Abstract
Aims/hypothesis
Sugar-sweetened beverages (SSBs) are a major dietary contributor to fructose intake. A molecular pathway involving the carbohydrate responsive element-binding protein (ChREBP) and the metabolic hormone fibroblast growth factor 21 (FGF21) may influence sugar metabolism and, thereby, contribute to fructose-induced metabolic disease. We hypothesise that common variants in 11 genes involved in fructose metabolism and the ChREBP-FGF21 pathway may interact with SSB intake to exacerbate positive associations between higher SSB intake and glycaemic traits.
Methods
Data from 11 cohorts (six discovery and five replication) in the CHARGE (Cohorts for Heart and Aging Research in Genomic Epidemiology) Consortium provided association and interaction results from 34,748 adults of European descent. SSB intake (soft drinks, fruit punches, lemonades or other fruit drinks) was derived from food-frequency questionnaires and food diaries. In fixed-effects meta-analyses, we quantified: (1) the associations between SSBs and glycaemic traits (fasting glucose and fasting insulin); and (2) the interactions between SSBs and 18 independent SNPs related to the ChREBP-FGF21 pathway.
Results
In our combined meta-analyses of discovery and replication cohorts, after adjustment for age, sex, energy intake, BMI and other dietary covariates, each additional serving of SSB intake was associated with higher fasting glucose (β ± SE 0.014 ± 0.004 [mmol/l], p = 1.5 × 10−3) and higher fasting insulin (0.030 ± 0.005 [log e pmol/l], p = 2.0 × 10−10). No significant interactions on glycaemic traits were observed between SSB intake and selected SNPs. While a suggestive interaction was observed in the discovery cohorts with a SNP (rs1542423) in the β-Klotho (KLB) locus on fasting insulin (0.030 ± 0.011 log e pmol/l, uncorrected p = 0.006), results in the replication cohorts and combined meta-analyses were non-significant.
Conclusions/interpretation
In this large meta-analysis, we observed that SSB intake was associated with higher fasting glucose and insulin. Although a suggestive interaction with a genetic variant in the ChREBP-FGF21 pathway was observed in the discovery cohorts, this observation was not confirmed in the replication analysis.
Trial registration
Trials related to this study were registered at clinicaltrials.gov as NCT00005131 (Atherosclerosis Risk in Communities), NCT00005133 (Cardiovascular Health Study), NCT00005121 (Framingham Offspring Study), NCT00005487 (Multi-Ethnic Study of Atherosclerosis) and NCT00005152 (Nurses’ Health Study).
Similar content being viewed by others
Introduction
Epidemiological evidence suggests that sugar-sweetened beverage (SSB) intake is associated with increased risk of the metabolic syndrome [1, 2] and type 2 diabetes [3]. Sucrose (table sugar) and high-fructose corn syrup are the most common forms of sugar in SSBs, composed of nearly equal amounts of glucose and fructose [4]. Evidence from some [5, 6], but not all [7, 8], human intervention studies suggests that it is the fructose moiety which elicits adverse cardiometabolic effects. Currently, an estimated 9.4% of adults in the USA have type 2 diabetes, while 34% have elevated blood glucose levels [9], a condition associated with insulin resistance and increased risk for type 2 diabetes. Excess sugar intake, particularly in the form of SSBs, is one aspect of the diet that may impair glucose homeostasis and contribute to greater insulin resistance [3, 10, 11].
Carbohydrate responsive element-binding protein (ChREBP, also known as MLX interacting protein like or MLXIPL) is a transcription factor that responds to intracellular carbohydrate metabolites and is a principal mediator of carbohydrate-induced gene expression in key metabolic tissues, including the liver [12,13,14]. Recent data indicate that hepatic ChREBP is particularly responsive to fructose intake [15] and contributes to fructose-induced lipid and glycaemic abnormalities in animals and humans [16, 17]. Variants in the CHREBP (also known as MLXIPL) locus associate with hypertriacylglycerolaemia and low HDL-cholesterol at genome-wide significance levels [18, 19]. We have also demonstrated that fructose ingestion in humans acutely increases circulating levels of the novel metabolic hormone fibroblast growth factor 21 (FGF21), and ChREBP is required for its activation [20, 21]. Pharmacological administration and genetic manipulation of FGF21 has pleiotropic effects on carbohydrate and lipid metabolism [22, 23]. We and others have recently reported that SNPs in the FGF21 locus are associated with higher circulating FGF21 concentrations and higher carbohydrate relative to fat intake in humans [24, 25]. Together, these data suggest that the ChREBP-FGF21 hormonal axis may mediate an adaptive metabolic response to sugar consumption.
Given a role for ChREBP in contributing to sugar-induced derangements in both lipid and glucose homeostasis [26, 27], we sought to test the hypothesis that variants associated with hypertriacylglycerolaemia in the ChREBP pathway might interact with SSB consumption to regulate glycaemic traits (see the electronic supplementary material [ESM] for further details about SNP selection). Aside from variants in CHREBP, we selected SNPs that have previously showed significant (i.e. p < 5 × 10−8) or suggestive (i.e. p < 5 × 10−6) associations with hypertriacylglycerolaemia or low HDL-cholesterol in human genes important for hepatic fructose and glucose metabolism (KHK, ALDOB, GCK, SLC2A2, SLC2A5) [17, 28,29,30,31,32,33,34,35,36]. We also included other genes implicated in the regulation of both ChREBP and blood triacylglycerol levels (FADS1 and TRIB1) [37,38,39,40,41]. Finally, we included variants in the loci that code for ChREBP-regulated metabolic hormone FGF21 and its obligate receptor KLB [14, 20, 42, 43].
We hypothesised that common (minor allele frequency [MAF] ≥5%) SNPs in these 11 genes may interact with SSB intake to regulate glycaemic traits and, in particular, that risk allele SNPs may exacerbate the positive associations between SSB intake and glycaemic traits. The aims of the current investigation were to: 1) evaluate the relationship between SSB intake and glycaemic traits, i.e. circulating levels of fasting glucose and fasting insulin, in studies from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium; and 2) examine whether these associations are modified by SNPs related to ChREBP function.
Methods
Discovery and replication cohorts
The present cross-sectional meta-analyses included up to 34,748 participants of European descent from 11 US and European cohort studies in the CHARGE Consortium Nutrition Working Group (ESM Table 1). Of those cohorts, six formed our discovery cohorts. Five additional cohorts (replication cohorts) were later invited to join the study to verify a suggestive interaction observed in analyses of the discovery cohorts. Participants provided written informed consent. The research protocol was approved by each institutional review board and/or oversight committee.
Dietary assessment, glycaemic trait measurements and other relevant variables
Dietary intake data were collected by validated food-frequency questionnaires (FFQs) in all cohorts, except in the Malmö Diet and Cancer [MDC] Study, which estimated intake using FFQs in combination with a 7 day food record for prepared/cooked meals (ESM Table 2). The type of FFQ used in each cohort differed slightly to capture the dietary habits of the specific population. SSB intake included regular caffeinated, caffeine-free and carbonated non-cola soft drinks (soda), and fruit-flavoured drinks, e.g. lemonade, Hawaiian punch. Fruit juice (100%) was not included in the estimation of SSBs with the exception of one study (Western Australian Pregnancy Cohort Study [Raine]) that could not distinguish fruit juice from other beverages based on their dietary assessment tool. One serving of SSB was defined as 360 ml (12 fl oz.; the volume in one standard soft drink can). Fruit intake, vegetable intake, whole grain intake and fish intake in servings/day, alcohol intake in grams/day and saturated fatty acid as percentage of total energy intake (where 1 g saturated fat has 37 kJ) were further quantified and used as covariates in the present analysis. SSB intake was considered continuously and further dichotomised into low (<1 serving [<360 ml/day]) and high (≥1 serving [≥360 ml/day]) intakes, whereas all remaining dietary variables were considered continuously only.
Glycaemic biomarkers were typically measured after ≥8 h fasting. Cohort-specific assessment methods for fasting glucose and fasting insulin were quantified using similar procedures, primarily by enzymatic methods and radioimmunoassay, respectively. Fasting glucose was not measured in one cohort (Nurses’ Health Study [NHS]). BMI was calculated from measured weight (kg) divided by height squared (m2). A description of cohort-specific methodologies for all relevant variables is provided in ESM Table 3.
Genotyping
Based on the hypothesis that genetic determinants of fasting hypertriacylglycerolaemia and insulin insensitivity may be shared, we used publicly available genotype-phenotype data [44] to select SNPs that have previously showed significant (i.e. p < 5 × 10−8) or suggestive (i.e. p < 5 × 10−6) associations with hypertriacylglycerolaemia or low HDL-cholesterol in humans, and were found within the CHREBP gene or genes predicted to regulate either ChREBP or the biological response to ChREBP activation. A total of 18 independent common (MAF ≥5%) SNPs in 11 genes in the ChREBP-FGF21 pathway were included in the present analysis (ESM Table 4).
In this analysis, SNPs were previously directly genotyped or imputed by participating cohorts before inclusion (ESM Table 5). SNPs were assessed for quality control using multiple metrics (see ESM Methods [Genotype exclusion criteria]). Not all SNPs were available in all participating cohorts (ESM Table 6).
Cohort-specific analyses
All discovery and replication cohorts followed a uniform, pre-specified analysis plan. Natural logarithmic transformation was applied to fasting insulin. Participants within each cohort were excluded from the present analysis when they had type 2 diabetes (prevalent or self-reported), were taking medication for type 2 diabetes, had fasting glucose ≥7 mmol/l (≥126 mg/dl) or were not fasting at blood draw. Participants were also excluded if they had implausible dietary data based on cohort-specific cut-points or missing genotype data.
The main associations between SSB intake and fasting glucose and insulin concentrations in the discovery and replication cohorts were estimated using linear regression models or linear mixed-effects models for family data, adjusted for the following covariates: model 1 adjusted for age, sex, energy intake (kJ) and study site for multi-centred cohorts (in the Cardiovascular Health Study [CHS], Multi-Ethnic Study of Atherosclerosis [MESA], Cardiovascular Risk in Young Finns Study [YFS] and Atherosclerosis Risk In Communities [ARIC] Study); model 2 adjusted for model 1 covariates plus smoking status, education status, alcohol intake and physical activity (except where unavailable: Rotterdam Study I [RS1], Rotterdam Study II [RS2] and Raine); model 3 adjusted for model 2 covariates plus BMI; model 4 adjusted for model 3 covariates plus fruit intake, vegetable intake, whole grain intake (except where unavailable: Netherlands Epidemiology in Obesity Study [NEO]), fish intake and saturated fatty acids (percentage of total energy). As satiety responses and energy compensation differ between men and women following SSB intake [45, 46], further analyses were conducted using stratification by sex.
The main associations between selected SNPs and glycaemic outcomes, as well as interaction analyses between SSB intake and SNPs, were also investigated (see ESM Methods [Genetic analyses]). In discovery cohorts, the interaction tests were performed for all selected SNPs on glycaemic outcomes using linear regression analyses or linear mixed-effects models for family data adjusted for age, sex, energy intake, BMI, study site for multi-centred cohorts and population structure where applicable. Suggestive interaction results (i.e. p < 0.05) for one SNP from the discovery cohorts was examined in the replication cohorts and further examined in sex-specific analyses, and in analyses whereby SSB intake was dichotomised into low (<1 serving/day) and high (≥1 serving/day) intakes. Secondary analyses examining sex-stratified associations for all selected SNPs and interactions were also pursued (see ESM Methods [Genetic analyses]).
Meta-analyses
For the discovery cohorts, we conducted inverse-variance weighted, fixed-effect meta-analyses using the ‘metafor’ R package (https://cran.r-project.org) for the main associations of SSB intake on fasting glucose and insulin, selected SNPs on outcomes, interactions between SSB intake and selected SNPs on outcomes, and sex-stratified main associations and interactions of the selected SNPs on outcomes. Statistical significance for the association/interaction tests was defined at a level of 0.001, based on Bonferroni correction for 36 (18 independent SNPs × 2 glycaemic outcomes) total tests. We performed post hoc power calculations using Quanto version 1.2.4 (http://biostats.usc.edu/Quanto.html) (see ESM Methods [Power calculations]).
For suggestive interaction results (i.e. unadjusted p < 0.05) from the discovery analyses, we further investigated: (1) the main effect associations of SSB intake with fasting glucose and insulin concentrations; (2) the main associations between the nominally significant SNPs and glycaemic outcomes; and (3) the interactions between SSB intake and nominally significant SNPs on glycaemic outcomes in replication cohorts. These suggestive interactions were further explored in sex-specific meta-analyses and in meta-analyses with SSB intake dichotomised into high and low intakes. In addition, we conducted a combined meta-analysis (combined discovery and replication cohorts) for the described analyses.
Heterogeneity across studies was examined and, when detected, followed by meta-regression and sensitivity analysis described in the ESM Methods (Sensitivity analyses). Finally, we ran random-effect meta-analyses for: (1) the main associations of SSB intake on fasting glucose and insulin; and (2) the interactions between SSB intake and selected SNPs on outcomes for the combined meta-analyses.
Results
Study characteristics
General demographic characteristics and dietary intake of participants in the discovery and replication cohorts are provided in Table 1 (ESM Table 7 for sex-stratified characteristics). The mean age ranged from 20.1 to 72.3 years and women comprised 52% to 100% within each cohort. The mean SSB intake ranged from 0.10 servings/day (RS1) to 0.98 servings/day (Raine). Mean BMI ranged from 24.5 to 29.6 kg/m2.
Associations of SSB intake with glycaemic traits
Associations between SSB intake and each glycaemic trait are presented in Table 2 (Figs 1, 2). Results are presented for the fully adjusted model (model 4), with other model results presented when findings varied. In the discovery cohort analyses, we observed a positive association between SSB intake and fasting insulin: each additional serving of SSB intake was associated with higher fasting insulin (β ± SE 0.027 ± 0.008 [log e pmol/l], p = 1.4 × 10−3). No statistically significant associations were observed between SSB intake and fasting glucose. In the replication cohort analyses, we observed a positive association between SSB intake and both fasting glucose (β ± SE 0.015 ± 0.005 (mmol/l), p = 2.3 × 10−3) and fasting insulin (β ± SE 0.032 ± 0.006 [log e pmol/l], p = 3.3 × 10−8). In combined meta-analyses, associations for both fasting glucose (β ± SE 0.014 ± 0.004 [mmol/l], p = 1.5 × 10−3) and fasting insulin (β ± SE 0.030 ± 0.005 [log e pmol/l], p = 2.0 × 10−10] were also observed.
In sex-stratified analyses, we observed a positive association between SSB intake and fasting glucose among women only (men: β ± SE 0.001 ± 0.006 mmol/l, p = 0.82 [all cohorts]; women: β ± SE 0.026 ± 0.006 mmol/l, p = 5.5 × 10−5 [all cohorts]), and between SSB intake and fasting insulin among both men and women (men: β ± SE 0.029 ± 0.006 log e pmol/l, p = 4.5 × 10−6; women: β ± SE 0.031 ± 0.007 log e pmol/l, p = 1.7 × 10−5 [all cohorts]) (Table 3). Overall, low heterogeneity (I 2 < 30%) was observed in fasting glucose-related analyses (model 4). Higher heterogeneity was observed in fasting insulin analyses, particularly among replication cohorts (I 2 69%).
Associations of SNPs with glycaemic traits
The main associations of selected SNPs on glycaemic traits are presented in ESM Tables 8 and 9. In the meta-analysis, we replicated associations between fasting glucose and GCK-rs4607517 [47], GCKR-rs1260326 [48] and SLC2A2-rs11920090 [47] variants. The association between fasting glucose and GCKR-rs1260326 was observed among women only (ESM Table 10). We also replicated the association between fasting insulin and GCKR-rs1260326 in the entire population and in sex-stratified analyses (ESM Table 11) [48]. We found a novel, statistically significant association between fasting glucose and FADS1-rs174546, and observed nominally significant associations (p < 0.05) for fasting glucose with KLB-rs1542423 and fasting glucose with SLC2A2-rs11924032 (ESM Table 8).
Interactions between SSB intake and selected SNPs on glycaemic traits
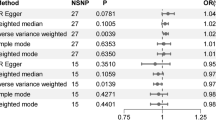
Meta-analysed estimates of the interactions between SSB intake and selected SNPs on glycaemic traits are presented in Table 4 (for sex-stratified results see ESM Tables 12, 13). In the discovery cohort analyses, we did not observe a statistically significant interaction between SSB intake and any candidate SNP, even in sex-stratified interaction analyses. We did, however, observe a suggestive interaction between SSB intake and KLB-rs1542423, an intronic SNP in the β-Klotho gene (KLB), for fasting insulin (β ± SE 0.0302 ± 0.011 log e pmol/l, p = 0.006). The effect of the interaction suggests a 0.0302 log e pmol/l higher fasting insulin with each additional serving of SSB intake per copy of the KLB-rs1542423 T allele (Fig. 3). This nominal interaction between SSB intake and KLB-rs1542423 for fasting insulin was not supported in the replication cohort analyses (β ± SE -0.0109 ± 0.0082 log e pmol/l, p = 0.18). The suggestive interaction for this SNP was also lacking in combined meta-analyses, sex-stratified analyses and analyses with dichotomised SSB intake (Table 5, Fig. 3). See ESM Results (Sex-stratified interaction analyses and Meta-regression and sensitivity analyses) for results of sex-stratified analyses for additional SNPs (ESM Tables 13, 14), as well as results from meta-regression and sensitivity analyses (ESM Tables 15, 16).
Discussion
In this meta-analysis involving more than 34,748 participants free of type 2 diabetes in 11 cohort studies from the CHARGE Consortium, we observed significant associations between SSB intake and fasting glucose and insulin concentrations, independent of demographics, overall adiposity, total energy intake and other dietary factors. We adjusted for BMI to consider whether obesity may be in the causal pathway between SSB and fasting insulin or glucose, since consuming SSB may lead to a higher BMI, and a higher BMI is associated with worsening glycaemic traits. We observed that the results remained largely the same without attenuation after accounting for BMI, suggesting that although SSBs may increase body weight and adiposity, the relationship with glycaemic traits is independent from adiposity. For each additional serving of SSBs, fasting insulin was 3% higher. The SSB association with fasting glucose was less consistent. Significant associations were observed only in the replication cohorts, and in the meta-analysis of all cohorts only in women. There was no evidence of SNP–SSB interactions in the meta-analysis of all cohorts or in the sex-stratified analysis.
This is the first meta-analysis to assess the association of SSB intake with measures of diabetic risk factors and confirms the positive association between SSB consumption and insulin resistance (HOMA-IR or fasting insulin) suggested by cross-sectional studies in adults [10, 11], young children [49] and adolescents [50, 51]. However, in well-controlled, short-term intervention studies in healthy adults, the evidence is less consistent with some studies reporting that consumption of fructose-containing sugars for 3–10 weeks has a detrimental effect on insulin sensitivity [5, 6, 52], whereas others observed no significant detrimental effect on insulin resistance [53, 54]. Nevertheless, given the observed associations between SSB intake and risk of diabetes [2, 3, 55], our results further favour efforts to assess the potential beneficial effects of reducing SSB consumption on cardiometabolic risk factors in human populations.
In this meta-analysis, we confirmed the previously reported SNP associations with fasting glucose and insulin in GCK-rs4607517 [47], GCKR-rs1260326 [48], SLC2A2-rs11920090 [47] and GCKR-rs1260326 in women only [48]. We also observed a positive association between fasting glucose and the FADS1-rs174546 variant, which is in linkage disequilibrium with FADS1-rs174550, recognised as having an association with fasting glucose [47] and also in linkage disequilibrium with the FADS1-rs174547 variant, which has been associated with atherogenic dyslipidaemia. While we have not formally investigated the relationship between SSB intake and our selected SNPs, our lookup in a large macronutrient intake genome-wide association study from the CHARGE Consortium indicates an association between the FGF21-rs8381.33 variant and carbohydrate intake (ESM Table 17).
To date, few studies have considered whether genetic variation impacts the susceptibility to the detrimental effects of SSB intake on key cardiometabolic traits. In a large cohort of men and women in the USA [56], as well as two large Swedish [57] and Finnish cohorts [58], SSB intake significantly interacted with underlying genetic predisposition for weight gain and obesity risk. More recently, daily SSB intake was observed to interact with variants in the 9p21 region to exacerbate the genetic predisposition effects on coronary artery disease in Hispanics living in Costa Rica [59]. In Hispanic children, the effects of PNPLA3 on liver fat were exacerbated under conditions of a high carbohydrate diet, in particular high sugar intake [60]. Although limited to a few studies, the findings indicate that SSB intake may interact with genetic variants to increase cardiometabolic risk in susceptible individuals.
Here, we pursued a candidate approach to examine whether SNPs in a ChREBP-FGF21 pathway might interact with SSB intake to regulate glycaemic traits. In the discovery phase of our analysis, we identified a promising interaction between the KLB SNP (rs1542423) and SSB for fasting insulin. We observed that individuals who carried a T allele in this SNP consistently had a higher level of fasting insulin in response to high SSB intake in five of the six discovery cohorts. Because these data were consistent with our hypothesis that variants in a ChREBP-FGF21 signalling axis might regulate metabolic traits in response to SSB intake, we sought out replication cohorts to further test this suggestive interaction. The interaction between SSB intake and KLB-rs1542423 for fasting insulin was not significant in the replication cohorts, or in the combined meta-analysis of all 11 participating cohorts, suggesting a false-positive finding. Because we observed a sex-specific main association between SSB intake and fasting glucose, we pursued sex-stratified interaction analyses of SSB intake by selected SNPs as a secondary analysis. We observed a suggestive interaction between SSB intake with one SNP in men (FGF21-rs838133) and one SNP (GCK-rs4607517) in women for fasting insulin.
There are several limitations to our study. One limitation is the focus on a small number of SNPs in a hypothesised candidate gene pathway. While this excludes many other genes and regulatory regions, the focus provides a testable hypothesis and reduces the penalty for genome-wide testing. Sufficiently large populations with the requisite genotyping, phenotyping and dietary information do not yet exist to achieve statistical power sufficient for a genome-wide approach. A second limitation is the heterogeneity within the discovery cohorts as well as heterogeneity between the discovery and replication cohorts. Although cohort inclusion is based upon European ancestry, each cohort has unique characteristics in terms of location, age, sex and covariate structure. For example, participants were, on average, younger in the replication cohorts compared with the discovery cohorts (mean age 54.2 vs 57.6 years). We have also observed additional significant differences in fasting insulin and SSB intake, among other general characteristics including BMI, smoking, education and energy intake (ESM Table 18). Although we attempted to adjust for age in our regression models, this difference in age could have a non-linear impact on the effects of the variant on SSB-induced insulin resistance, thereby contributing to residual confounding. The difference in age could contribute to the difference in SSB intake as the mean SSB intake was higher in the replication cohorts (0.19 to 0.98 servings/day) compared with the discovery cohorts (0.10 to 0.32 servings/day) (p < 0.0001). Furthermore, meta-regression findings suggest differences in the magnitude, but not significance, of the associations between SSB intake and fasting insulin. This may be a result of differences in the moderator in the analysis, such as age and BMI, or as a result of other trait differences in the cohorts. Despite those differences, the associations still remain in subgroup analyses, with the exception of subgroup analyses by sample size, possibly as a result of low power in the analyses with smaller cohorts.
It is important to note that these analyses use a cross-sectional design, incorporating the phenotypic measures (fasting glucose and fasting insulin) and SSB intake at one point in time. Although many of the cohorts contributing to the analyses are longitudinal in nature, not all have measures of outcome and exposure longitudinally. Thus, we did not capture long-term SSB intake patterns, which probably change with age, and thus misclassification of dietary exposure may vary across cohorts. Furthermore, SSB intake was significantly associated with fasting glucose among women, but not men. It has been noted that the effects of excessive sugars on glycaemic traits in animal models are sexually dimorphic although the pattern is not the same as observed here [61]. Though we have no mechanistic explanation for the difference at this time, our results support the need for future studies concerning the metabolic effects of SSBs to carefully consider sex-based stratification.
Finally, the use of self-reported data in our assessment of dietary intake may be susceptible to reporting bias, such as under-reporting, and the validity of questionnaires may vary across cohorts, thereby potentially attenuating associations. Strengths of the study include the large sample size attained by our meta-analytic approach necessary to detect gene–environment interactions. Our collaborative approach also enabled us to standardise our analyses across cohorts. The observed interaction regression coefficients were small compared with the magnitude of interaction observed in other studies looking at gene–environment interactions between SSBs and cardiometabolic outcomes [56, 59]. Thus, even with the large sample size in this study, it is possible that we were insufficiently powered to detect and replicate a small gene–SSB interaction. If such interactions did exist, but are too small to be detected in this analysis, the clinical relevance of such small interactions might be questioned. Nevertheless, our candidate gene approach was suggestive of interaction at one locus, and the ChREBP-FGF21 pathway remains mechanistically interesting.
Variants within the CHREBP locus associate with hypertriacylglycerolaemia [18, 19]. For this analysis, SNPs within candidate genes in a putative ChREBP-FGF21 signalling axis (ESM Table 4) were selected based on genome-wide or sub-genome-wide association with fasting triacylglycerol levels, and not on the basis of glycaemic traits. This approach was pursued because excess sugar consumption is thought to cause hypertriacylglycerolaemia, and hypertriacylglycerolaemia and insulin resistance are linked epidemiologically and may share common pathogenic mechanisms [26, 62, 63]. Thus, implicit to this strategy is the hypothesis that genetic determinants of fasting hypertriacylglycerolaemia may be linked to insulin sensitivity. One limitation of this approach is that mechanisms mediating sugar-induced hypertriacylglycerolaemia and insulin resistance may be distinct. A second limitation is that our analyses were limited to 18 lead SNPs, and it is possible that SNPs that interact with the environment to associate with a trait are distinct from the variants that associate with a trait unconditioned on the environment, particularly those associating with a trait at genome-wide significance threshold. Thus, it may be necessary to examine all SNPs within a locus of interest as opposed to a lead SNP, although this would further increase the burden of multiple testing. Future studies should undertake a more comprehensive testing for interactions between SSB intake and key genes like KLB on glycaemic outcomes.
In summary, the present observational study from 11 cohorts is the largest investigation of the relationship between SSB intake, genetics and glycaemic traits. We observed that SSB intake was positively associated with higher fasting insulin and glucose. Although a suggestive interaction with a genetic variant in the ChREBP-FGF21 signalling axis was observed in the discovery cohorts, this observation was not confirmed in the replication analysis. In conclusion, our results suggest that SSB consumption may unfavourably impact glucose homeostasis in different populations, regardless of genotypes at loci within the ChREBP-FGF21 signalling axis.
Abbreviations
- ARIC:
-
Atherosclerosis Risk In Communities
- CHARGE:
-
Cohorts for Heart and Aging Research in Genomic Epidemiology
- ChREBP:
-
Carbohydrate responsive element-binding protein
- CHS:
-
Cardiovascular Health Study
- FFQ:
-
Food-frequency questionnaire
- FGF21:
-
Fibroblast growth factor 21
- FHS:
-
Framingham Heart Study
- MAF:
-
Minor allele frequency
- MDC:
-
Malmö Diet and Cancer
- MESA:
-
Multi-Ethnic Study of Atherosclerosis
- NEO:
-
Netherlands Epidemiology in Obesity Study
- NHS:
-
Nurses’ Health Study
- RS1:
-
Rotterdam Study I
- RS2:
-
Rotterdam Study II
- SSB:
-
Sugar-sweetened beverage
- YFS:
-
Cardiovascular Risk in Young Finns Study
References
Ma J, McKeown NM, Hwang S-J et al (2016) Sugar-sweetened beverage consumption is associated with change of visceral adipose tissue over 6 years of follow-up. Circulation 133:370–377
Malik VS, Popkin BM, Bray GA et al (2010) Sugar-sweetened beverages and risk of metabolic syndrome and type 2 diabetes: a meta-analysis. Diabetes Care 33:2477–2483
Imamura F, O’Connor L, Ye Z et al (2015) Consumption of sugar sweetened beverages, artificially sweetened beverages, and fruit juice and incidence of type 2 diabetes: systematic review, meta-analysis, and estimation of population attributable fraction. BMJ. https://doi.org/10.1136/bmj.h3576
Welsh JA, Sharma A, Cunningham SA, Vos MB (2011) Consumption of added sugars and indicators of cardiovascular disease risk among US adolescents. Circulation 123:249–257
Stanhope KL, Schwarz JM, Keim NL et al (2009) Consuming fructose-sweetened, not glucose-sweetened, beverages increases visceral adiposity and lipids and decreases insulin sensitivity in overweight/obese humans. J Clin Invest 119:1322–1334
Aeberli I, Hochuli M, Gerber PA et al (2013) Moderate amounts of fructose consumption impair insulin sensitivity in healthy young men. Diabetes Care 36:150–156
Kuzma JN, Cromer G, Hagman DK et al (2016) No differential effect of beverages sweetened with fructose, high-fructose corn syrup, or glucose on systemic or adipose tissue inflammation in normal-weight to obese adults: a randomized controlled trial. Am J Clin Nutr 104:306–314
Silbernagel G, Machann J, Häring H-U et al (2014) Plasminogen activator inhibitor-1, monocyte chemoattractant protein-1, e-selectin and C-reactive protein levels in response to 4-week very-high-fructose or -glucose diets. Eur J Clin Nutr 68:97–100
Centers for Disease Control 2017. Centers for Disease Control National Diabetes Statistics Report. www.cdc.gov/diabetes/data/statistics/statistics-report.html. Accessed 17 October 2017
Lana A, Rodríguez-Artalejo F, Lopez-Garcia E (2014) Consumption of sugar-sweetened beverages is positively related to insulin resistance and higher plasma leptin concentrations in men and nonoverweight women. J Nutr 144:1099–1105
Yoshida M, McKeown NM, Rogers G et al (2007) Surrogate markers of insulin resistance are associated with consumption of sugar-sweetened drinks and fruit juice in middle and older-aged adults. J Nutr 137:2121–2127
Ogawa Y, Kurosu H, Yamamoto M et al (2007) β-Klotho is required for metabolic activity of fibroblast growth factor 21. Proc Natl Acad Sci 104:7432–7437
Uyeda K, Repa JJ (2006) Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab 4:107–110
Iizuka K, Takeda J, Horikawa Y (2009) Glucose induces FGF21 mRNA expression through ChREBP activation in rat hepatocytes. FEBS Lett 583:2882–2886
Koo H-Y, Miyashita M, Simon Cho BH, Nakamura MT (2009) Replacing dietary glucose with fructose increases ChREBP activity and SREBP-1 protein in rat liver nucleus. Biochem Biophys Res Commun 390:285–289
Erion DM, Popov V, Hsiao JJ et al (2013) The role of the carbohydrate response element-binding protein in male fructose-fed rats. Endocrinology 154:36–44
Kim M-S, Krawczyk SA, Doridot L et al (2016) ChREBP regulates fructose-induced glucose production independently of insulin signaling. J Clin Invest 126:4372–4386
Kooner JS, Chambers JC, Aguilar-Salinas CA et al (2008) Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat Genet 40:149–151
Kathiresan S, Melander O, Guiducci C et al (2008) Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet 40:189–197
Dushay JR, Toschi E, Mitten EK et al (2015) Fructose ingestion acutely stimulates circulating FGF21 levels in humans. Mol Metab 4:51–57
Fisher FM, Kim M, Doridot L et al (2017) A critical role for ChREBP-mediated FGF21 secretion in hepatic fructose metabolism. Mol Metab 6:14–21
Emanuelli B, Vienberg SG, Smyth G et al (2014) Interplay between FGF21 and insulin action in the liver regulates metabolism. J Clin Invest 124:515–527
Gimeno RE, Moller DE (2014) FGF21-based pharmacotherapy—potential utility for metabolic disorders. Trends Endocrinol Metab 25:303–311
Tanaka T, Ngwa JS, van Rooij FJ et al (2013) Genome-wide meta-analysis of observational studies shows common genetic variants associated with macronutrient intake. Am J Clin Nutr 97:1395–1402
Chu AY, Workalemahu T, Paynter NP et al (2013) Novel locus including FGF21 is associated with dietary macronutrient intake. Hum Mol Genet 22:1895–1902
Grundy SM (1999) Hypertriglyceridemia, insulin resistance, and the metabolic syndrome. Am J Cardiol 83:25–29
Olefsky JM, Farquhar JW, Reaven GM (1974) Reappraisal of the role of insulin in hypertriglyceridemia. Am J Med 57:551–560
Santer R, Rischewski J, von Weihe M et al (2005) The spectrum of aldolase B (ALDOB) mutations and the prevalence of hereditary fructose intolerance in Central Europe. Hum Mutat 25:594
van Schaftingen E (1989) A protein from rat liver confers to glucokinase the property of being antagonistically regulated by fructose 6-phosphate and fructose 1-phosphate. Eur J Biochem 179:179–184
Agius L (2008) Glucokinase and molecular aspects of liver glycogen metabolism. Biochem J 414:1–18
Helliwell PA, Richardson M, Affleck J, Kellett GL (2000) Stimulation of fructose transport across the intestinal brush-border membrane by PMA is mediated by GLUT2 and dynamically regulated by protein kinase C. Biochem J 350:149–154
Bonthron DT, Brady N, Donaldson IA, Steinmann B (1994) Molecular basis of essential fructosuria: molecular cloning and mutational analysis of human ketohexokinase (fructokinase). Hum Mol Genet 3:1627–1631
Corpe CP, Basaleh MM, Affleck J et al (1996) The regulation of GLUT5 and GLUT2 activity in the adaptation of intestinal brush-border fructose transport in diabetes. Pflugers Arch 432:192–201
Burant CF, Takeda J, Brot-Laroche E et al (1992) Fructose transporter in human spermatozoa and small intestine is GLUT5. J Biol Chem 267:14523–14526
Dentin R, Pégorier J-P, Benhamed F et al (2004) Hepatic glucokinase is required for the synergistic action of ChREBP and SREBP-1c on glycolytic and lipogenic gene expression. J Biol Chem 279:20314–20326
Iizuka K, Bruick RK, Liang G et al (2004) Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc Natl Acad Sci U S A 101:7281–7286
Dentin R, Benhamed F, Pégorier J-P et al (2005) Polyunsaturated fatty acids suppress glycolytic and lipogenic genes through the inhibition of ChREBP nuclear protein translocation. J Clin Invest 115:2843–2854
Tanaka T, Shen J, Abecasis GR et al (2009) Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI study. PLoS Genet 5:e1000338
Chambers JC, Zhang W, Sehmi J et al (2011) Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet 43:1131–1138
Ishizuka Y, Nakayama K, Ogawa A et al (2014) TRIB1 downregulates hepatic lipogenesis and glycogenesis via multiple molecular interactions. J Mol Endocrinol 52:145–158
Jump DB (2011) Fatty acid regulation of hepatic lipid metabolism. Curr Opin Clin Nutr Metab Care 14:115–120
Talukdar S, Owen BM, Song P et al (2016) FGF21 regulates sweet and alcohol preference. Cell Metab 23:344–349
Adams AC, Cheng CC, Coskun T, Kharitonenkov A (2012) FGF21 requires βklotho to act in vivo. PLoS One 7:e49977
Global Lipids Genetics Consortium (2013) Discovery and refinement of loci associated with lipid levels. Nat Genet 45:1274–1283
Ranawana DV, Henry CJK (2010) Are caloric beverages compensated for in the short-term by young adults? An investigation with particular focus on gender differences. Appetite 55:137–146
Gadah NS, Kyle LA, Rogers PJ (2012) Gender differences in the satiety effects of sugar-containing drinks. Appetite 59:626
Dupuis J, Langenberg C, Prokopenko I et al (2010) New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 42:105–116
Saxena R, Hivert M-F, Langenberg C et al (2010) Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet 42:142–148
Wang J, Light K, Henderson M et al (2014) Consumption of added sugars from liquid but not solid sources predicts impaired glucose homeostasis and insulin resistance among youth at risk of obesity. J Nutr 144:81–86
Bel-Serrat S, Mouratidou T, Santaliestra-Pasías AM et al (2013) Clustering of multiple lifestyle behaviours and its association to cardiovascular risk factors in children: the IDEFICS study. Eur J Clin Nutr 67:848–854
Bremer AA, Auinger P, Byrd RS (2009) Relationship between insulin resistance-associated metabolic parameters and anthropometric measurements with sugar-sweetened beverage intake and physical activity levels in US adolescents: findings from the 1999–2004 National Health and Nutrition Examination Survey. Arch Pediatr Adolesc Med 163:328–335
Rezvani R, Cianflone K, McGahan JP et al (2013) Effects of sugar-sweetened beverages on plasma acylation stimulating protein, leptin and adiponectin: relationships with metabolic outcomes. Obesity 21:2471–2480
Angelopoulos TJ, Lowndes J, Sinnett S, Rippe JM (2016) Fructose containing sugars at normal levels of consumption do not effect adversely components of the metabolic syndrome and risk factors for cardiovascular disease. Nutrients 8:179
Black RNA, Spence M, McMahon RO et al (2006) Effect of eucaloric high- and low-sucrose diets with identical macronutrient profile on insulin resistance and vascular risk. Diabetes 55:3566–3572
Wang M, Yu M, Fang L, Hu R-Y (2015) Association between sugar-sweetened beverages and type 2 diabetes: a meta-analysis. J Diabetes Investig 6:360–366
Qi Q, Chu AY, Kang JH et al (2012) Sugar-sweetened beverages and genetic risk of obesity. N Engl J Med 367:1387–1396
Brunkwall L, Chen Y, Hindy G et al (2016) Sugar-sweetened beverage consumption and genetic predisposition to obesity in 2 Swedish cohorts. Am J Clin Nutr 104:809–815
Olsen NJ, Ängquist L, Larsen SC et al (2016) Interactions between genetic variants associated with adiposity traits and soft drinks in relation to longitudinal changes in body weight and waist circumference. Am J Clin Nutr 104:816–826
Zheng Y, Li Y, Huang T et al (2016) Sugar-sweetened beverage intake, chromosome 9p21 variants, and risk of myocardial infarction in Hispanics. Am J Clin Nutr 103:1179–1184
Davis JN, Lê K-A, Walker RW et al (2010) Increased hepatic fat in overweight Hispanic youth influenced by interaction between genetic variation in PNPLA3 and high dietary carbohydrate and sugar consumption. Am J Clin Nutr 92:1522–1527
Horton TJ, Gayles EC, Prach PA et al (1997) Female rats do not develop sucrose-induced insulin resistance. Am J Phys 272:R1571–R1576
Reaven GM (1988) Role of insulin resistance in human disease. Diabetes 37:1595–1607
Haffner SM, Stern MP, Hazuda HP et al (1990) Cardiovascular risk factors in confirmed prediabetic individuals: does the clock for coronary heart disease start ticking before the onset of clinical diabetes? JAMA 263:2893–2898
Acknowledgements
Infrastructure for the CHARGE Consortium is supported in part by the National Heart, Lung, and Blood Institute grant HL105756. Cohort-specific sources of support and acknowledgements are presented in ESM Table 1. We thank J. C. Florez (Diabetes Unit, Massachusetts General Hospital, USA) for his help in the genesis of this project. Preliminary results were presented as an abstract at the ADA 75th Scientific Sessions in 2015.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Funding
NMM received funding from the Boston Area Diabetes, Endocrinology Research Center Feasibility Program (P30 DK057521) to support part of this research, and she was funded in part by the US Department of Agriculture, under agreement No. 58-1950-0-014. MAH is supported by R01 DK100425. CES is supported by K08 HL112845. JBM is supported by K24DK080140 and U01DK078616. KLY is supported by KL2TR001109.
Duality of interest
BP serves on the Data and Safety Monitoring Board of a clinical trial funded by the manufacturer (Zoll LifeCor) and on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. All other authors declare no conflict of interest.
Contribution statement
The authors’ responsibilities were as follows: NMM, HSD, JM and MAH: designed the study; NMM, HSD, JM, DEH, JCK-dJ, CES, TT, MG, RNL, DR, ES, ACF-W, DOM-K, YL, CAW, ETML, VM, KLY, KJM, LAC, C-AS, T-AC, RL-G, TH, WHO, OR, KR, JBM, UE, LMS, FRR, AH, MK, BMP, LB, AGU, JV, DSS, IS, KEN, DM, JD, MO-M, SSR, RdM, LQ, CEP, OHF, TL and MAH: played a role in acquisition of the data and critical revision of the manuscript for important intellectual content; NMM, HSD, JM, DEH, JCK-dJ, CES, TT, MG, RNL, DR, ES, ACF-W, DOM-K, YL, CAW, ETML, VM and MAH: contributed to statistical analyses; NMM, HSD, JM, DEH, JCK-dJ, CES, TT and MAH: interpreted data; NMM, HSD, JM, DEH, JCK-dJ, CES, TT, MG, RNL, DR, ES, ACF-W, DOM-K, YL, CAW, ETML, VM, JBM and MAH: contributed to writing of the manuscript; all authors read and approved the final version of the manuscript. NMM and HSD (joint co-first authors) are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Electronic supplementary material
ESM
(PDF 550 kb)
Rights and permissions
About this article
Cite this article
McKeown, N.M., Dashti, H.S., Ma, J. et al. Sugar-sweetened beverage intake associations with fasting glucose and insulin concentrations are not modified by selected genetic variants in a ChREBP-FGF21 pathway: a meta-analysis. Diabetologia 61, 317–330 (2018). https://doi.org/10.1007/s00125-017-4475-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-017-4475-0