Abstract
Aims/hypothesis
There is an extensive body of literature suggesting the involvement of multiple loci in regulating the action of metformin; most findings lack replication, without which distinguishing true-positive from false-positive findings is difficult. To address this, we undertook evidence-based, multiple data integration to determine the validity of published evidence.
Methods
We (1) built a database of published data on gene–metformin interactions using an automated text-mining approach (n = 5963 publications), (2) generated evidence scores for each reported locus, (3) from which a rank-ordered gene set was generated, and (4) determined the extent to which this gene set was enriched for glycaemic response through replication analyses in a well-powered independent genome-wide association study (GWAS) dataset from the Genetics of Diabetes and Audit Research Tayside Study (GoDARTS).
Results
From the literature search, seven genes were identified that are related to the clinical outcomes of metformin. Fifteen genes were linked with either metformin pharmacokinetics or pharmacodynamics, and the expression profiles of a further 51 genes were found to be responsive to metformin. Gene-set enrichment analysis consisting of the three sets and two more composite sets derived from the above three showed no significant enrichment in four of the gene sets. However, we detected significant enrichment of genes in the least prioritised category (a gene set in which their expression is affected by metformin) with glycaemic response to metformin (p = 0.03). This gene set includes novel candidate genes such as SLC2A4 (p = 3.24 × 10−04) and G6PC (p = 4.77 × 10−04).
Conclusions/interpretation
We have described a semi-automated text-mining and evidence-scoring algorithm that facilitates the organisation and extraction of useful information about gene–drug interactions. We further validated the output of this algorithm in a drug-response GWAS dataset, providing novel candidate loci for gene–metformin interactions.
Similar content being viewed by others
Introduction
Metformin has been used for 60 years by more than 150 million people worldwide. It is the first-line monotherapy prescribed at diagnosis in people with type 2 diabetes [1], and also slows progression to type 2 diabetes in people with elevated but non-diabetic glucose levels who are unable or unwilling to adhere to lifestyle modification [2,3,4].
Despite the popularity of metformin in diabetes treatment, its mechanisms of action are poorly understood; suppression of endogenous glucose production via activation of AMP-kinase (AMPK) has been hypothesised [5]. However, a preserved glucose-lowering effect has been reported in AMPK knockout mice [6]. Alternative, non-AMPK-dependent, mechanisms include inhibition of mitochondrial glycerophosphate dehydrogenase activity [7] and adenylate cyclase-mediated inhibition of the gluconeogenic pathway in favour of glycolysis [8]. In a recent study performed on Caenorhabditis elegans and extended to human cell lines, Wu et al identified two new targets of metformin action: the nuclear pore complex and the gene encoding acyl-CoA dehydrogenase 10 [9].
Diabetes treatment guidelines adopt a one-size-fits-all approach, and do not take into account interindividual variation in response. Yet there is considerable between-patient variation in treatment effects, with some responding poorly or not at all and others being highly sensitive to the drug or experiencing extreme adverse drug reactions [10]. Up to 30% of individuals treated with metformin develop nausea, bloating, abdominal pain and/or diarrhoea, and 5–10% are unable to continue with metformin treatment [11]. Heritability studies indicate that genetic variation underlies around 34% of the variability in metformin response [12].
Previous candidate gene-based pharmacogenetic studies of metformin have largely focused on loci encoding transporter proteins; little emphasis has been placed on genes in the pharmacodynamics (PD) domain, and much of the published data are inconclusive and sometimes controverted [10]. Hypothesis-free genome-wide association studies (GWASs) on metformin have identified a genome-wide significant variant, rs11212617, near the ATM gene for metformin-induced glycaemic response [13]. Given that this SNP lies in a large block of genes that are in linkage disequilibrium, the authors performed cellular work and suggested ATM to be the causal gene.
AMPK, the energy sensor, is the downstream target of metformin and is believed to be involved in the PD of metformin. Selective inhibition of ataxia telangiectasia mutated (ATM) protein by KU-55933 resulted in a marked reduction in metformin-induced AMPK activation, suggesting involvement of ATM in AMPK activation. However, cellular studies showed marked inhibition of organic cation transporter (OCT)1, an important mediator of metformin uptake by the liver, by KU-55933, suggesting that the observed attenuated AMPK phosphorylation could also be due to inhibition of OCT1 [14]. A recent GWAS study from the MetGen consortium reported an association between an intronic SLC2A2 variant, rs8192675, and the glycaemic response to metformin [15].
Owing to the vast literature on gene–metformin interactions, obtaining an unbiased overview of the evidence is extremely difficult. While meta-analysis delivers trustworthy findings if well conducted, heterogeneity in study designs, analytic strategies, population characteristics and data selection biases present challenges to such analyses [16]. Thus, to facilitate this process, automated approaches to integrate evidence from multiple sources, cataloguing the levels of evidence, validating in a real-world dataset, and using this to prioritise genes for follow-up are increasingly favoured [17, 18].
Here, we established a semi-automated text-mining pipeline to prioritise biological candidate genes that show evidence of interaction with metformin based on strength of evidence from published studies. We then evaluated the prioritised gene sets by examining their enrichment using a well-powered external dataset.
Methods
Data collection
Selection and download of articles
Articles that make reference to studies of genes and metformin in humans, identified through PubMed, were identified using the Fast Automated Biomedical Literature Extraction (FABLE) tool [19]. Accordingly, 13,914 articles were identified, of which 5963 reported independent information (Fig. 1). PubMed article identifiers (PMIDs) were collected for automated download of full text articles using Batch Entrez and EndNote. These tools permit access to articles from journals that are either open access or to which our institution (Lund University) subscribes. In most cases, PDFs are the default source of information from published articles. Thus, batch conversion of PDF to text format was done using Xpdf 3.04 (ftp.foolabs.com, accessed from 1 February to 30 June 2014).
Gene and drug dictionary construction
Gene and drug names are often described using more than one naming convention, abbreviation and/or synonym in the biomedical literature. Therefore, we compiled a comprehensive dictionary of gene names and abbreviations by extracting gene synonyms from NCBI Gene (www.ncbi.nlm.nih.gov/gene/), UCSC Genome Brower (www.genome.ucsc.edu/), SymAtlas (www.biogps.org/), Google (www.google.com/), GeneCards (www.genecards.org/) and iLINCS (www.ilincs.org/ilincs/), which was subsequently used to standardise data for a given gene. A drug dictionary capturing generic name, brand names, synonyms and International Union of Pure and Applied Chemistry (IUPAC) names of metformin was also developed from drug cards of the Drug Bank (www.drugbank.ca/) (see electronic supplementary material [ESM] Table 1). All these databases were accessed from 1 February to 30 June 2014.
Sentence extraction
Sentence extraction involves text segmentation, tokenisation and named entity recognition. Sentence segmentation and tokenisation were achieved using the Lingua::EN::Sentence module in the Perl software package, which is freely available from the Comprehensive Perl Archive Network (CPAN) (http://search.cpan.org/~shlomoy/Lingua-EN-Sentence-0.14/lib/Lingua/EN/Sentence.pm, accessed from 1 February to 30 June 2014). Gene and drug names were tagged using a Perl-based mark-up algorithm that uses a set of hashes and regular expressions. Sentences that contain a drug and a gene (i.e. a gene–drug dyad) were extracted from the corpus of each article (e.g., from titles, abstracts or main body of texts).
Annotation of extracted sentences
Analyses are based on the assumption that gene–drug dyads coalesce within a single sentence. Thus, each sentence was manually annotated to describe relationships between genes and metformin according to the annotation guideline given from the gene–drug interaction corpus and comparative evaluation by the Discovery through Integration and Extraction of Genomic and Clinical Knowledge (http://diego.asu.edu/, accessed 15 August 2014) [20]. ‘Interaction’ words are those that describe the presence of an interaction. For the purpose of these annotations, interactions refer to the action, effect or influence of the gene on a clinical outcome, pharmacokinetics (PK) or PD of the drug. Furthermore, the action, effect or influence of the drug on gene expression is also included as a component of interaction.
Annotation categories
Main annotations used to confirm the presence or absence of interaction between genes and metformin can be direct or indirect, and explicit or inferred. For the current analysis, three categories of data about interactions were documented: direct explicit, indirect explicit and indirect inferred. Only direct and explicit interactions were taken forward for further analysis. Different annotation subcategories were also used, along with interactions if they existed in sentences. These include ‘increased interaction’, ‘decreased interaction’ or ‘negation’. Negation indicates an absence of interaction and is usually represented by negative words such as ‘not’ or ‘never’ (ESM Table 2).
Developing the evidence-scoring algorithm
An iterative ranking algorithm was developed based on the pharmacogenomic relatedness, frequency and consistency of evidence for co-occurring gene–drug pairs. Each gene was given a score based on the strength of evidence for interaction with metformin. The scoring algorithm is adapted from the Coriell Personalized Medicine Collaborative pharmacogenomics appraisal [17]. Accordingly, each gene was given a score consisting of seven categories, ranging from 1 (representing the strongest evidence; presence of consistent clinical data) to 7 (the weakest evidence; published evidence showing lack of effect of the gene on drug response). See the ESM Methods for further details.
Once all the evidence for a given gene had been gathered, a single score was assigned based on the greatest strength of evidence. For evidence scores 1–3, the drug–phenotype association should be consistent across different studies. If the data were found to be inconsistent, the clinical relevance of the gene was considered unknown and a score of 4–6, as appropriate, was returned. A score of 7 was given if the clinical relevance was clearly refutable based on the available evidence. Table 1 outlines criteria for each score with their assessment outcomes.
Genome-wide association data
Cohort description
The validation cohort was from the Genetics of Diabetes and Audit Research Tayside Study (GoDARTS) consisting of 2568 adults of European ancestry diagnosed with type 2 diabetes who had been on stable metformin treatment for at least 6 months with no history of insulin use before or during the study period [13]. All participants had a baseline HbA1c > 7% (53 mmol/mol) and < 14% (129.5 mmol/mol).
Genotyping and quality control
Genotyping and quality control procedures for the GoDARTS cohort are described elsewhere [13, 14, 21]. See ESM Methods for further details.
Glycaemic response definition and model
A linear regression model of glycaemic response was fitted as the maximum HbA1c reduction recorded within 1–18 months of the metformin index date adjusted for baseline HbA1c, creatinine clearance, adherence, dose, drug group and baseline gap (the number of days between baseline HbA1c measure and metformin index date). The final glycaemic response model was: HbA1c reduction = baseline HbA1c + creatinine clearance + adherence + average daily dose + drug group + baseline gap + genotype.
Each SNP was tested for association with a continuous measure of glycaemic response (HbA1c reduction) with SNPTEST v2.5 (https://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html) [22] using multiple linear regression assuming an additive model. Association test results were combined with fixed-effects inverse-variance-weighted meta-analysis using Genome-Wide Association Meta Analysis software v2.1.34 (www.geenivaramu.ee/en/tools/gwama) [23]. Software was accessed from 1 February to 30 June 2014.
Gene-set enrichment analysis
We carried out enrichment analysis using Meta-Analysis Gene-set Enrichment of variaNT Associations (MAGENTA v2.4) (https://software.broadinstitute.org/mpg/magenta/, accessed from 1 February to 30 June 2014) [24] to test whether literature-identified gene sets were enriched with glycaemic response to metformin in a well-powered GWAS from the GoDARTS consisting of 2568 individuals with type 2 diabetes treated with metformin. Five sets of genes identified from the literature were used for gene-set enrichment analysis (GSEA): (1) genes directly related to clinical outcomes of metformin; (2) genes associated with either the PK or PD of metformin but not directly related to the clinical outcome; (3) genes whose expression is affected by the presence of metformin and not included in either (1) or (2) above; (4) genes related to clinical outcome, PK or PD (1 + 2); and (5) genes related to a clinical outcome and/or PK/PD/expression (1 + 2 + 3). See ESM Methods for further details.
Results
Data retrieval and extraction
From our screen of articles with a key word ‘metformin’ in FABLE, we identified 5963 unique articles published from 1968 to January 2014 (Fig. 1). Among these, 3575 (60%) were accessed as full text articles (ESM Fig. 1) and the remaining 2388 (40%) as abstracts (ESM Fig. 2). Although other parts may contain biologically relevant information, the best keyword per total word was obtained from abstracts [25]. ESM Fig. 3 shows the distribution of full text articles and abstracts by year of publication. A total of 2009 sentences were extracted with 3063 co-occurrences of metformin and genes. After removing non-interaction shared entities, and hypothetical, possible and indirect interactions, 1074 direct and explicit co-occurrences were annotated.
Genes related to clinical outcomes as a consequence of metformin treatment
From the search outlined above, seven genes were identified that appear to modify the effects of metformin on diabetes-related clinical outcomes. These genes were assigned evidence code 1 and thus found to be clinically relevant (ESM Table 3). These genes included encoding proteins that affect metformin transport (SLC22A1, SLC22A2, SLC47A1). While SLC22A1 and SLC22A2 encode OCT1 and OCT2, respectively, SLC47A1 encodes multi drug and toxin extrusion (MATE)1.
OCT1 transports metformin in the gut and facilitates its hepatic uptake [26, 27]. OCT2 and MATE1, expressed in the kidney, are widely reported to be involved in the renal excretion of metformin [28, 29]. Multiple variants in these genes are reported to affect functionality and therapeutic response to metformin [10]. STK11, PRKAA2, ATM and SHBG genes also showed consistent evidence of interactions with metformin on clinical outcomes. ATM encodes for serine/threonine protein kinase that belongs to the PI3/PI4-kinase family. This gene is primarily involved in DNA damage response but also involved in insulin signalling and beta cell dysfunction [30].
The liver kinase beta 1 (LKB1)–AMPK pathway controls hepatic glucose homeostasis and may play a role in the therapeutic effects of insulin-sensitising glucose-lowering agents [31]. While STK11 encodes LKB1, PRKAA2 encodes AMPK alpha 2 subunit. LKB1 is the upstream kinase of AMPK, an element involved in cellular metabolism and energy homeostasis [32]. Zhou et al reported that AMPK could be a key molecular effector of metformin. Activation of AMPK by metformin was shown to be associated with a subsequent reduction in the production of glucose in the liver [33]. SHBG encodes the sex hormone binding protein (SHBG), and variation at this locus has been related to polycystic ovary syndrome [34]. According to Ding et al, the level of circulating SHBG is inversely related to insulin resistance and may be causally related to type 2 diabetes [35].
Genes related to PK and/or PD of metformin
Those genes that affected transport of the drug in the body or influenced metformin action but did not appear to consistently affect clinical outcomes were assigned a score of 2 (ESM Table 4). Of these, SLC47A2, SLC22A3 and SLC29A4 encode transporter proteins MATE2, OCT3 and plasma membrane monoamine transporter (PMAT), respectively. These genes were found to be predictive of the PK parameters of metformin. MATE2 is a transporter protein expressed in the brush border of the kidney [36]. Stocker et al reported an association of this protein with renal clearance and subsequent glucose-lowering effect of metformin [37]. OCT3, expressed in the brush border of the intestine and the basolateral hepatocyte membrane, could have a role in the gut absorption and hepatic intake of metformin [38, 39]. Significant interindividual variation in hepatic OCT3 mRNA levels and association of genetic variants in OCT3 (mRNA) with reduced OCT3 mRNA expression in the liver has previously been reported [40]. PMAT is mainly expressed in the luminal side of the intestine and is involved in the intestinal absorption of metformin [40]. The remaining 12 genes were associated with the PD of the drug.
Genes whose expression is influenced by metformin
Genes that encode proteins in which their cellular and molecular function is consistently affected in the presence of metformin may have potential clinical relevance. Accordingly, they were assigned a score of 3. ESM Table 5 shows a total of 51 genes that have potential relevance in predicting clinical outcome and/or PK or PD properties of the drug.
Gene-set enrichment analysis
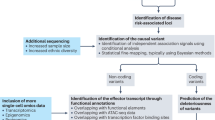
We performed GSEA to test the enrichment of literature-identified metformin-related gene sets in a hypothesis-free GWAS from the GoDARTS. Overall, five sets of genes were constructed (Table 2 and Fig. 2) and tested for enrichment. We obtained the nominal enrichment p value for each gene (ESM Table 6) and then gene set after running MAGENTA (Table 3).
Literature-identified gene sets used for MAGENTA analysis. A, genes directly related to clinical outcomes of metformin; B, genes associated with either the PK or PD of metformin; C, genes whose expression is affected by metformin; D, genes related to the clinical outcome, PK or PD; E, genes related to clinical outcome and/or PK/PD/expression
Four of the five gene sets showed no significant enrichment; the one that contained genes whose expression was affected by the presence of metformin showed significant enrichment (p < 0.05) (Table 3). In this gene set, six out of 17 genes above the 75th percentile enrichment cut-off (the expected number of genes above the cut-off being 11) were determined to have true associations with the glycaemic response to metformin. SLC2A4 (p = 3.24 × 10−04), G6PC (p = 4.77 × 10−04) and MAPK1 (p = 1.51 × 10−03) were among the top-ranking genes in this gene set. These genes encode GLUT4, glucose 6-phosphatase (G6PC) and mitogen-activated protein kinase 1 enzymes, respectively.
Discussion
Patients vary greatly in their glycaemic response, optimal dosage and adverse drug reactions following metformin therapy [41]. Genetics accounts for about 34% of this variability [12]. Hence, there is a case for more personalised therapy in diseases such as type 2 diabetes. Understanding how genetic variation impacts the effects of glucose-lowering drugs or helps to refine the characterisation of type 2 diabetes might improve treatment effectiveness and decrease adverse drug reactions, morbidity, mortality and cost of treatment.
Although there are publications in relation to the PK, PD and clinical outcomes of metformin, there is no database that concisely summarises the mechanisms describing gene–drug interactions. In most cases, specific evidence of interactions is buried deep within the literature, making it extremely difficult to comprehend the overall weight of evidence for given interactions. This problem is not specific to gene–metformin interactions, but is one that is common to the gene–drug and gene–environment interaction literature per se. In this paper, however, we focus on interactions between metformin and genes that impact clinical outcomes, PK and/or PD of the drug using a comprehensive text-mining strategy.
Our analyses identified seven genes ranked as ‘top priority’ to predict metformin-related clinical outcomes. These genes constituted three solute carrier family genes (SLC22A1, SLC22A2 and SLC47A1) that are related to the PK of metformin, and four PD-related genes (ATM, STK11, PRKAA2 and SHBG). Fifteen genes were found to affect the PK/PD of metformin without being consistently related to clinical outcomes. A third gene set in which expression or activation is affected by the presence of metformin has also been identified from the text-mining. A GSEA using GWAS data from GoDARTS showed significant enrichment of the third category for glycaemic response.
Genes that showed consistent changes in cellular and molecular functions in the presence of metformin may have potential clinical relevance in the search for biomarkers that predict the therapeutic outcome of metformin. This includes SLC2A4 and G6PC. SLC2A4 encodes GLUT4, which plays a crucial role in regulating blood glucose homeostasis by facilitating insulin-stimulated glucose transport into skeletal muscles [42]. Metformin is shown to modulate translocation of GLUT4 in skeletal muscle and adipocytes [43]. G6PC gene encodes G6PC, a rate-limiting enzyme in hepatic glucose production. It is involved in glucose production via the gluconeogenesis and glycogenolysis pathways. Cellular studies have shown metformin to suppress glucose phosphatase enzyme expression independent of AMPK [44]. While the top genes according to the GSEA were SLC2A4 and G6PC, there is a possibility that the true signal could be from neighbouring genes. ESM Fig. 4 shows regional association plots around SLC2A4 and G6PC.
The stringent significance threshold for a GWAS could overlook moderate-association signals that may have detrimental collective effects in certain pathways. Therefore, integrating GSEA guided by carefully curated evidence from the literature, as we did here, is likely to identify signals that are overlooked in a GWAS. In this study, we tested enrichment of text-mining-based prioritised gene sets on a GWAS of metformin response. Gene sets that were given high priority (those related to clinical outcome, PK or PD) showed no significant enrichment for multiple modest associations. This is probably due to the fact that our scoring system is subject to the publication bias that is well known to affect candidate gene association studies. For example, there are many small studies reporting the association between variants in metformin transporter genes and glycaemic response, resulting in a higher priority of PK genes in our gene set ranking. However, a meta-analysis of most published studies, in up to 8000 metformin-treated individuals with type 2 diabetes from the MetGen consortium, showed the putative pharmacogenetics variants in five transporter genes had no significant impact on the glycaemic response to metformin [45], which is in line with our GSEA. The ranking system prioritises published evidence dominated by PK studies. Yet PK variants do not seem to alter metformin response [45]. Therefore, this is largely due to candidate gene-based approaches. The molecular and cellular-based approaches looked at mostly PD studies and hence cast the net wider, so being less focused on the transporters.
The enrichment of association signals within the low-priority gene set highlights the need to follow up association signals for loci with multiple modest effects for glycaemic response that have been previously overlooked, such as SLC2A4 and G6PC, which rank high in the GWAS, with p values of 3.24 × 10−04 and 4.77 × 10−04, respectively.
Although the pipeline described here has generated novel evidence of gene–metformin interactions, it by no means renders a complete overview of all relevant literature. For example, only 60% of the index articles were accessible as full text, and conversion of the native PDF file into text format might have caused relevant data to be lost, as some articles used formats that are incompatible with the file templates used in our pipeline. Furthermore, data contained in tables and figures in the index papers were extracted in a semi-structured format, making data retrieved from tables and figures challenging to analyse and interpret. Thus, for papers in which important results were included in tables and figures, but not articulated in the title, abstract or main text of the article, evidence of interactions may have been overlooked.
Some genes are reported in papers in a non-standard form that may not be captured by the dictionary-based named entity recognition used in this study, and in some articles, non-standard abbreviations for metformin, instead of its original generic or brand name (e.g. the term ‘MET’), are occasionally used. Such abbreviations are not found in any standard drug abbreviation protocol. Sentences with such abbreviations are likely to be incompletely characterised using the text retrieval strategy adopted here. Other barriers to data assimilation include the extent to which anaphora (‘use of grammatical substitute [as pronoun or a pro-verb] to refer to the denotation of a preceding word or group of words’ [46]) and epiphora (‘the repetition of a word or words at the end of two or more successive verses, clauses, or sentences’ [46]) were resolved, which may also have resulted in loss of information; this might have occurred, for example, when a gene and a drug were found in separate sentences (i.e. not described in the same sentence).
Larger studies with regular iterations that use extensive literature coverage, SNP-level annotation and more intensively automated machine learning approaches are likely to extend the observations reported here. GSEA using a larger GWAS dataset for metformin response is likely to increase the power and produce convincing results. Replication using other GWAS datasets including SNP–SNP interaction analyses is also likely to extend the number of validated interactions, as some of those prioritised using our algorithm may have ethnically specific effects that are undetectable in the ethnically homogeneous GoDARTS cohort.
Conclusions
We have developed a semi-automated text-mining and evidence-scoring algorithm that could help to organise and extract useful information from the literature on gene–drug and gene–environment interactions. According to our analysis, genes that encode transporter proteins such as OCTs and MATEs yield the strongest evidence of modifying the therapeutic outcomes of metformin. In addition, genes in the LKB1–AMPK pathway are also found to be related to the therapeutic outcomes of the glucose-lowering drug. However, we did not detect enrichment for the highly prioritised gene sets using GWAS data from the GoDARTS cohort; instead it was the gene set that were ranked with lower evidence scores that showed statistically significant enrichment in the GoDARTS.
Given that the genomic architecture of drug response is complex and the mechanism of metformin action is still not clearly known, candidate gene studies investigating drug response to metformin have had limited success. In an alternative approach, here we have identified novel genes potentially associated with metformin action. Using a text-mining approach of the published literature, we have identified a gene set derived from cellular and functional studies associated with metformin response. The association of genetic variation in these genes, including SLC2A4 and G6PC, needs further replication and follow-up.
Abbreviations
- AMPK:
-
AMP-kinase
- ATM:
-
Ataxia telangiectasia mutated
- FABLE:
-
Fast Automated Biomedical Literature Extraction
- G6PC:
-
Glucose 6-phosphatase
- GoDARTS:
-
Genetics of Diabetes and Audit Research Tayside Study
- GSEA:
-
Gene-set enrichment analysis
- GWAS:
-
Genome-wide association study
- LKB:
-
Liver kinase beta
- MAGENTA:
-
Meta-Analysis Gene-set Enrichment of variaNT Associations
- MATE:
-
Multi drug and toxin extrusion
- OCT:
-
Organic cation transporter
- PD:
-
Pharmacodynamics
- PK:
-
Pharmacokinetics
- PMAT:
-
Plasma membrane monoamine transporter
- SHBG:
-
Sex-hormone binding protein
- STK:
-
Serine/threonine kinase
References
Inzucchi SE, Bergenstal RM, Buse JB et al (2012) Management of hyperglycaemia in type 2 diabetes: a patient-centered approach. Position statement of the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD). Diabetologia 55:1577–1596
Slama G (2003) The potential of metformin for diabetes prevention. Diabete Metab 29:6S104–6S111
Cleasby ME, Dzamko N, Hegarty BD, Cooney GJ, Kraegen EW, Ye JM (2004) Metformin prevents the development of acute lipid-induced insulin resistance in the rat through altered hepatic signaling mechanisms. Diabetes 53:3258–3266
Knowler WC, Barrett-Connor E, Fowler SE, Hamman RF, Lachin JM, Walker EA (2002) Diabetes prevention program research group. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med 346:393–403
Pernicova I, Korbonits M (2014) Metformin-mode of action and clinical implications for diabetes and cancer. Nat Rev Endocrinol 10:143–156
Foretz M, Hébrard S, Leclerc J et al (2010) Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state. J Clin Invest 120:2355–2369
Madiraju AK, Erion DM, Rahimi Y et al (2014) Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature 510:542–546
Miller RA, Chu Q, Xie J, Foretz M, Viollet B, Birnbaum MJ (2013) Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic AMP. Nature 494:256–260
Wu L, Zhou B, Oshiro-Rapley N et al (2016) An ancient, unified mechanism for metformin growth inhibition in C. elegans and cancer. Cell 167:1705–1718
Dawed AY, Zhou K, Pearson ER (2016) Pharmacogenetics in type 2 diabetes: influence on response to oral hypoglycemic agents. Pharmacogenomics Pers Med 9:17–29
Garber AJ, Duncan TG, Goodman AM, Mills DJ, Rohlf JL (1997) Efficacy of metformin in type II diabetes: results of a double-blind, placebo controlled, dose-response trial. Am J Med 103:491–497
Zhou K, Donnelly L, Yang J et al (2014) Heritability of variation in glycaemic response to metformin: a genome-wide complex trait analysis. Lancet Diabetes Endocrinol 2:481–487
Zhou K, Bellenguez C, Spencer CC et al (2011) Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat Genet 43:117–120
Yee SW, Chen L, Giacomini KM (2012) The role of ATM in response to metformin treatment and activation of AMPK. Nat Genet 44:359–360
Zhou K, Yee S, Seiser E et al (2016) Variation in the glucose transporter gene SLC2A2 is associated with glycemic response to metformin. Nat Genet 48:1055–1059
Bosker F, Hartman C, Nolte I et al (2011) Poor replication of candidate genes for major depressive disorder using genome-wide association data. Mol Psychiatry 216:516–532
Gharani N, Keller MA, Stack CB et al (2013) The Coriell personalized medicine collaborative pharmacogenomics appraisal, evidence scoring and interpretation system. Genome Med 5:1–19
Sun J, Jia P, Fanous AH et al (2009) A multi-dimensional evidence-based candidate gene prioritization approach for complex diseases-schizophrenia as a case. Bioinformatics 25:2595–2602
Fast Automated Biomedical Literature Extraction (FABLE). Available from http://itre.cis.upenn.edu/~myl/languagelog/archives/003656.html. Accessed 3 Jun 2014
Discovery Through Integration and Extraction of Genomic and Clinical Knowledge (DIEGO) lab. Biomedical informatics research lab at Arizona state university. Available from http://diego.asu.edu/. Accessed 15 Aug 2014
Morris AP, Voight BF, Teslovich TM et al (2012) Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 44:981–990
Marchini J, Howie B, Myers S, McVean G, Donnelly P (2007) A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 39:906–913
Mägi R, Morris AP (2010) GWAMA: software for genome-wide association meta-analysis. BMC Bioinforma 11:288
Segrè AV, DIAGRAM Consortium, MAGIC investigators et al (2010) Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet 6:e1001058. doi:10.1371/journal.pgen.1001058
Shah PM, Perez-Iratxeta C, Bork P et al (2003) Information extraction from full text scientific articles: where are the keywords? BMC Bioinforma 4:1–9
Han TH, Everett RS, Proctor WR et al (2013) Organic cation transporter 1 (OCT1/mOct1) is localized in the apical membrane of Caco-2 cell monolayers and enterocytes. Mol Pharmacol 84:182–189
Müller J, Lips KS, Metzner L, Neubert RH, Koepsell H, Brandsch M (2005) Drug specificity and intestinal membrane localization of human organic cation transporters (OCT). Biochem Pharmacol 70:1851–1860
Kimura N, Masuda S, Tanihara Y et al (2005) Metformin is a superior substrate for renal organic cation transporter OCT2 rather than hepatic OCT1. Drug Metab Pharmacokinet 20:379–386
Otsuka M, Matsumoto T, Morimoto R, Arioka S, Omote H, Moriyama Y (2005) A human transporter protein that mediates the final excretion step for toxic organic cations. Proc Natl Acad Sci U S A 102:17923–17928
Schneider JG, Finck BN, Ren J et al (2006) ATM-dependent suppression of stress signaling reduces vascular disease in metabolic syndrome. Cell Metab 4:377–389
Ding EL, Song Y, Manson JE et al (2008) Single nucleotide polymorphisms in genes encoding LKB1 (STK11), TORC2 (CRTC2) and AMPK alpha2-subunit (PRKAA2) and risk of type 2 diabetes. Mol Genet Metab 93:200–209
Rena G, Pearson ER, Sakamoto K (2013) Molecular mechanism of action of metformin: old or new insights? Diabetologia 56:1898–1906
Zhou G, Myers R, Li Y et al (2001) Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest 108:1167–1174
Chen C, Smothers J, Lange A, Nestler JE, Strauss III JF, Wickham Iii EP (2010) Sex hormone-binding globulin genetic variation: associations with type 2 diabetes mellitus and polycystic ovary syndrome. Minerva Endocrinol 35:271–280
Ding EL, Song Y, Manson JE et al (2009) Sex hormone–binding globulin and risk of type 2 diabetes in women and men. N Engl J Med 361:1152–1163
Becker ML, Pearson ER, Tkáč I (2013) Pharmacogenetics of oral antidiabetic drugs. Int J Endocrinol 2013:1–10
Stocker SL, Morrissey KM, Yee SW et al (2013) The effect of novel promoter variants in MATE1 and MATE2 on the pharmacokinetics and pharmacodynamics of metformin. Clin Pharmacol Ther 93:186–194
Nies AT, Koepsell H, Winter S et al (2009) Expression of organic cation transporters OCT1 (SLC22A1) and OCT3 (SLC22A3) is affected by genetic factors and cholestasis in human liver. Hepatology 50:1227–1240
Chen L, Pawlikowski B, Schlessinger A et al (2010) Role of organic cation transporter 3 (SLC22A3) and its missense variants in the pharmacologic action of metformin. Pharmacogenet Genomics 20:687–699
Zhou M, Xia L, Wang J (2007) Metformin transport by a newly cloned proton-stimulated organic cation transporter (plasma membrane monoamine transporter) expressed in human intestine. Drug Metab Dispos 35:1956–1962
Martono DP, Lub R, Heerspink HJ et al (2015) Systematic review or meta-analysis predictors of response in initial users of metformin and sulphonylurea derivatives: a systematic review. Diabet Med 32:853–864
Huang S, Czech MP (2007) The GLUT4 glucose transporter. Cell Metab 5:237–252
Meuillet EJ, Wiernsperger N, Mania-Farnell B, Hubert P, Cremel G (1999) Metformin modulates insulin receptor signaling in normal and cholesterol-treated human hepatoma cells (HepG2). Eur J Pharmacol 377:241–252
Ota S, Horigome K, Ishii T et al (2009) Metformin suppresses glucose-6-phosphatase expression by a complex I inhibition and AMPK activation-independent mechanism. Biochem Biophys Res Commun 388:311–316
Dujic T, Zhou K, Yee SW et al (2016) Variants in pharmacokinetic transporters and glycaemic response to metformin: a MetGen meta-analysis. Clin Pharmacol Ther. doi:10.1002/cpt.567
Merriam-Webster’s Collegiate Dictionary, 10th edn. (1999) Available from http://www.merriam-webster.com. Accessed 15 May 2017
Acknowledgements
AYD would like to acknowledge the Swedish Institute Study Scholarship (SISS), the Diabetes Research on Patient Stratification (DIRECT) and Y. G. Meskel for his support in sentence extraction. AA was funded under grant agreement no. 115317 (DIRECT).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Data availability
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
Funding
This work was supported in part by the Innovative Medicines Initiative Joint Undertaking under grant agreement no. 115317 (DIRECT), resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007-2013) and EFPIA companies in kind contribution.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
PWF and ERP conceived the study. AYD, AA and PWF designed the analyses. AYD, AA and KZ analysed the data. All the authors interpreted the data, and AYD, ERP and PWF wrote the manuscript. All the authors revised the manuscript and approved the final version submitted for publication. AYD and PWF are the guarantors of this work.
Electronic supplementary material
ESM
(PDF 552 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Dawed, A.Y., Ali, A., Zhou, K. et al. Evidence-based prioritisation and enrichment of genes interacting with metformin in type 2 diabetes. Diabetologia 60, 2231–2239 (2017). https://doi.org/10.1007/s00125-017-4404-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-017-4404-2