Gómez Gonzalez, Maria Luisa Gascón Castillo, José Garnacho-Montero, Joan Ramon Masclans, Ana Salazar Degracia, Judit Bigas, Rosana Muñoz-Bermúdez, Clara Vilà-Vilardel, Francisco Parrilla, Irene Dot, Ana Zapatero, Yolanda Díaz, María Pilar Gracia, Purificación Pérez, Andrea Castellví, Cristina Climent, Lidia Serra, Laura Barbena, Iosune Cano, Alba Herraiz, Pilar Marcos, Laura Rodríguez, Maria Teresa Sariñena, Ana Sánchez, Juan Fernando Masa Jimenez, Gemma Gomà, Mercedes Ibarz, Diego De Mendoza, Victoria Alcaraz-Serrano, Joan Ramon Badia, Manuel Castella, Leticia Bueno, Catia Cillóniz, Pamela Conde, Javier Fernández, Karsa Kiarostami, Alexandre López-Gavín, Cecilia L Mantellini, Carla Speziale, Nil Vázquez, Hua Yang, Minlan Yang, Carlos Ferrando, Pedro Castro, Marta Arrieta, Jose Maria Nicolas, Rut Andrea, Marta Barroso, Sergio Álvarez, Dario Garcia-Gasulla, Adrián Tormos, Cesar Aldecoa, Rubén Herrán-Monge, José Ángel Berezo García, Pedro Enríquez Giraudo, Pablo Cardinal Fernández, Alberto Rubio López, Orville Báez Pravia, Leire Pérez Bastida, Antonjo Alvarez Ruiz, Anna Parera Pous, Ana López Lago, Eva Saborido Paz, Patricia Barral Segade, Manuel Valledor Mendez, Luciano Aguilera, Esther López-Ramos, Ángela Leonor Ruiz-García, Belén Beteré, Rafael Blancas, Cristina Dólera, Gloria Perez Planelles, Enrique Marmol Peis, Maria Dolores Martinez Juan, Miriam Ruiz Miralles, Eva Perez Rubio, Maria Van der Hofstadt Martin-Montalvo, Tatiana Villada Warrington, Sara Guadalupe Moreno Cano, Federico Gordo, Basilisa Martinez Palacios, Maria Teresa Nieto, Sergio Ossa, Ana Ortega, Miguel Sanchez, Bitor Santacoloma.
Funding
Financial support was provided by the Instituto de Salud Carlos III de Madrid (COV20/00110, ISCIII); Fondo Europeo de Desarrollo Regional (FEDER); “Una manera de hacer Europa”; Centro de Investigación Biomedica En Red–Enfermedades Respiratorias (CIBERES); and Donation program “estar preparados” UNESPA, Madrid, Spain. DdGC has received financial support from the Instituto de Salud Carlos III (Miguel Servet 2020: CP20/00041), co-funded by European Social Fund (ESF)/ “Investing in your future”. Adrian Ceccato acknowledges receiving financial support from Instituto de Salud Carlos III (ISCIII; Sara Borrell 2021: CD21/00087).
Author information
Authors and Affiliations
Consortia
Contributions
Conception and design of the study: AT, FG, FB, AM, LFB, EB. Data acquisition: all authors. Statistical analysis: AG. Data analysis and interpretation: AT, AM, CC, AC, LFB, JBM, FB. Manuscript drafting: AT, FG, FB. Critical revision for important intellectual content: all authors. Final approval of the submitted version: all authors. CiberesUCICOVID consortium participated in data collection.
Corresponding authors
Ethics declarations
Conflicts of interest
The authors have disclosed that they do not have any conflicts of interest.
Role of the funder/sponsor
The funding sources had no role in the design or conduct of the study; collection, management, analysis, or interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The CIBERESUCICOVID Project investigators are listed in the Acknowledgement section.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Galli, F., Bindo, F., Motos, A. et al. Procalcitonin and C-reactive protein to rule out early bacterial coinfection in COVID-19 critically ill patients. Intensive Care Med 49, 934–945 (2023). https://doi.org/10.1007/s00134-023-07161-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00134-023-07161-1