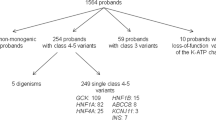
Abstract
Aims/hypothesis
Monogenic forms of diabetes (MODY, neonatal diabetes mellitus and syndromic forms) are rare, and affected individuals may be misclassified and treated suboptimally. The prevalence of type 1 diabetes is high in Finnish children but systematic screening for monogenic diabetes has not been conducted. We assessed the prevalence and clinical manifestations of monogenic diabetes in children initially registered with type 1 diabetes in the Finnish Pediatric Diabetes Register (FPDR) but who had no type 1 diabetes-related autoantibodies (AABs) or had only low-titre islet cell autoantibodies (ICAs) at diagnosis.
Methods
The FPDR, covering approximately 90% of newly diagnosed diabetic individuals aged ≤15 years in Finland starting from 2002, includes data on diabetes-associated HLA genotypes and AAB data (ICA, and autoantibodies against insulin, GAD, islet antigen 2 and zinc transporter 8) at diagnosis. A next generation sequencing gene panel including 42 genes was used to identify monogenic diabetes. We interpreted the variants in HNF1A by using the gene-specific standardised criteria and reported pathogenic and likely pathogenic findings only. For other genes, we also reported variants of unknown significance if an individual’s phenotype suggested monogenic diabetes.
Results
Out of 6482 participants, we sequenced DNA for 152 (2.3%) testing negative for all AABs and 49 (0.8%) positive only for low-titre ICAs (ICAlow). A monogenic form of diabetes was revealed in 19 (12.5%) of the AAB-negative patients (14 [9.2%] had pathogenic or likely pathogenic variants) and two (4.1%) of the ICAlow group. None had ketoacidosis at diagnosis or carried HLA genotypes conferring high risk for type 1 diabetes. The affected genes were GCK, HNF1A, HNF4A, HNF1B, INS, KCNJ11, RFX6, LMNA and WFS1. A switch from insulin to oral medication was successful in four of five patients with variants in HNF1A, HNF4A or KCNJ11.
Conclusions/interpretation
More than 10% of AAB-negative children with newly diagnosed diabetes had a genetic finding associated with monogenic diabetes. Because the genetic diagnosis can lead to major changes in treatment, we recommend referring all AAB-negative paediatric patients with diabetes for genetic testing. Low-titre ICAs in the absence of other AABs does not always indicate a diagnosis of type 1 diabetes.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.

Introduction
Large-scale genetic screening for monogenic diabetes in children and adolescents has not been performed. A prevalence of 2.1% was shown in an unselected childhood diabetes cohort (n=821) from Australia, while paediatric patients without type 1 diabetes-related autoantibodies (AABs) have a higher prevalence (4–15%) [1,2,3] especially in combination with residual C-peptide secretion (8–24%) [4, 5]. Monogenic diabetes includes very rare forms (neonatal diabetes [NDM], syndromic diabetes, monogenic autoimmune diabetes) but the most common form is MODY, which follows an autosomal dominant inheritance pattern. Individuals with MODY are often diagnosed with diabetes during late childhood, adolescence or early adulthood [6, 7]. While pathogenic variants in the genes encoding hepatocyte nuclear factor 1α (HNF1A) and glucokinase (GCK) are carried by more than 60% of the affected individuals [8], variants in more than ten genes have been conclusively associated with MODY [9].
A diagnosis of monogenic diabetes often enables personalised treatment. Some individuals may be switched from insulin to oral medication [10,11,12], especially those with pathogenic variants in HNF1A, HNF4A or genes encoding subunits of the ATP-sensitive potassium (KATP) channel of the pancreatic beta cell (KCNJ11 or ABCC8). The genetic diagnosis motivates familial testing and, in some cases, guides screening for comorbidities associated with the genetic finding.
Our main objective was to assess the prevalence and clinical manifestations of monogenic diabetes in individuals who were initially registered with type 1 diabetes in the Finnish Pediatric Diabetes Register (FPDR) but had no type 1 diabetes-related autoantibodies or had only low-titre islet cell autoantibodies (ICAs) at diagnosis. We also present two patients who underwent a successful switch from insulin to oral glucose-lowering agents years after the diagnosis of diabetes.
Methods
Study population
The FPDR covers more than 90% of children and adolescents aged ≤15 years diagnosed with diabetes in Finland since June 2002 [13]. In this study, we included 6482 participants of the FPDR initially registered with type 1 diabetes before November 2018 (mean±SD age at diagnosis 8.3±4.1 years, 43% female sex, estimated adult BMI [ISO-BMI, predicted by the BMI at registration, age and sex according to the Finnish growth centiles] 20.6±4.3 kg/m2). Most participants were ≤15 years at registration, but 23 had turned 16. For most patients, DNA samples and general information on the diagnosis of diabetes were available through the register (electronic supplementary material [ESM] Table 1). We studied the parents of those with gene variants putatively associated with diabetes if the parents had joined the register.
The sequencing and the mixed-meal tolerance test (MMTT) in an individual with a pathogenic variant in the KCNJ11 gene were performed in collaboration with the FINNMODY study [14], which identified and characterised individuals with a suspected or established diagnosis of monogenic diabetes in Finland from 2014 onwards (www.botnia-study.org/finnmody). The results of the MMTT were compared with 45 control individuals without diabetes from the Botnia Study described previously [15]. The ethics committees of the Helsinki and Uusimaa Hospital District approved the studies which were conducted according to the Principles of the Declaration of Helsinki. All FPDR participants and/or their guardians signed an informed consent declaration. Individuals whose case reports are presented also agreed to their publication. Clinical details include minor modifications to maintain the participants’ anonymity.
Laboratory analyses
Autoantibody status
Samples taken at diagnosis were tested for ICAs with indirect immunofluorescence or for autoantibodies against insulin, GAD, islet antigen 2 and zinc transporter 8 (IAA, GADA, IA-2A, ZnT8A) with specific radiobinding assays, as previously described [16]. The thresholds for positivity were as follows: ≥2.5 JDRF units (JDFU) for ICA; ≥1.57 relative units (RU) for IAA; ≥0.77 RU for IA-2A; ≥5.36 RU for GADA and ≥0.50 RU for ZnT8A.
HLA genotyping
The HLA class II genotyping was performed as previously described [17]. The HLA risk classification for type 1 diabetes based on the HLA-DR/DQ genotype frequencies observed in the Finnish population rests on disease risk associations of each haplotype, taking into consideration the synergistic effects of the DR3-DQ2 and DR4-DQ8 haplotypes. Each haplotype is classified as protective, neutral or susceptible. The highest risk was associated with the DRB1*04:01-DQA1*03-DQB1*03:02 haplotype, whereas the other common DR4-DQ8 haplotype in Finland, DRB1*04:04-DQA1*03-DQB1*03:02, had a less strong disease association similar to the DRB1*03-DQA1*05-DQB1*02 haplotype [18].
MMTT
An MMTT composed of carbohydrates, fat and protein was consumed by participants within 10 min. Serial blood sampling was performed before and 15, 30, 60, 90, 120, 150 and 180 min after commencing the meal. See ESM Methods for details of the MMTT.
Next generation sequencing
In collaboration with the sequencing unit of the Institute for Molecular Medicine Finland, University of Helsinki, a next generation sequencing (NGS) gene panel was designed to include 42 genes reported to be associated with monogenic forms of diabetes, lipodystrophy or other glycaemic traits: ABCC8, AKT2, APPL1, BLK, CEL, CISD2, DCAF17, DNAJC3, DYRK1B, EIF2AK3, FOXP3, GATA4, GATA6, GCK, GLIS3, HNF1A, HNF1B, HNF4A, IER3IP1, INS, INSR, KCNJ11, KLF11, LMNA, NEUROD1, NEUROG3, PAX4, PCBD1, PDX1, PIK3R1, PLIN1, POLD1, PPARG, PPP1R15B, PTF1A, RFX6, SLC19A2, SLC2A2, TRMT10A, WFS1, ZBTB20 and ZFP57. For HNF4A, the pancreatic transcript NM_175914.4 was used instead of the canonical transcript. The gene list from Ellard et al [19] was complemented from other sources such as the gene panel for monogenic diabetes from the leading laboratory in Exeter (https://www.diabetesgenes.org/, accessed 12 May 2022; previous versions of the panel assessed for the panel design). After excluding repetitive elements from the initial target region of 2,600,298 bp (the exons, flanking intronic regions [~±50 bp] and ~1 kbp region upstream of each gene), 19,084 probes were designed to cover a region of 2,285,454 bp. The samples, prepared with the Twist Enzymatic Fragmentation (EF) library (Twist Bioscience, South San Francisco, CA, USA), were run on the NovaSeq S2 platform with NovaSeq reagents (Illumina, San Diego, California, USA). The panel was successfully tested against known pathogenic or benign variants (GCK c.45+3A>G, c.544G>A, c.556C>T, c.563C>T, c.781G>A, c.823C>T, c.1198del, c.398_399insACATCTCTGAGTGCATCTCCGACT; HNF1A c.431T>C, c.779C>T, c.872dup, c.1501G>A, c.824_826del; HNF1B c.443C>T, c.721G>A, c.1474G>A; HNF4A c.225-8C>G, c.421C>T, c.691C>T, c.926G>A, c.256_257del; RFX6 c.878_879del and PDX1 c.226G>A, c.634G>A, c.716C>A) but it was not able to detect the CEL gene (encoding carboxyl ester lipase) variable number tandem repeat (VNTR) in the samples provided by A. Molven (University of Bergen). The mean read depth (the mean of the exonic mean depth per gene across samples) was 293, and for HNF1A, HNF1B, HNF4A and GCK it was 310, with 99.99% of the exonic targets covered by the read depth of 20 or more.
We confirmed the genetic findings reported in Table 1 in an accredited commercial laboratory (Blueprint Genetics, Espoo, Finland or HUSLAB Laboratory of Genetics, Helsinki, Finland).
Variant interpretation
We interpreted the variants in HNF1A according to the gene-specific criteria (https://clinicalgenome.org/affiliation/50016/, accessed 6 November 2021), applying the American College of Medical Genetics and Genomics (ACMG) criteria [20], and reported only likely pathogenic or pathogenic variants. For all other genes, we applied the general ACMG criteria [21], with modifications adopted from the HNF1A-specific criteria that either downgrade the weight of evidence or upgrade it especially for criteria relating to co-segregation [PP1] and for phenotype [PP4]. However, to apply HNF1A-specific rules for other genes is potentially conservative, as the modifications are not counterbalanced by additional gene-specific evidence. Therefore, we also report the variants of unknown significance (VUS) in non-HNF1A genes if the following criteria were met: (1) the individual’s phenotype was suggestive of monogenic diabetes; and (2) the variants had a low allele frequency in the gnomAD population database and were either protein-truncating variants, splice site variants predicted with a high SpliceAI score to alter splicing or a non-conservative missense variant with a REVEL score >0.7.
The statistical and graphical analyses were performed using SPSS Statistics V25.0 (IBM, USA) and R V4.1.2 [22].
Results
Of 6482 participants in the FPDR, 162 (2.5%) were negative for all tested AABs at diagnosis, and 57 (0.9%) had only low-titre ICA (2.5–10 JDFU), referred to as the ICAlow group. DNA samples were available for 152 (94%) individuals in the AAB-negative group (2.3% of all participants) and for 49 (86%) individuals in the ICAlow group (0.8% of all participants).
In total, we report a gene finding associated with monogenic diabetes in 19/152 AAB-negative individuals (12.5% of the screened AAB-negative individuals and 0.3% of all 6482 children). Of these 19 patients, 14 had a gene variant classified as pathogenic or likely pathogenic by the ACMG criteria (Table 1). Four of these variants resided in GCK, five in HNF1A, HNF4A or HNF1B, four in the insulin gene (INS) and one in KCNJ11. Four individuals with variants in the laminin A/C (LMNA, n=1) or wolframin ER transmembrane glycoprotein (WFS1, n=3) genes were diagnosed with syndromic forms of monogenic diabetes. One individual had a protein-truncating variant in the regulatory factor X6 gene (RFX6), recently shown to be associated with MODY with reduced penetrance [23]. Interestingly, only four of these 19 individuals had a known family history of diabetes. Two in the ICAlow group had pathogenic variants in HNF1A and GCK, resulting in a monogenic diabetes prevalence of 4.1% (2/49) (Table 1).
The 19 AAB-negative individuals diagnosed with monogenic diabetes had a median (range) age at diagnosis of 9.6 (2.1–15.7) years and ISO-BMI of 19.6 (13.3–35) kg/m2. None had presented with significant ketosis or ketoacidosis at the time of diagnosis or carried a high-risk HLA genotype associated with type 1 diabetes (p=1.4965×10−7, χ2 test, compared with the AAB-negative individuals without a monogenic finding, with 15% having a high-risk HLA genotype and 16% presenting with ketoacidosis at diagnosis). The two individuals in the ICAlow group carried neutral HLA genotypes and were diagnosed as teenagers with mild hyperglycaemia without ketosis. Overall, the protective and neutral HLA genotypes regarding type 1 diabetes risk were more frequent in individuals without AABs in the FPDR than in those with AABs (protective, 12% vs 3%; neutral, 23% vs 15%) and risk genotypes were less common (high risk, 14% vs 25%) (p=0.000356, χ2 test). In the ICAlow group, protective HLA genotypes were found only in 2% of individuals. Further clinical details are presented in ESM Table 1.
We contacted all 21 patients with monogenic findings and contacted their treating hospitals. Eight (38%) individuals (GCK [four], WFS1 [two], HNF1B [one] and LMNA [one]) had been diagnosed with monogenic diabetes during their clinical follow-up, irrespective of our study. In others, our register-based revision of diagnosis led to treatment changes. A diagnosis of GCK-MODY resulted in discontinuation of metformin in a patient misclassified as having type 2 diabetes. Despite initially presenting with insulin deficiency, a switch from insulin treatment to oral medication had been successful in four of five patients with findings in HNF1A, HNF4A or KCNJ11 genes by the time of publication. We describe the process for two of them below.
Patient with a VUS in HNF4A: transfer from insulin pump to oral glucose-lowering agent
A 13-year-old girl without a family history of diabetes presented with fatigue, obesity and hyperglycaemia (fasting plasma glucose 16.8 mmol/l, HbA1c 103 mmol/mol [11.6%], no ketosis or acidosis). Relatively low C-peptide (0.6 nmol/l) in relation to hyperglycaemia had led to a diagnosis of type 1 diabetes. The girl was negative for AABs but the HLA genotype suggested a moderately increased genetic risk for type 1 diabetes. Insulin treatment was initiated. Four years after diagnosis, the total daily insulin administered via insulin pump was fairly low (0.6–0.7 U/kg) considering her age and ISO-BMI (28 kg/m2) and her C-peptide was surprisingly good (0.47 nmol/l at glucose 12.1 mmol/l).
The sequencing revealed a heterozygous de novo variant in HNF4A. There was no information from the neonatal phase. Although the variant was classified as VUS, the bioinformatics (see Table 1) and clinical clues (no AABs, persisting C-peptide, low total daily insulin requirement) supported an empiric trial with repaglinide, a meglitinide drug with the same mechanism of action as sulfonylureas. Within 25 days, the girl was gradually switched from insulin to repaglinide (Fig. 1). Because the girl was overweight with suspected accompanying insulin resistance, metformin was initiated to further improve glycaemic control. After these treatment changes, she experienced fewer episodes of hypoglycaemia and displayed improved glycaemic control (decrease in HbA1c from 74 to 53 mmol/mol [8.9 to 7.0%]).
Patient with the HNF4A variant who was transferred from insulin pump to an oral glucose-lowering agent. (a) Bar plot showing the total daily dose of basal (dark blue bars) and bolus insulin (light blue bars) administered by the insulin pump, as well as the sequential initiation and dose increase of repaglinide at the major three meals of the day (indicated by arrows). Within 25 days, the patient gradually switched from insulin treatment to repaglinide. The vertical dashed line represents the initiation of the treatment transfer. (b) The 14 day mean level of sensor glucose measured by continuous glucose monitoring (Dexcom G6) during the treatment with the insulin pump (red line) and after the switch to repaglinide (blue line), with IQRs illustrated by the shaded area. Not only could the individual discontinue the insulin treatment but she also experienced fewer episodes of mild hypoglycaemia and improved her glycaemic control. The horizontal dotted lines represent the target range of glycaemia (3.9–10 mmol/l)
Transfer from insulin to sulfonylurea in a patient with KCNJ11 p.(Arg201His)
A 2-year-old boy, born small for gestational age, with no hypoglycaemia in the neonatal phase, was admitted to hospital for hyperglycaemia during infection (no ketosis, HbA1c 101 mmol/mol [11.4%], C-peptide 0.07 nmol/l). No AABs were detected, the HLA genotype risk group was neutral and he had no family history of diabetes. Insulin therapy was initiated. Within 2 years following diagnosis, he developed an absolute insulin deficiency with undetectable C-peptide. Fourteen years after the diagnosis, he was on an insulin pump with an average insulin requirement of 1.2 U/kg/day.
The gene panel revealed a heterozygous activating variant in the KCNJ11 gene encoding for the Kir6.2 subunit of the KATP channel (Table 1), preventing its closure. In most cases, this results in permanent or transient NDM. This variant p.(Arg201His) can also cause KCNJ11-MODY with variable onset and severity of diabetes [24, 25].
As suggested by Pearson et al [26], we introduced treatment with an oral sulfonylurea (glibenclamide). The first dose of 0.03 mg/kg (1.75 mg), given 1 h before a standard MMTT, resulted in a hypoglycaemic event (requiring i.v. treatment with glucose) 30 min after the start of the meal. The insulin level increased dramatically after the first dose of sulfonylurea, indicating a rapid activation of endogenous insulin secretion from the beta cells. The C-peptide and proinsulin levels increased to a level similar to that seen in individuals without diabetes [15] (over 80 min, insulin increased from <1.4 to 1043.1 pmol/l, C-peptide increased from <0.003 to 2.07 nmol/l and proinsulin increased from <1.56 to 12.5 pmol/l) (Table 2). A minimal dose of 0.875 mg glibenclamide twice daily was first continued, then was doubled while the individual was an inpatient, and subsequently increased over several weeks up to 0.6 mg/kg per day. In parallel, the insulin dose was gradually reduced and discontinued 80 days later (Fig. 2).
Patient with the KCNJ11 variant who was transferred from insulin to a sulfonylurea. (a) Bar plot showing the total daily doses of basal (dark blue bars) and bolus insulin (light blue bars) and glibenclamide doses (black line, right-hand y-axis), along with increasing doses of glibenclamide. The insulin dose was gradually reduced and eventually the insulin treatment was discontinued after 80 days. The vertical dashed line represents the initiation of the treatment transfer. (b) The 14 day mean level of sensor glucose measured by continuous glucose monitoring (Dexcom G6) before the initiation of glibenclamide (red line) and after the switch to glibenclamide (blue line), with IQRs illustrated by the shaded area. The glycaemic control was remarkably improved by glibenclamide treatment. The horizontal dotted lines represent the target range of glycaemia (3.9–10 mmol/l)
On the sulfonylurea, the glycaemic control improved remarkably (time in range 3.9–10 mmol/l improved from 42% before the sulfonylurea to 98% with the sulfonylurea; CV for glucose changed from 46% to 17%).
Discussion
Using a sequencing panel of 42 genes in the FPDR covering most Finnish paediatric patients diagnosed with type 1 diabetes since 2002, we identified a monogenic cause for diabetes in 12.5% of the AAB-negative individuals (9.2% with findings classified as pathogenic or likely pathogenic and 3.3% with VUS findings considered relevant). In addition, 4.1% of those positive only for low-titre ICAs had a monogenic cause.
The observed prevalence among individuals negative for five AABs was twice as high as the 6.5% reported in the Norwegian Childhood Diabetes Registry [2]. The difference is partly explained by only two AABs (GADA and IA-2A) measured in the Norwegian Registry but mainly by the included genes, as 8.5% of our AAB-negative individuals had pathogenic variants in GCK, HNF1A, HNF4A, HNF1B and INS, which were screened for in Norway. In addition, INS p.(Glu37Lys), a VUS reported in this study, is not a definitive finding (ESM Table 2). To interpret the relevance of any VUS in INS without conclusive prior functional variant-level evidence calls for caution as there is no unambiguous phenotype associated with heterozygous INS variants. However, as the variant turned out to be de novo, we included it on our list. If we focus on the most common MODY genes GCK, HNF1A and HNF4A, the prevalence in both our study (4%) and the Norwegian study (6%) is lower than the 15% seen in the Swedish National Cohort [3], likely explained by the latter including patients with all types of diabetes from paediatric clinics, with age at diagnosis of up to 18 years. In addition, a Lithuanian study including young adults up to 25 years of age showed a higher prevalence [27]. However, all these cohorts are demographically different, and the classification of variants has been variable. The inclusion of less symptomatic individuals would also increase the proportion of GCK-MODY that is associated with mild and life-long hyperglycaemia, as seen in the Lithuanian study.
Since the identification of ICAs [28], screening for type 1 diabetes-associated AABs has become routine in many countries. While the other AABs target specific proteins, the ICAs bind to various intracellular structures in the islets [29]. At the time of diagnosis, most insulin-deficient paediatric patients are positive for ICAs, although low ICA titres (≤10 JDFU) are also found in non-diabetic family members and in 4% of the Finnish general paediatric population [30, 31]. Hence, we extended the screening to the ICAlow individuals to reveal a monogenic cause for diabetes in 4.1% of this group. To date, it is not known whether pathogenic variants could potentially contribute to diabetes in some AAB-positive individuals.
Besides AAB negativity, clinical features in support of genetic testing have included low HbA1c, lack of type 1 diabetes-predisposing HLA genotypes, family history of diabetes and absence of ketoacidosis at diagnosis [32]. In support, and similar to the Swedish cohort, none of our studied individuals with monogenic diabetes had presented with severe ketosis or ketoacidosis. Further, most of the 19 individuals carried a protective or neutral HLA genotype regarding type 1 diabetes risk and none carried a high-risk genotype, whereas less than 20% in the whole FPDR carried a protective or neutral HLA genotype [33]. While a lack of risk genotypes should lead to considering alternative diagnosis to type 1 diabetes, we do not suggest using HLA typing to exclude patients from genetic testing for monogenic diabetes, considering the overall prevalence of risk HLA genotypes in the Finnish general population. On the other hand, 12 out of the 19 individuals would not have met the suggested HbA1c criterion of <58 mmol/mol (7.5%) [3] (data not shown). We also noted that few of the individuals had reported a positive family history for diabetes and one-third had a confirmed de novo variant. All in all, only one individual scored high (75%) in the MODY probability calculator [34], whereas ten individuals with sufficient data for the calculation scored low (<20%). However, the prevalence of de novo genetic findings in our study is likely high, as those with a known family history of monogenic diabetes would presumably not have been registered as having type 1 diabetes in this register.
Variants in GCK and, surprisingly, INS were the most common causes for monogenic diabetes in our study, followed by variants in HNF1A, HNF4A and HNF1B. However, we are likely to underestimate the prevalence of GCK-MODY, as individuals with stable and mild hyperglycaemia might remain undiagnosed or they might be misclassified as having type 2 diabetes [35] or just not be registered in the FPDR. In the HNF4A gene, we report a novel missense variant, c.112T>C p.(Cys38Arg), graded as a VUS (Table 1). The successful transfer of the individual carrying this variant from insulin pump therapy to an oral short-acting meglitinide proves that she did not have type 1 diabetes but does not exclude the possibility of type 2 diabetes. However, the relatively low C-peptide level in relation to the degree of hyperglycaemia suggested mild insulin deficiency rather than resistance at diagnosis at the age of 13 years. We speculate that the loss of a cysteine (Cys) residue might destabilise the protein structure of hepatic nucleocyte factor 4α because the formation of a Cys-Cys disulfide bridge is hampered [36]. We also identified an individuals with an RFX6 protein-truncating variant previously associated with MODY with reduced penetrance [23]. However, the variant p.(His293fs) might be far more prevalent in Finland than first assumed, as the FinnGen Study reported an allele frequency of 2.0×10−3 in Finland (https://r7.finngen.fi/variant/6-116916217-TAC-T, accessed 1 July 2022).
Identification of a monogenic cause for diabetes enables personalisation of the treatment. Dietary treatment involving balancing the carbohydrate intake can be sufficient in some forms of MODY. Individuals with HNF1A- and HNF4A-MODY or who carry KCNJ11 gene variants [26, 37] can successfully be treated with a sulfonylurea [10, 38] or with sulfonylurea-like meglitinides [12]. However, after having witnessed a severe hypoglycaemic event in our patient with the KCNJ11 variant upon the initiation of the sulfonylurea therapy, despite a lower first dose than suggested in published treatment protocols [26], we would advise starting sulfonylurea treatment under inpatient care. In our patient, the first dose of sulfonylurea triggered an immediate significant increase in proinsulin, C-peptide and insulin concentrations indicating their rapid release from beta cells. This was possibly enhanced by the suppression of glucagon secretion, suggested to be mediated though the paracrine effect of somatostatin, resulting in the loss of appropriate counter-regulation during insulin-induced hypoglycaemia [39]. The rapid insulin response with moderate C-peptide and proinsulin response preceding the severe hypoglycaemic event could indicate an uncontrolled release of previously produced insulin from a readily releasable pool of granules in beta cells, and not the newly synthesised hormone [40]. No hypoglycaemia occurred when the MMTT was repeated during sulfonylurea therapy, and the increase in proinsulin, insulin and C-peptide levels was tapered. The stimulation by incretin hormones was similar during both MMTTs, suggesting that these hormones have minimal impact in the development of hypoglycaemia.
In addition to allowing tailored treatment, a correct genetic diagnosis is crucial for further diagnostics and follow-up of possible comorbidities associated with syndromic forms of diabetes. HNF1B is associated with a multisystem disorder including renal manifestations, genital tract abnormalities, abnormal liver function, biliary cysts and neurological features [41]. LMNA is associated with familial partial lipodystrophy and, therefore, insulin resistance and diabetes as well as muscular diseases [42] (the individual with an LMNA variant in this study had a pre-established diagnosis of muscular disease). Wolfram syndrome (DIDMOAD) is a very rare disease involving diabetes mellitus, diabetes insipidus, blindness, deafness and progressive brainstem degeneration [43]. However, there is phenotypic variation in syndromic monogenic diseases, as also suggested by the individual with two WFS1 variants and partial phenotype of Wolfram syndrome including diabetes mellitus and opticus atrophy (Table 1).
The study has some limitations. We screened only those paediatric patients included in the FPDR and registered initially with type 1 diabetes, who were negative for five different AABs or who had only marginally elevated ICAs. Although the FPDR reaches 90% of newly diagnosed children and adolescents with diabetes, 10% of the individuals with diabetes are lost and DNA samples were not available for all. Additionally, regardless of the register welcoming all kinds of diabetes, most research involving the FPDR concerns type 1 diabetes. Thus, paediatricians may be more likely to refer individuals with type 1 diabetes than those with a suspicion of other types of diabetes. Presumably, many individuals with known family history for monogenic diabetes are not registered in the FPDR and, in this study, we did not include samples from individuals who participated in a previously published study from Finland, in which participants were diagnosed with transient or permanent monogenic forms of diabetes before the age of 1 year between years 1980 and 2015 [44]. These facts, and the globally highest incidence of type 1 diabetes in Finland [45], explains to some extent the lower overall prevalence of monogenic diabetes found in our study (0.3%) compared with the published prevalence in other countries [3, 5, 46]. Specific repetitive targets such as CEL VNTR were beyond the performance of our gene panel. In addition, the panel did not include the genes for mitochondrial diabetes for technical reasons. Therefore, the true prevalence of monogenic forms among all paediatric patients diagnosed with diabetes in Finland might be somewhat higher than found here. In future, further investigation of the AAB-negative individuals will include whole exome sequencing with both nuclear and mitochondrial genes.
Early screening of monogenic diabetes in children with AAB-negative diabetes can have a major impact on the choice of treatment, enabling oral glucose-lowering treatment instead of insulin injections, with benefits on glycaemic control and long-term complications. Early targeted follow-up can be organised for individuals with syndromic forms of diabetes. Cost-effectiveness analyses, summarised recently by Naylor [47], have also shown that genetic testing for monogenic diabetes can be cost-effective or cost-saving in neonatal diabetes [48], in all paediatric patients presumed to have type 1 diabetes [49] and even young adult patients presumed to have type 2 diabetes [50].
In conclusion, our findings demonstrate the importance of determining the AAB status at diagnosis of diabetes in children and adolescents and justify testing for monogenic causes of diabetes in AAB-negative individuals and those with low-titre ICA regardless of family history of diabetes, especially if HLA genotypes conferring increased risk for type 1 diabetes are not detected.
Data availability
The de-identified datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AAB:
-
Autoantibody
- ACMG:
-
American College of Medical Genetics and Genomics
- FPDR:
-
Finnish Pediatric Diabetes Register
- GADA:
-
GAD autoantibody
- IA-2A:
-
Islet antigen 2 autoantibody
- IAA:
-
Insulin autoantibody
- ICA:
-
Islet cell autoantibody
- ISO-BMI:
-
Estimated adult BMI in children
- JDFU:
-
JDRF units
- KATP :
-
ATP-sensitive potassium
- MMTT:
-
Mixed-meal tolerance test
- NDM:
-
Neonatal diabetes
- NGS:
-
Next generation sequencing
- RU:
-
Relative unit
- VNTR:
-
Variable number tandem repeat
- VUS:
-
Variant of unknown significance
- ZnT8A:
-
Zinc transporter 8 autoantibody
References
Johnson SR, Ellis JJ, Leo PJ et al (2019) Comprehensive genetic screening: The prevalence of maturity-onset diabetes of the young gene variants in a population-based childhood diabetes cohort. Pediatr Diabetes 20(1):57–64. https://doi.org/10.1111/pedi.12766
Johansson BB, Irgens HU, Molnes J et al (2017) Targeted next-generation sequencing reveals MODY in up to 6.5% of antibody-negative diabetes cases listed in the Norwegian Childhood Diabetes Registry. Diabetologia 60(4):625–635. https://doi.org/10.1007/s00125-016-4167-1
Carlsson A, Shepherd M, Ellard S et al (2020) Absence of islet autoantibodies and modestly raised glucose values at diabetes diagnosis should lead to testing for MODY: lessons from a 5-year pediatric Swedish national cohort study. Diabetes Care 43(1):82–89. https://doi.org/10.2337/dc19-0747
Pihoker C, Gilliam LK, Ellard S et al (2013) Prevalence, characteristics and clinical diagnosis of maturity onset diabetes of the young due to mutations in HNF1A, HNF4A, and glucokinase: results from the search for diabetes in youth. J Clin Endocrinol Metab 98(10):4055–4062. https://doi.org/10.1210/jc.2013-1279
Shepherd M, Shields B, Hammersley S et al (2016) Systematic population screening, using biomarkers and genetic testing, identifies 2.5% of the U.K. pediatric diabetes population with monogenic diabetes. Diabetes Care 39(11):1879–1888. https://doi.org/10.2337/dc16-0645
Fajans SS, Bell GI (2011) MODY: history, genetics, pathophysiology, and clinical decision making. Diabetes Care 34(8):1878–1884. https://doi.org/10.2337/dc11-0035
Riddle MC, Philipson LH, Rich SS et al (2020) Monogenic diabetes: from genetic insights to population-based precision in care. Reflections from a diabetes care editors’ expert forum. Diabetes Care 43(12):3117–3128. https://doi.org/10.2337/dci20-0065
Naylor R, Knight Johnson A, del Gaudio D (1993) Maturity-onset diabetes of the young overview. In: Adam MP, Ardinger HH, Pagon RA et al (eds) GeneReviews®. University of Washington, Seattle, Seattle (WA)
Laver TW, Wakeling MN, Knox O et al (2022) Evaluation of evidence for pathogenicity demonstrates that BLK, KLF11, and PAX4 should not be included in diagnostic testing for MODY. Diabetes 71(5):1128–1136. https://doi.org/10.2337/db21-0844
Zhang H, Colclough K, Gloyn AL, Pollin TI (2021) Monogenic diabetes: a gateway to precision medicine in diabetes. J Clin Invest 131(3):142244. https://doi.org/10.1172/JCI142244
Delvecchio M, Pastore C, Giordano P (2020) Treatment options for MODY patients: a systematic review of literature. Diabetes Ther 11(8):1667–1685. https://doi.org/10.1007/s13300-020-00864-4
Tuomi T, Honkanen EH, Isomaa B, Sarelin L, Groop LC (2006) Improved prandial glucose control with lower risk of hypoglycemia with nateglinide than with glibenclamide in patients with maturity-onset diabetes of the young type 3. Diabetes Care 29(2):189–194. https://doi.org/10.2337/diacare.29.02.06.dc05-1314
Hekkala A, Reunanen A, Koski M, Knip M, Veijola R (2010) Age-related differences in the frequency of ketoacidosis at diagnosis of type 1 diabetes in children and adolescents. Diabetes Care 33(7):1500–1502. https://doi.org/10.2337/dc09-2344
Kettunen JLT, Rantala E, Dwivedi OP et al (2022) A multigenerational study on phenotypic consequences of the most common causal variant of HNF1A-MODY. Diabetologia 65(4):632–643. https://doi.org/10.1007/s00125-021-05631-z
Dwivedi OP, Lehtovirta M, Hastoy B et al (2019) Loss of ZnT8 function protects against diabetes by enhanced insulin secretion. Nat Genet 51(11):1596–1606. https://doi.org/10.1038/s41588-019-0513-9
Knip M, Virtanen SM, Seppä K et al (2010) Dietary intervention in infancy and later signs of beta-cell autoimmunity. N Engl J Med 363(20):1900–1908. https://doi.org/10.1056/NEJMoa1004809
Mikk M-L, Kiviniemi M, Laine A-P et al (2014) The HLA-B*39 allele increases type 1 diabetes risk conferred by HLA-DRB1*04:04-DQB1*03:02 and HLA-DRB1*08-DQB1*04 class II haplotypes. Hum Immunol 75(1):65–70. https://doi.org/10.1016/j.humimm.2013.09.008
Ilonen J, Kiviniemi M, Lempainen J et al (2016) Genetic susceptibility to type 1 diabetes in childhood - estimation of HLA class II associated disease risk and class II effect in various phases of islet autoimmunity. Pediatr Diabetes 17(Suppl 22):8–16. https://doi.org/10.1111/pedi.12327
Ellard S, Lango Allen H, De Franco E et al (2013) Improved genetic testing for monogenic diabetes using targeted next-generation sequencing. Diabetologia 56(9):1958–1963. https://doi.org/10.1007/s00125-013-2962-5
Harrison SM, Biesecker LG, Rehm HL (2019) Overview of specifications to the ACMG/AMP variant interpretation guidelines. Curr Protoc Hum Genet 103(1):e93. https://doi.org/10.1002/cphg.93
Richards S, Aziz N, Bale S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17(5):405–424. https://doi.org/10.1038/gim.2015.30
R. Core Team (2013) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Httpwww R-Proj Org
Patel KA, Kettunen J, Laakso M et al (2017) Heterozygous RFX6 protein truncating variants are associated with MODY with reduced penetrance. Nat Commun 8(1):888. https://doi.org/10.1038/s41467-017-00895-9
Gloyn AL, Pearson ER, Antcliff JF et al (2004) Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N Engl J Med 350(18):1838–1849. https://doi.org/10.1056/NEJMoa032922
Bonnefond A, Philippe J, Durand E et al (2012) Whole-exome sequencing and high throughput genotyping identified KCNJ11 as the thirteenth MODY gene. PLoS One 7(6):e37423. https://doi.org/10.1371/journal.pone.0037423
Pearson ER, Flechtner I, Njølstad PR et al (2006) Switching from insulin to oral sulfonylureas in patients with diabetes due to Kir6.2 mutations. N Engl J Med 355(5):467–477. https://doi.org/10.1056/NEJMoa061759
Stankute I, Verkauskiene R, Blouin J-L et al (2020) Systematic genetic study of youth with diabetes in a single country reveals the prevalence of diabetes subtypes, novel candidate genes, and response to precision therapy. Diabetes 69(5):1065–1071. https://doi.org/10.2337/db19-0974
Bottazzo GF, Florin-Christensen A, Doniach D (1974) Islet-cell antibodies in diabetes mellitus with autoimmune polyendocrine deficiencies. Lancet 2(7892):1279–1283. https://doi.org/10.1016/s0140-6736(74)90140-8
Månsson L, Törn C, Landin-Olsson M (2001) Islet cell antibodies represent autoimmune response against several antigens. Int J Exp Diabetes Res 2(2):85–90. https://doi.org/10.1155/edr.2001.85
Atkinson MA, Maclaren NK (1994) The pathogenesis of insulin-dependent diabetes mellitus. N Engl J Med 331(21):1428–1436. https://doi.org/10.1056/NEJM199411243312107
Karjalainen JK (1990) Islet cell antibodies as predictive markers for IDDM in children with high background incidence of disease. Diabetes 39(9):1144–1150. https://doi.org/10.2337/diab.39.9.1144
Hattersley AT, Greeley SAW, Polak M et al (2018) ISPAD clinical practice consensus guidelines 2018: the diagnosis and management of monogenic diabetes in children and adolescents. Pediatr Diabetes 19(Suppl 27):47–63. https://doi.org/10.1111/pedi.12772
Taka A-M, Härkönen T, Vähäsalo P et al (2022) Heterogeneity in the presentation of clinical type 1 diabetes defined by the level of risk conferred by human leukocyte antigen class II genotypes. Pediatr Diabetes 23(2):219–227. https://doi.org/10.1111/pedi.13300
Shields BM, McDonald TJ, Ellard S, Campbell MJ, Hyde C, Hattersley AT (2012) The development and validation of a clinical prediction model to determine the probability of MODY in patients with young-onset diabetes. Diabetologia 55(5):1265–1272. https://doi.org/10.1007/s00125-011-2418-8
Chakera AJ, Spyer G, Vincent N, Ellard S, Hattersley AT, Dunne FP (2014) The 0.1% of the population with glucokinase monogenic diabetes can be recognized by clinical characteristics in pregnancy: the Atlantic Diabetes in Pregnancy cohort. Diabetes Care 37(5):1230–1236. https://doi.org/10.2337/dc13-2248
Wiedemann C, Kumar A, Lang A, Ohlenschläger O (2020) Cysteines and disulfide bonds as structure-forming units: insights from different domains of life and the potential for characterization by NMR. Front Chem 8:280. https://doi.org/10.3389/fchem.2020.00280
Bowman P, Sulen Å, Barbetti F et al (2018) Effectiveness and safety of long-term treatment with sulfonylureas in patients with neonatal diabetes due to KCNJ11 mutations: an international cohort study. Lancet Diabetes Endocrinol 6(8):637–646. https://doi.org/10.1016/S2213-8587(18)30106-2
Pearson ER, Starkey BJ, Powell RJ, Gribble FM, Clark PM, Hattersley AT (2003) Genetic cause of hyperglycaemia and response to treatment in diabetes. Lancet 362(9392):1275–1281. https://doi.org/10.1016/S0140-6736(03)14571-0
Vergari E, Knudsen JG, Ramracheya R et al (2019) Insulin inhibits glucagon release by SGLT2-induced stimulation of somatostatin secretion. Nat Commun 10(1):139. https://doi.org/10.1038/s41467-018-08193-8
Fu Z, Gilbert ER, Liu D (2013) Regulation of insulin synthesis and secretion and pancreatic beta-cell dysfunction in diabetes. Curr Diabetes Rev 9(1):25–53. https://doi.org/10.2174/157339913804143225
Kettunen JLT, Parviainen H, Miettinen PJ et al (2017) Biliary anomalies in patients with HNF1B diabetes. J Clin Endocrinol Metab 102(6):2075–2082. https://doi.org/10.1210/jc.2017-00061
Mory PB, Crispim F, Freire MBS et al (2012) Phenotypic diversity in patients with lipodystrophy associated with LMNA mutations. Eur J Endocrinol 167(3):423–431. https://doi.org/10.1530/EJE-12-0268
Reschke F, Rohayem J, Maffei P et al (2021) Collaboration for rare diabetes: understanding new treatment options for Wolfram syndrome. Endocrine 71(3):626–633. https://doi.org/10.1007/s12020-021-02622-3
Huopio H, Miettinen PJ, Ilonen J et al (2016) Clinical, genetic, and biochemical characteristics of early-onset diabetes in the finnish population. J Clin Endocrinol Metab 101(8):3018–3026. https://doi.org/10.1210/jc.2015-4296
Parviainen A, But A, Siljander H, Knip M, Finnish Pediatric Diabetes Register (2020) Decreased incidence of type 1 diabetes in young finnish children. Diabetes Care 43(12):2953–2958. https://doi.org/10.2337/dc20-0604
Irgens HU, Molnes J, Johansson BB et al (2013) Prevalence of monogenic diabetes in the population-based Norwegian childhood diabetes registry. Diabetologia 56(7):1512–1519. https://doi.org/10.1007/s00125-013-2916-y
Naylor R (2019) Economics of genetic testing for diabetes. Curr Diab Rep 19(5):23. https://doi.org/10.1007/s11892-019-1140-7
Greeley SAW, John PM, Winn AN et al (2011) The cost-effectiveness of personalized genetic medicine. Diabetes Care 34(3):622–627. https://doi.org/10.2337/dc10-1616
Johnson SR, Carter HE, Leo P et al (2019) Cost-effectiveness analysis of routine screening using massively parallel sequencing for maturity-onset diabetes of the young in a pediatric diabetes cohort: reduced health system costs and improved patient quality of life. Diabetes Care 42(1):69–76. https://doi.org/10.2337/dc18-0261
Naylor RN, John PM, Winn AN et al (2014) Cost-effectiveness of MODY genetic testing: translating genomic advances into practical health applications. Diabetes Care 37(1):202–209. https://doi.org/10.2337/dc13-0410
Acknowledgements
We thank all participants, hospitals and personnel of the Finnish Pediatric Diabetes Register and FINNMODY Study as well as the collaborators at FIMM Genomics NGS Sequencing unit at University of Helsinki supported by HiLIFE and Biocenter Finland. Some of the data were presented as an abstract at the 58th EASD Annual Meeting in 2022.
Authors’ relationships and activities
The authors declare that there are no relationships or activities that might bias, or be perceived to bias, their work.
Contribution statement
All authors have made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data, have participated in drafting the article or revising it critically for important intellectual content, and have approved the final version to be published. TT is responsible for the integrity of the work as a whole.
Funding
Open Access funding provided by University of Helsinki including Helsinki University Central Hospital. The FPDR has been supported by the Academy of Finland (grant no. 292538 to MK), the Sigrid Juselius Foundation, Finska Läkaresällskapet, and the Liv and Hälsa Fund. The Botnia and FINNMODY Study have been supported by grants from the Folkhälsan Research Foundation, the Sigrid Juselius Foundation, the Academy of Finland (grants 263401, 267882 and 312063 to LG; grant no. 312072 to TT), University of Helsinki, Nordic Center of Excellence in Disease Genetics, EU (EXGENESIS, MOSAIC FP7-600914), Ollqvist Foundation, Swedish Cultural Foundation in Finland, Novo Nordisk Foundation, Finnish Diabetes Research Foundation, Foundation for Life and Health in Finland, Signe and Ane Gyllenberg Foundation, Finnish Medical Society, Paavo Nurmi Foundation, State Research Funding via the Helsinki University Hospital, Perklén Foundation, Närpes Health Care Foundation and Ahokas Foundation. The study has also been supported by the Ministry of Education in Finland, Municipal Health Care Center and Hospital in Jakobstad and Health Care Centers in Vasa, Närpes and Korsholm.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM
(PDF 169 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Harsunen, M., Kettunen, J.L.T., Härkönen, T. et al. Identification of monogenic variants in more than ten per cent of children without type 1 diabetes-related autoantibodies at diagnosis in the Finnish Pediatric Diabetes Register. Diabetologia 66, 438–449 (2023). https://doi.org/10.1007/s00125-022-05834-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-022-05834-y