Abstract
Aims/hypothesis
The proinflammatory cytokine IL-1β induces apoptosis in pancreatic beta cells via pathways dependent on nuclear factor-κB (NF-κB), mitogen-activated protein kinase, and protein kinase C. We recently showed suppressor of cytokine signalling (SOCS)-3 to be a natural negative feedback regulator of IL-1β- and IFN-γ-mediated signalling in rat islets and beta cell lines, preventing their deleterious effects. However, the mechanisms underlying SOCS-3 inhibition of IL-1β signalling and prevention against apoptosis remain unknown.
Methods
The effect of SOCS-3 expression on the global gene-expression profile following IL-1β exposure was microarray-analysed using a rat beta cell line (INS-1) with inducible SOCS-3 expression. Subsequently, functional analyses were performed.
Results
Eighty-two known genes and several expressed sequence tags (ESTs) changed expression level 2.5-fold or more in response to IL-1β alone. Following 6 h of IL-1β exposure, 23 transcripts were up-regulated. Of these, several, including all eight transcripts relating to immune/inflammatory response pathways, were suppressed by SOCS-3. Following 24 h of IL-1β exposure, secondary response genes were detected, affecting metabolism, energy generation, protein synthesis and degradation, growth arrest, and apoptosis. The majority of these changes were prevented by SOCS-3 expression. Multiple IL-1β-induced NF-κB-dependent proapoptotic early response genes were inhibited by SOCS-3 expression, suggesting that SOCS-3 inhibits NF-κB-mediated signalling. These observations were experimentally confirmed in functional analyses.
Conclusions/interpretation
This study suggests that there is an unexpected cross-talk between the SOCS/IFN and the IL-1β pathways of signalling in pancreatic beta cells, which could lead to a novel perspective of blocking two important proapoptotic pathways in pancreatic beta cells by influencing a single signalling molecule, namely SOCS-3.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The proinflammatory cytokines IL-1β, IFN-γ and TNF-α are released during insulitis, an early event in the pathogenesis of type 1 diabetes. The consequent triggering of beta cell apoptosis is the result of a complex interaction between protective and destructive mechanisms depending on de novo gene and protein expression [1]. The combination of genome, transcriptome and proteome analyses reveals molecular mechanisms that are responsible for the destructive pathways and that are thus potential new targets for intervention in disease progression or recurrence in grafted beta cells [2].
IL-1β contributes to beta cell destruction and potentially to type 1 diabetes via induction of proapoptotic proteins and pathways that are dependent on NF-κB, mitogen-activated protein kinase and protein kinase C. This induction is, in part, mediated through induced production of nitric oxide (NO) and different reactive oxygen species [1, 3, 4, 5]. In parallel, several protective mechanisms are activated, e.g. the free radical scavengers manganese superoxide dismutase (MnSOD), glutathione peroxidase and catalase [6, 7], and elements with anti-apoptotic properties, e.g. thioredoxin, calbindin, bcl-2 and A20 [8, 9, 10, 11]. In general, however, the ability of the beta cell to activate a sufficient protective response is impaired, which could, in part, explain their sensitivity to the toxic effect of cytokines [12].
Tight regulation of the signal transduction pathways activated by proapoptotic cytokines is required to prevent potentially devastating effects of inflammation on host cells. Members of the SOCS family were originally reported as immediate–early response genes, preventing IL-6 and IFN signalling through the inhibition of the phosphorylation of the signal transducers and activators of transcription (STAT) family of transcription factors mediated by the Janus kinases (JAKs) [13]. Regulated themselves by the JAK–STAT pathway, SOCS-1 and SOCS-3 therefore define negative feedback loops controlling duration and magnitude of the cellular responses to IFN [14].
Using rat islets and beta cell lines we recently showed SOCS-3 to be a particularly effective defence mechanism against cytokine-mediated beta cell destruction [15]. Surprisingly, we found that SOCS-3, in addition to regulation of IFN signalling, also constitutes a negative feedback regulator of IL-1β-mediated signalling, capable of inhibiting the toxic effect of IL-1β. This effect, not previously described in other tissue, was associated with, but probably not exclusively caused by, inhibition of IL-1β-induced iNOS (inducible nitric oxide synthase) transcription and NO production [15].
The aim of the present study was to further elucidate the mechanisms behind the inhibitory effects of SOCS-3 on IL-1β-mediated signalling, gene transcription and beta cell destruction. For these analyses we used insulin-producing cells with doxycycline-inducible SOCS-3 expression (INS-1r3#2), allowing characterisation of the global expression profile of IL-1β-affected early (6-h) and late (24-h) response genes in the presence or absence of induced SOCS-3 expression.
This approach revealed several transcripts known to be regulated in expression by NF-κB (including iNOS) to be inhibited by induced SOCS-3 expression. In beta cells, IL-1β-induced NF-κB signalling is known to be associated with several proapoptotic pathways, and NF-κB has been shown to be essential for iNOS transcription [16]. Inhibition of NF-κB activation, by overexpression of a non-degradable I-κB super-repressor, significantly reduces apoptosis induced by IL-1β and IFN [17, 18]. By different functional analyses we demonstrate in this study that SOCS-3 expression indeed inhibits NF-κB-dependent transcription and inhibits IL-1β-induced beta cell toxicity in a dose-dependent fashion through inhibited transcription of several proapoptotic genes.
Materials and methods
Cell culture and cytokines
The INS-r3#2 cells with tetracycline/doxycycline-inducible SOCS-3 expression were generated and cultured as previously described [15]. To analyse the effects of SOCS-3 overexpression, doxycycline (Sigma Aldrich, Saint Louis, Mo., USA) was added and the cells cultured for 1 day to allow for SOCS-3 expression. For analysis of the IL-1β effects, the cells were cultured for an additional 6 h, 24 h or 3 days in the presence or absence of IL-1β.
The concentrations of doxycycline and IL-1β were chosen based on titrations performed previously [15] and in the present study. Recombinant mouse IL-1β (5–15×105 U/µg) was purchased from BD Pharmingen (San Diego, Calif., USA). In the previous study [15] we used recombinant human IL-1β (4×105 U/µg) from Novo Nordisk, Bagsværd, Denmark [19]. Mass titration of IL-1β from the two different sources revealed identical toxicity profiles in the beta cell lines used (data not shown).
Functional analyses
To measure the viability of the cells the mitochondrial activity was determined using the CellTiter 96 Non-Radioactive Cell Proliferations Assay (Promega, Madison, Wis., USA), also termed the MTT (dimethylthiazol-diphenyltetrazolium bromide) assay as described [20]. The MTT activity, measured at an optical density of A578 nm, is based on the ability of living cells to convert the tetrazolium salt into formazan via the mitochondrial enzyme succinate dehydrogenase. We previously demonstrated this to be directly proportional to the number of viable cells in the beta cells exposed to IL-1β [20].
We measured NO production as accumulated nitrite by the Griess reagent [21] in a fraction of the culture medium used for the MTT assay. Absorbance was measured at 540 nm and nitrite concentration was calculated from an NaNO2 standard curve.
GeneChip analysis
In order to analyse the global gene expression following IL-1β exposure in the presence or absence of SOCS-3 expression, the INS-r3#2 cells were seeded at a density of 106 cells per well in 6-well culture plates (Nalge Nunc International, Rochester, N.Y., USA). RNA extraction was carried out, following 6 or 24 h of IL-1β exposure, by the RNAzol method according to the manufacturer’s instructions (Cinna Biotecx, Houston, Tex., USA), and was dissolved in H2O. To minimise experimental and biological variation in the analysis, six independent experiments for each condition were performed. An equal amount of total RNA from experiments one to three and four to six was then pooled (pool 1 and pool 2 respectively). Micro-array analysis was performed on each pool as described previously [22] by hybridising 20 µg of biotinylated cRNA prepared from 10 µg of total RNA from each condition to Affymetrix GeneChip RG U34A Arrays (Affymetrix, Santa Clara, Calif., USA). Each array contains gene probe sets for approximately 7000 known genes and 1000 expressed sequence tags (ESTs). The differential expression was determined by comparison of the two arrays from each condition. For both control arrays the GeneChip Suite software MAS4.0 (Affymetrix) was used. Only differentially expressed genes that were changed in the same direction in all four comparisons and that had a mean change of 2.5-fold or more were scored as significant.
The effects of SOCS-3 on the expression level of the IL-1β-affected genes were calculated and given as percentages.
Semi-quantitative RT-PCR analysis
To verify the regulation of a selected number of genes found by the GeneChip analysis, cDNA synthesis was performed using the TaqMan Gold RT-PCR Kit (Perkin-Elmer, Boston, Mass., USA) according to the manufacturer’s instructions, and the samples were subjected to a semi-quantitative PCR analysis. Amplification with specific primers was performed for an optimised (22–27) number of PCR cycles. Each PCR sample contained template cDNA, 1× Taq DNA polymerase buffer, 50 mmol/l dNTP, 1.5 mmol/l MgCl2, 10 µmol/l of each primer pair, 2.5 U Taq DNA polymerase (Promega), and 0.1 MBq α33dCTP (Amersham Pharmacia, San Francisco, Calif., USA). The primers and their respective amplicon length were as follows: IRF-1 forward 5′-CCAGCTCTGTCACCGTGCGTCG-3′, reverse 5′-CCTTGCCATCCACGTGCGTCG-3′ (223 bp); NF-κB p105 forward 5′-GATGAGGGAGTAGTGCCAGGCACC-3′, reverse 5′-CCCAGACCTAACTTCTGCGCCAG-3′ (230 bp); I-κBα forward 5′-CCTCTGGCTGCCGAGCCCTG-3′, reverse 5′-CTACACTGGCCAGGCAGCCCTG-3′ (304 bp); iNOS forward 5′-CAGCAATGGGCAGACTCT-3′, reverse 5′-CACAGGCTGCCCGGAAGGTTTG-3′ (247 bp); iCAM forward 5′-CCTGGAGGTGGGCACCCAGC-3′, reverse 5′-TTCTGAGACCTCCGGCTGGCTC-3′ (295 bp); MnSOD forward 5′-GGCTGGCTTGGCTTCAATAAG-3′, reverse 5′-ACACATCAATCCCCAGCAGTG-3′ (112 bp); GADD153 forward 5′-GCATGAAGGAGAAGGAGCAG-3′, reverse 5′-CAGCATGTGCACTGGAGATT-3′ (242 bp); c-myc forward 5′-AAATTCGAGCTGCTGCCCACC-3′, reverse 5′-CCAGTTTGGCAGCGGCCGAG-3′ (283 bp); SOCS-3 forward 5′-ATCCAGGAACTCCCGAATG-3′, reverse 5′-GGACCCCCTCCTTTTCTTTGC-3′ (261 bp). Following separation on a 6% polyacrylamide gel, the amplified products were identified and quantified by ImageQuant hardware and software (Amersham Pharmacia).
Western blotting
For analysis of expressed iNOS and SOCS-3 protein the INS-r3#2 cells were cultured as described for the GeneChip analysis and western blotting was performed as described [23] with the following antibodies: rabbit polyclonal anti-human/murine iNOS IgG (BD Pharmingen), and murine monoclonal anti-Flag (Sigma Aldrich) for SOCS-3 (Flag-tagged). The secondary antibodies were rat anti-rabbit IgG and rabbit anti-murine IgG (Cell Signalling Technologies, Cambridge, Mass., USA), both conjugated with horseradish peroxidase. Binding was visualised using X-ray films (Amersham) and the LumiGlo enhanced chemiluminescence detection kit (Cell Signalling Technologies).
iNOS and NF-κB promoter activity
In order to investigate the inhibitory effect of SOCS-3 at the transcriptional level of IL-1β signalling, the INS-r3#2 cells were seeded in duplicate in 24-well dishes and exposed to doxycycline (0.5 µg/ml) for 1 day, prior to transfection, at approximately 75% confluence. Transient transfection was performed overnight with a total of 2 µg of plasmid DNA (0.2 µg of an internal control [pRL-TK], 0.4 µg of iNOS or NF-κB promoter construct [pGL3-Basic] and 1.4 µg of empty vector [pcDNA3; Invitrogen, Carlsbad, Calif., USA]) using SuperFect (Qiagen, Valencia, Calif., USA) according to the manufacturer’s manual. The cells were cultured for an additional 6 h (NF-κB promoter analysis) or 24 h (iNOS promoter analysis) in the presence or absence of 150 pg/ml IL-1β and doxycycline. All of the iNOS promoter constructs contained the proximal 1002 bp of the rat wild-type (wt) iNOS promoter fused to a luciferase reporter gene in the pGL3-Basic vector [24]. With the exception of the wt-iNOS-1002luc construct, several promoter binding sites were individually mutated: a distal NF-κB site (iNOS-1002-dNFmut), a proximal NF-κB site (iNOS-1002-pNFmut), a γ-IFN-activated site (GAS; iNOS-1002-GASmut) and an IFN-stimulated response element (ISRE; iNOS-1002-ISRE-mut) as previously described [24].
For analysis of NF-κB-dependent gene transcription, the PathDetect NF-κB cis-Reporting System (Stratagene, La Jolla, Calif., USA) was used, with the luciferase reporter gene selectively regulated by 5× synthetic NF-κB promoter enhancer elements. The luciferase activities were assayed using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions and measured using a luminometer (Berthold Technologies, Bad Wildbad, Germany).
Nuclear extract and electrophoretic mobility shift assays of NF-κB DNA binding
To investigate whether SOCS-3 inhibits NF-κB DNA binding, the INS-r3#2 cells were precultured in 100-mm dishes for 2 days in medium containing 10% FCS. Culture was continued with 0.5% FCS with or without 1 µg/ml doxycycline, and after 20 h the cells were incubated with or without 75 pg/ml of IL-1β for 15 min. Nuclear extracts were isolated and electrophoretic mobility shift assay (EMSA) was performed as described [25]. A double-stranded oligonucleotide (5′-agctAGCTTCAGAGGGGACTTTCCGAGAGG-3′; DNA Technology, Aarhus, Denmark) was used to detect NF-κB DNA binding. In supershift analysis, nuclear extracts were preincubated for 30 min at 4 °C with NF-κB or STAT-1 antibodies (Santa Cruz Biotechnology, Santa Cruz, Calif., USA).
Statistical analysis
Results are given as means ± SD. Comparisons versus the respective control groups were carried out using the Student’s paired t test. A p value of less than 0.05 was considered significant.
Results
IL-1β dose–response characterisation of the INS-r3#2 cells
Dose–response experiments were performed to establish the protective window of SOCS-3 on IL-1β-induced beta cell toxicity and to find the optimal concentration of IL-1β to be used in the GeneChip analysis. Consistent with our previous analyses using IL-1β from a different supplier [15], a 50% reduced viability, as determined by the MTT assay, was obtained using approximately 150 pg/ml of recombinant mouse IL-1β and this effect was fully prevented by induced SOCS-3 expression (Fig. 1a). Analysis of NO production demonstrated that the protective effect of SOCS-3 was accompanied by a dose-dependent reduction in IL-1β-induced NO production (Fig. 1b).
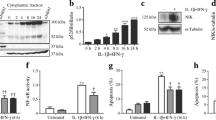
Effect of SOCS-3 on IL-1β-induced toxicity and NO production. The INS-1r3#2 cells were cultured in the absence (filled symbols) or presence (open symbols) of induced SOCS-3 expression (induced by 0.5 µg/ml doxycycline) and exposed to increasing IL-1β concentrations for 3 days followed by (a) measurement of viability at an optical density (OD) of 578 nm, and (b) measurement of accumulated NO at 540 nm. n=3
Influence of SOCS on the IL-1β-activated gene expression profile
Using 150 pg/ml mouse IL-1β, based on the titration in Figure 1, GeneChip analyses were performed to characterise the early (6-h) and late (24-h) gene expression profiles associated with the protective effect of SOCS-3 expression on IL-1β-mediated beta cell destruction.
Based on the criteria described in the Materials and methods, a total of 97 transcripts of fully annotated genes revealed differential expression patterns under the experimental conditions, i.e. a change of 2.5-fold or more in expression levels relative to the control condition. They were sorted into eight groups according to function (Table 1). Following 6 h of exposure to IL-1β, 25 of the genes were altered in expression level (23 up-regulated and two down-regulated), whereas 24 h of IL-1β exposure resulted in altered expression levels for 64 genes (35 up-regulated and 29 down-regulated). The large number of down-regulated genes after 24 h suggests activation of several secondary response pathways. Of the 48 differentially expressed ESTs (Table 2), 29 showed more than 97% homology to known transcripts in GeneBank. Several of these were also influenced by IL-1β in a SOCS-3-dependent fashion. However, since the ESTs are not yet fully annotated, they are not discussed further.
The majority of early (6-h) changes falls within a group of immune/inflammatory response genes (group 8) including several encoding membrane proteins known to activate and amplify the cellular immune response. In contrast, after 24 h of IL-1β exposure, mainly secondary response genes were up-regulated, encoding proteins with effects on metabolism and energy generation (group 5), protein synthesis/degradation (group 4), and growth and apoptosis (group 6).
The influence of SOCS-3 expression on gene expression in the absence of IL-1β was addressed in control experiments. In 6-h experiments (doxycycline present in the 24-h preculture period + 6 h in the absence of IL-1β) the only significant change when compared with control cells cultured in the absence of doxycycline was a nine-fold up-regulation of SOCS-3 gene expression. This suggests that SOCS-3 and doxycycline had little or no influence on gene expression under these conditions. In the 24-h experiments (24-h preculture + 24 h in the absence of IL-1β) only five genes were significantly altered in expression level (lactogen receptor −3.1-fold [down-regulated]; islet amyloid polypeptide [IAPP] 2.7-fold [up-regulated]; cell cycle protein p55 [CDC] 3.6-fold; distal-less 3 [Dlx-3] homeobox −3.7-fold; cell adhesion regulator [CAR1] −2.7-fold). No effect of SOCS-3 expression was observed on the expression level of transcripts encoding proteins known to influence the level of IL-1β signalling, e.g. the IL-1β receptor and the IL-1β receptor antagonist, both present on the array.
In contrast, striking regulatory effects of SOCS-3 were seen on multiple genes influenced by IL-1β (Table 1). A clear pattern was observed following 6 h of IL-1β exposure, reflecting a direct interaction of SOCS-3 with IL-1β-activated gene transcription. Thus, SOCS-3 inhibited the transcription by 23 to 62% in 16 of the 23 known transcripts significantly up-regulated after 6 h of IL-1β exposure (Table 1). Several of these early response genes are known to encode proteins involved in activation or amplification of an immune/inflammatory response (group 8), e.g. ICAM, proteasome and complement components, chemokines, mob-1 and MIP-2. Furthermore, nine of the genes have previously been shown to be dependent on NF-κB signalling (see Discussion for details). One of these, the IL-1β-induced iNOS gene expression, was clearly inhibited (49%), in line with the functional data in Figure 1. In contrast, SOCS-3 overexpression had only minor effects on the few IL-1β-influenced genes within signal transduction (group 2) and gene transcription (group 3) after 6 and 24 h of IL-1β exposure. Finally, SOCS-3 potentiated IL-1β-induced down-regulation of some receptors, e.g. the nuclear receptor (RNR-1), lactogen receptor and prolactin receptor (group 1).
The pattern appearing after 24 h of exposure to IL-1β in the presence or absence of induced SOCS-3 expression was more complex, reflecting activation of several secondary pathways resulting from the IL-1β-mediated cellular impairment. For 28 of the 35 fully annotated genes up-regulated following 24 h of IL-1β exposure, the up-regulation was reduced by 22 to 100%. Furthermore, SOCS-3 expression reduced down-regulation by 26 to 100% of 25 of the 29 transcripts significantly down-regulated following 24 h of exposure to IL-1β. This reflects the protective effect of SOCS-3 against the effects of IL-1β (Fig. 1). For example, an approximately two-fold up-regulation of all the genes encoding the ribosomal subunits (group 4) was seen in response to IL-1β alone, whereas the expression of these transcripts was only marginally affected in the presence of SOCS-3. More strikingly, genes involved in metabolism and energy generation (group 5) fluctuated after 24 h of IL-1β exposure. This was almost fully prevented in the presence of SOCS-3. Several genes involved in growth arrest and apoptosis, e.g. GADD 153 and c-myc (group 6), and in the defence–repair mechanisms, e.g. heme oxygenase, MnSOD and HSP 70 (group 7), were up-regulated after 24 h of IL-1β exposure. This was also prevented by SOCS-3 expression. Interestingly, SOCS-3 also fully prevented the IL-1β-induced down-regulation of islet amyloid polypeptide gene expression (group 1).
To summarise, SOCS-3 overexpression did not lead to complete inhibition of IL-1β signalling, nor did it lead to equal inhibition of all affected transcripts. Nevertheless, overexpression of SOCS-3 “constructively” modulated the expression of multiple IL-1β-regulated genes, which may favour beta cell survival.
Semi-quantitative RT-PCR analysis of mRNA expression
To validate the array analysis, a subset of eight genes with variable regulation by IL-1β and SOCS-3 were selected for semi-quantitative PCR analysis (Fig. 2). Due to the very low levels of basal expression of some transcripts, data are displayed as percentages of expression relative to the condition inducing the highest response to IL-1β exposure, e.g. after 6 h (iNOS, IRF1, MnSOD, NF-κB-p105, iCAM, and I-κBα) or 24 h (GADD153 and c-myc). Consistent with the results of the GeneChip analysis, we observed that the mRNAs for iNOS, IRF-1 and iCAM were all induced after 6 h of IL-1β exposure. This induction was significantly inhibited by SOCS-3 overexpression. In contrast, the mRNAs encoding for NF-κB-p105 and I-κBα, although equally induced by IL-1β, were not affected significantly by SOCS-3 after 6 h of IL-1β stimulation. After 24 h, the IL-1β-induced expression of GADD153, c-myc, IRF-1 and I-κBα was significantly inhibited by SOCS-3. In summary, the results of the quantitative analyses were concordant with the results obtained in the array analysis, with the exception of the 24-h inhibitory effect of SOCS-3 on IRF-1 and I-κBα not detected in the array analysis, and on MnSOD not confirmed in the RT-PCR analysis.
Semi-quantitative RT-PCR of selected genes shown to be IL-1β regulated by the gene array analyses. Eight selected transcripts shown in the gene array analyses to be up-regulated by IL-1β were analysed. Culture conditions were the same as described in Table 1, i.e. exposure for 6 or 24 h to the following conditions: −Dox (control; open bars), +Dox (0.5 µg/ml; filled bars), +IL-1β (150 pg/ml) −Dox (dotted bars), and IL-1β (150 pg/ml) +Dox (0.5 µg/ml; hatched bars). a. Data presented as percentages of the IL-1β −Dox condition with the highest induction being either 6 h or 24 h for the respective genes. * p>0.05. b. Intensity of the individual PCR products. Data are presented as means ± SD of three individual experiments
SOCS-3 inhibits IL-1β-induced iNOS gene and protein expression through NF-κB signalling
The inhibitory effect of SOCS-3 on IL-1β-induced iNOS protein expression (Fig. 3) confirmed the transcript (Fig. 2) and NO analyses (Fig. 1b). Thus, at the maximal level of SOCS-3 expression obtainable in the INSr3#2 clone, a full inhibition of iNOS protein expression was seen at 6 h of exposure to IL-1β, whereas iNOS expression was partially suppressed following 24 h of IL-1β exposure. To characterise the mechanisms behind SOCS-3-inhibited iNOS gene expression, the influence of SOCS-3 on transcription factors known to be involved in IL-1β-induced iNOS gene activation was addressed. This was accomplished by measuring the ability of SOCS-3 to inhibit IL-1β-induced iNOS promoter activation using constructs with mutations in the corresponding transcription factor binding sites. SOCS-3 expression inhibited IL-1β-induced promoter activity regardless of the mutation in a proximal NF-κB site (iNOS-1002-pNFmut/pNF-κB), a γ-IFN-activated site (GAS; iNOS-1002-GASmut), or an IFN-stimulated response element (ISRE; iNOS-1002-ISRE-mut), demonstrating that the effect of SOCS-3 was not mediated through any of these sites (Fig. 4). Mutation in the ISRE element resulted in a significantly (p=0.03) increased IL-1β-induced activation of the iNOS promoter as compared with the wild-type construct, suggesting a more global suppressive effect on IL-1β-induced iNOS transcription in general although it was not influenced by SOCS-3 expression (Fig. 4). Mutation in the distal NF-κB binding site (iNOS-1002-dNFmut/dNF-κB) fully abolished IL-1β-induced iNOS promoter activity, making it impossible to evaluate a potential inhibitory role of SOCS-3 on NF-κB activation through this site (Fig. 4).
Suppression of IL-1β-induced iNOS protein expression by SOCS-3 expression. INSr3#2 cells were preincubated for 24 h with or without Dox (0.5 µg/ml) before exposure for either 6 or 24 h to IL-1β (150 pg/ml). Cell lysates (10 µg/lane) were subjected to western blot analyses for iNOS (upper band) and Flag-tagged SOCS-3 (lower band). The figure represents four individual experiments
Inhibition of IL-1β-induced iNOS promoter activity by SOCS-3. After preincubation for 24 h in the absence (white or horizontally hatched bars) or presence of Dox (0.5 µg/ml; black or diagonally hatched bars), the INSr3#2 cells were transiently transfected overnight. This was followed by 24 h of culture in the absence (white and black bars) or presence (hatched bars) of 150 pg IL-1β/ml. The figure shows the iNOS promoter activities measured by a luciferase gene reporter assay. Wt, GAS, ISRE, dNF-κB and pNF-κB refer to the wild-type construct, and the constructs mutated in the binding sites for STAT-1, IRF-1 and NF-κB (a distal and a proximal) respectively. Data are shown as the ratio of luciferase : renilla and are presented as means ± SD of four individual experiments. * p>0.02
Based on these observations and the large number of IL-1β-induced NF-κB-dependent early response genes inhibited by induced SOCS-3 expression identified through the GeneArray analysis, subsequent analyses were focused on the NF-κB-dependent pathway. The inhibitory effect of SOCS-3 on NF-κB DNA binding activity was first investigated by EMSA. The induction of NF-κB DNA binding upon stimulation with IL-1β (Fig. 5a, lane 2) was significantly reduced in the presence of SOCS-3 (lane 4). Specific binding of the probe was verified by competition with non-labelled oligonucleotides (lanes 5 and 6) and not with a non-specific oligo (lane 7). Furthermore, supershift was seen with a NF-κB-specific antibody (lane 8), whereas a non-relevant antibody (directed against STAT-1) was unable to bind the complex (lane 9).
Inhibition of IL-1β-induced NF-κB binding and transcriptional activity by SOCS-3. a. Nuclear extracts were isolated from INSr3#2 cells exposed to IL-1β and doxycycline as illustrated in Figure 4. First, the cells were cultured for 20 h in the absence or presence of 1 µg/ml doxycycline and subsequently stimulated for 15 min with IL-1β. EMSA was performed using an NF-κB binding probe. Data are representative of three independent experiments. b. INSr3#2 cells were cotransfected overnight with an NF-κB-dependent luciferase reporter gene construct in the absence (white and horizontally hatched bars) or presence (black or diagonally hatched bars) of Dox (0.5 ug/ml), and then stimulated for 6 h with IL-1β (horizontally and diagonally hatched bars). Cell lysates were subjected to luminometric analysis. Data are shown as the ratio of luciferase : renilla and presented as means ± SD of four individual experiments. * p>0.02
The fact that the inhibitory effect of SOCS-3 was, at least in part, mediated through the inhibition of NF-κB activity was further demonstrated using a specific NF-κB-dependent luciferase reporter gene construct. A two-fold induction of the NF-κB transcription in response to IL-1β was observed (Fig. 5a, lane 2). In contrast, following SOCS-3 induction, no significant IL-1β-induced NF-κB-dependent transcription was observed (lane 4).
Discussion
The ultimate goal of proteome and transcriptome analyses of cytokine-exposed beta cells is the identification of novel targets for the preservation of beta cell mass in prediabetic subjects, or the generation of genetically manipulated beta cells able to resist the immune assault after transplantation [26]. Here we focus on SOCS-3 to elucidate mechanisms underlying the unexpected inhibition of IL-1β-induced signalling and cytotoxicity to beta cells. By systematically evaluating the effects of SOCS-3 on both global gene expression and the expression of a key gene (iNOS) at the protein, mRNA and promoter activity level, we demonstrate that SOCS-3 acts at a proximal stage, preventing NF-κB activation and subsequent induction of several primary and secondary response genes. These genes encode proteins involved in intracellular beta cell destructive and defensive responses as well as proteins relevant for direct or chemokine-mediated interaction with immune cells. Interestingly, we observed low impact of SOCS-3 overexpression on the steady-state homeostatic gene expression pattern in cells cultured in the absence of IL-1β.
We have previously shown that SOCS-3 inhibited IL-1β-induced iNOS gene expression [15], a finding confirmed and extended here using protein data, promoter studies and gene array analysis. Exposure to IL-1β in the absence of induced SOCS-3 expression resulted in up-regulation of iNOS and in NO production. It is well known that cytokines may be toxic to beta cells through different mechanisms. Indeed, our previous analysis of the effect of SOCS-3 on the toxicity induced in INS-1 cells exposed to IL-1β or IFN supported this notion [15]. Here, toxicity in response to IL-1β was associated with induced NO production, previously shown to be a key component in IL-1β-induced toxicity, whereas exposure to IFN alone resulted in an NO-independent toxicity, suggested to be associated with induced ICE expression [27]. The fact that two independent mechanisms may be activated in a dose- and time-dependent fashion is further supported by the finding that although NO was induced in INS-1 cells following 24 h of exposure to a combination of IL-1β (10 U/ml) and IFN (100 U/ml), the toxic effect was not prevented by inhibition of NO by the iNOS blocker N G-monomethyl-L-arginine [28]. In contrast, in other studies exposing INS-1 for a longer time and/or at a higher concentration to IL-1β alone revealed a clear NO-dependent toxicity that was prevented by blocking NO production with the same L-arginine analogue [6, 29, 30]. Similar NO-dependent impairment of beta cell function and/or induced toxicity has been demonstrated following IL-1β exposure of isolated rodent or human islets of Langerhans as well as by the protective effects of NOS inhibitors on the development of type 1 diabetes in animal models (reviewed in [4, 31] and [32, 33, 34]).
Experiments addressing the combined effect of IL-1β and IFN on the gene expression profile using INS-1 cells revealed an NO-dependent effect on the expression of approximately 50% of the late response genes (8–24 h), affecting protein synthesis/degradation, metabolism and energy generation, growth arrest, apoptosis, redox regulation and chaperone functions [28]. No NO-dependent genes were observed among the early response genes, reflecting the fact that production of NO was first detected after 12 h of cytokine exposure [28]. Here we demonstrate that in the presence of SOCS-3, several of these late effects of IL-1β were prevented. In contrast, SOCS-3 overexpression had little effect on IL-1β-mediated up-regulation of the early response genes involved in signal transduction and gene transcription. Within the group of immune/inflammatory response genes, SOCS-3 expression clearly prevented transcription of the majority of the early response genes (ICAM, proteasome and complement components, chemokines, mob-1 and MIP-2) without any effect on the late response genes.
Focusing on the early response genes, it appeared that 16 of the 23 IL-1β-up-regulated transcripts were affected (>23%) by SOCS-3 overexpression. Among these, nine genes, namely iNOS, ICAM, complement C3, Mob-1, MIP-1, CX3C, NF-κB-p105, IRF-1 and fibrinogen-γ are usually regulated in an NF-κB-dependent manner [3, 18, 35, 36, 37, 38, 39, 40, 41], suggesting that inhibition of IL-1β signalling by SOCS-3 affects the NF-κB pathway. Furthermore, all of these transcripts were previously shown to be independent of induced NO production [28]. Sequential mutations in a proximal NF-κB binding site, a GAS site and an ISRE site in the iNOS promoter did not influence the inhibitory effect of SOCS-3 on IL-1β, indicating that the effect was primarily mediated by interference with NF-κB binding to the distal NF-κB binding site. Indeed, NF-κB binding to the distal NF-κB binding site of the iNOS promoter was shown to be essential for iNOS gene transcription [3].
As demonstrated by our EMSA analysis, SOCS-3 inhibited the DNA binding activity of NF-κB following 15 min of IL-1β exposure, suggesting that SOCS-3-mediated inhibition of IL-1β is an early event, possibly exerted via inhibition of post-translational modification, e.g. phosphorylations, known to be necessary for NF-κB-mediated signal transduction [42]. The inhibitory effect of SOCS-3 on NF-κB was furthermore detected by our NF-κB-dependent reporter assay following 6 h of IL-1β exposure, a time point at which several NF-κB-regulated genes, identified by the array, were suppressed by SOCS-3. This demonstrates that SOCS-3 inhibits IL-1β signalling and gene regulation, at least in part, through inhibition of NF-κB signalling. To our knowledge, this is the first demonstration of NF-κB as a target for SOCS-mediated inhibition of IL-1β-activated signalling.
The ability of both SOCS-1 and SOCS-3 to inhibit IFN-mediated signalling by inhibiting phosphorylation and resulting activation of the JAK–STAT pathway is well documented [14] and both SOCS-1 and SOCS-3 have been shown to inhibit IFN-mediated signalling in beta cells [15, 43, 44]. Until now, however, SOCS-3 is the only member of the SOCS family that has been shown to also interfere in the IL-1β signalling associated with beta cell destruction [15]. Thus, further analyses are needed to clarify in detail the molecular mechanisms involved in this novel effect of SOCS-3 and to investigate to what extent other members of the SOCS family possess the same ability.
During the evaluation of the gene expression profile following 24 h of IL-1β exposure, nearly all transcripts were partly suppressed (range 30–50%) by SOCS-3, in line with the partial inhibition of iNOS. This is sufficient to fully suppress apoptosis (Fig. 1) [15]. Interestingly, a few of the IL-1β-induced genes (24 h, group 6) were completely suppressed by SOCS-3, namely the proto-oncogene c-myc, the long interspersed repetitive DNA sequences (LINE)-3 and -4, the 2.4-kb long interspersed repeat DNA, and the L1 retroposon ORF2. They have all been implicated in the induction of apoptosis [45, 46, 47]. Since initiation of apoptosis is a point of no return, we speculate that the induction of these transcripts in IL-1β-exposed cells defines a threshold after which the apoptotic program is initiated.
In primary beta cells, c-myc has been shown to be a cytokine-up-regulated NF-κB-dependent gene [18] and overexpression of c-myc in beta cells in transgenic mice increased both beta cell proliferation and apoptosis, with eventual predominance of the apoptotic effect leading to beta cell loss and hyperglycaemia [45]. The ability of c-myc to mediate cytokine-induced apoptosis appears to be dependent on site-specific phosphorylation of this proto-oncogene by MAPK or c-Jun NH2-terminal kinase (JNK) [48, 49]. Interestingly, JNK is highly activated by IL-1β in beta cells and JNK inhibition has been shown to prevent IL-1β-mediated beta cell apoptosis [50, 51].
Less well known is the group of genes (termed LINEs) represented by LINE-3, LINE-4, the 2.4-kb long interspersed repeat DNA and the L1 retroposon ORF2. They all represent transposable elements that replicate via an RNA intermediate (retro-transposons) similar to that of retroviruses, and they are present in thousands of copies throughout the genome. Usually, LINEs are termed silent “fossil DNA” without transcriptional potential and are widely used as markers in genetic and phylogenetic studies [52]. However, several studies have reported functional features of the LINE elements [46, 47, 53], including a strong association with apoptosis. The relevance of these DNA elements in beta cell destruction is at present unknown, and it cannot be ruled out that our findings represent a clone-specific phenomenon.
We did not observe up-regulation of Fas, which is a known cytokine-induced gene in the beta cell. However, consistent with previous findings [22, 28], this gene has not been shown to be regulated in either purified primary rat beta cells or INS cells subjected to mRNA array analysis by arrays similar to ours. Still, the gene has been found to be regulated by more traditional mRNA approaches [36, suggesting that the Fas probes located on the array may display poor affinity/hybridisation to this particular mRNA.
Likewise, we did not observe increased SOCS-3 transcription in the INS-1r3#2 cells in response to the IL-1β concentration used. In our previous RT-PCR analysis of rat islets, we observed a small but significant induction of SOCS-3 mRNA expression using a similar IL-1β concentration [15]. However, IL-1β-induced SOCS-3 transcription was dose dependent and SOCS-3 mRNA expression was much higher at a higher dose of IL-1β. Indeed, SOCS-3 was also found to be up-regulated by IL-1β in previous gene array analyses using a rat insulinoma RIN cell line [54]. In the present gene array analysis, SOCS-3 was shown to be down-regulated. However, as evident from the RT-PCR analysis, this reflects calculations based on a very low basal expression level. The difference in IL-1β-induced SOCS-3 expression between the different systems may reflect differences in kinetics, well known among different beta cell lines as well as among native islets of different species.
Proteome analyses of IL-1β-exposed rat islets revealed a complex pattern of altered protein expression involving both protective and deleterious mechanisms, as well as many posttranslational modifications reflecting activation/deactivation of multiple pathways [26]. Subsequently, transcriptome-based analyses have confirmed and expanded these observations [55]. The combination of experimental genome, transcriptome and proteome analyses with bioinformatics may help to elucidate molecular mechanisms underlying beta cell destruction mediated by proinflammatory cytokines, and may provide valuable information about the pathogenesis of type 1 diabetes as well as targets for the development of preventive or curative strategies [26]. The possibility of intervening in this process by overexpressing protective proteins in islets transplanted to diabetic animals has recently been demonstrated. Overexpression of A20, a zinc finger protein inhibiting NF-κB activation and subsequent cytokine- or Fas-mediated apoptosis, protected transplanted islets and reduced the number of islets required to obtain normoglycaemia by 50% in a rat model of type 1 diabetes [11]. Furthermore, the potential of pharmacological modulation of protective processes was substantiated by the finding that a vitamin D analogue inhibited cytokine-induced beta cell apoptosis through induction of A20 expression [56]. That members of the SOCS family may indeed represent interesting targets in the aim to prevent beta cell destruction is supported by the recent demonstration that overexpression of SOCS-1 under the insulin promoter prevented the development of diabetes in one of two transgenic NOD lines established [44].
In conclusion, we have demonstrated that SOCS-3 overexpression not only prevents IL-1β-induced beta cell destruction but also prevents IL-1β-induced up-regulation of transcripts encoding several surface markers and chemokines that could activate or potentiate an immune assault against the beta cells. SOCS-3 represents an attractive target for the development of novel means to prevent immune-mediated beta cell destruction associated with type 1 diabetes through simultaneous inhibition of signal transduction at proximal stages of IFN-induced JAK–STAT-induced and IL-1β-induced NF-κB-dependent proapoptotic pathways.
Abbreviations
- EMSA:
-
electrophoretic mobility shift assay
- EST:
-
expressed sequence tag
- iNOS:
-
inducible nitric oxide synthase
- JAK:
-
Janus kinase
- JNK:
-
c-Jun NH2-terminal kinase
- MnSOD:
-
manganese superoxide dismutase
- NF-κB:
-
nuclear factor-κB
- NO:
-
nitric oxide
- SOCS:
-
suppressor of cytokine signalling
- STAT:
-
signal transducers and activators of transcription
References
Nerup J, Mandrup-Poulsen T, Helqvist S et al. (1994) On the pathogenesis of IDDM. Diabetologia 37:S82–S89
Pociot F, Karlsen AE (2002) Combined genome and proteome approach to identify new susceptibility genes. Am J Med Genet 115:55–60
Eizirik DL, Flodström M, Karlsen AE, Welsh N (1996) The harmony of the spheres: inducible nitric oxide synthase and related genes in pancreatic beta cells. Diabetologia 39:875–890
Eizirik DL, Mandrup-Poulsen T (2001) A choice of death—the signal-transduction of immune-mediated beta cell apoptosis. Diabetologia 44:2115–2133
Rabinovitch A, Suarez-Pinzon WL (2003) Role of cytokines in the pathogenesis of autoimmune diabetes mellitus. Rev Endocr Metab Disord 4:291–299
Hohmeier HE, Thigpen A, Tran VV, Davis R, Newgard CB (1998) Stable expression of manganese superoxide dismutase (MnSOD) in insulinoma cells prevents IL-1beta-induced cytotoxicity and reduces nitric oxide production. J Clin Invest 101:1811–1820
Lortz S, Tiedge M, Nachtwey T et al. (2000) Protection of insulin-producing RINm5F cells against cytokine-mediated toxicity through overexpression of antioxidant enzymes. Diabetes 49:1123–1130
Hotta M, Tashiro F, Ikegami H et al. (1998) Pancreatic beta cell-specific expression of thioredoxin, an antioxidative and antiapoptotic protein, prevents autoimmune and streptozotocin-induced diabetes. J Exp Med 18:1445–1451
Rabinovitch A, Suarez-Pinzon WL, Sooy K, Strynadka K, Christakos S (2001) Expression of calbindin-D-28k in a pancreatic islet beta cell line protects against cytokine-induced apoptosis and necrosis. Endocrinology 142:3649–3655
Rabinovitch A, Suarez-Pinzon W, Strynadka K et al. (1999) Transfection of human pancreatic islets with an anti-apoptotic gene (bcl-2) protects beta cells from cytokine-induced destruction. Diabetes 48:1223–1229
Grey ST, Longo C, Shukri T et al. (2003) Genetic engineering of a suboptimal islet graft with A20 preserves beta cell mass and function. J Immunol 170:6250–6256
Tiedge M, Lortz S, Drinkgern J, Lenzen S (1996) Relation between antioxidant enzyme gene expression and antioxidative defence status of insulin-producing cells. Diabetes 46:1733–1742
Starr R, Willson T, Viney E et al. (1997) A family of cytokine-inducible inhibitors of signalling. Nature 387:917–921
Krebs D, Hilton D (2000) SOCS: physiological suppressors of cytokine signalling. J Cell Sci 113:2813–2819
Karlsen AE, Rønn SG, Lindberg K et al. (2001) Suppressor of cytokine signaling 3 (SOCS-3) protects beta cells against interleukin-1β- and interferon-γ-mediated toxicity. Proc Natl Acad Sci USA 98:12191–12196
Kwon G, Corbett JA, Rodi CP, Sullivan P, McDaniel ML (1995) Interleukin-1β induced nitric oxide synthase expression by rat pancreatic β-cells: Evidence for the involvement of nuclear factor κB in the signaling mechanism. Endocrinology 136:4790–4795
Giannoukakis N, Rudert WA, Trucco M, Robbins PD (2000) Protection of human islets from the effects of interleukin-1-beta; by adenoviral gene transfer of an I-kappa B repressor. J Biol Chem 275:36509–36513
Cardozo AK, Heimberg H, Heremans Y et al. (2001) A comprehensive analysis of cytokine-induced and nuclear factor-kappa B-dependent genes in primary rat pancreatic beta cells. J Biol Chem 276:48879–48886
Dalboge H, Bayne S, Christensen T, Hejnaes KR (1989) Cloning and expression of an interleukin-1beta; precursor and its conversion to interleukin-1beta. FEBS Lett 246:89–93
Nielsen K, Karlsen AE, Deckert M et al. (1999) Beta-cell maturation leads to in vitro sensitivity to cytotoxins. Diabetes 48:2324–2332
Green IC, Wagner DA, Glogowski J et al. (1982) Analysis of nitrate, nitrite and [12N]nitrate in biological fluids. Anal Biochem 126:131–138
Cardozo AK, Kruhoeffer M, Leeman R, Ørntoft TF, Eizirik DL (2001) Identification of novel cytokine-induced genes in pancreatic beta cells by high-density oligonucleotide arrays. Diabetes 50:909–920
Hansen L, Madsen B, Teisner B, Nielsen J, Billestrup N (1998) Characterization of the inhibitory effect of growth hormone on primary preadipocyte differentiation. Mol Endocrinol 12:1140–1149
Darville M, Eizirik D (1998) Regulation by cytokines of the inducible nitric oxide synthase promoter in insulin-producing cells. Diabetologia 41:1101–1108
Rønn SG, Hansen JA, Lindberg K, Karlsen AE, Billestrup N (2002) The effect of suppressor of cytokine signaling 3 on GH signaling in beta cells. Mol Endocrinol 16:2124–2134
Karlsen AE, Sparre T, Nielsen K, Nerup J, Pociot F (2001) Proteome analysis—a novel approach to understand the pathogenesis of type 1 diabetes mellitus. Dis Markers 17:205–216
Karlsen AE, Pavlovic D, Nielsen K et al. (2000) Interferon-gamma induces interleukin-1 converting enzyme expression in pancreatic islets by an interferon regulatory factor-1-dependent mechanism. J Clin Endocrinol Metab 85:830–836
Kutlu B, Cardozo AK, Darville MI et al. (2003) Discovery of gene networks regulating cytokine-induced dysfunction and apoptosis in insulin-producing INS-1 cells. Diabetes 54:2701–2719
Aarnes M, Schonberg S, Grill V (2002) Fatty acids potentiate interleukin-1 beta toxicity in the beta cell line INS-1E. Biochem Biophys Res Commun 296:189–193
Li LX, Yoshikawa H, Egeberg KW, Grill V (2003) Interleukin-1 beta swiftly down-regulates UCP-2 mRNA in beta cells by mechanisms not directly coupled to toxicity. Cytokine 23:101–107
McDaniel ML, Corbett JA, Kwon G, Hill JR (1997) A role for nitric oxide and other inflammatory mediators in cytokine-induced pancreatic beta-cell dysfunction and destruction. Adv Exp Med Biol 426:313–319
Lindsay RM, Smith W, Rossiter SP et al. (1995) N(omega)-nitro-L-arginine methyl ester reduces the incidence of IDDM in BB/E rats. Diabetes 44:365–368
Wu G (1995) Nitric oxide synthesis and the effect of aminoguanidine and NG-monomethyl-L-arginine on the onset of diabetes in the spontaneously diabetic BB rat. Diabetes 44:360–364
Kato Y, Miura Y, Yamamoto N, Ozaki N, Oiso Y (2003) Suppressive effects of a selective inducible nitric oxide synthase (iNOS) inhibitor on pancreatic beta-cell dysfunction. Diabetologia 46:1228–1233
Heimberg H, Heremans Y, Jobin C et al. (2001) Inhibition of cytokine-induced NF-kappa B activation by adenovirus-mediated expression of a NF-kappa B super-repressor prevents beta-cell apoptosis. Diabetes 50:2219–2224
Darville MI, Eizirik DL (2001) Cytokine induction of Fas gene expression in insulin-producing cells requires the transcription factors NF-kappa B and C/EBP. Diabetes 50:1741–1748
Garcia GE, Xia YY, Chen SZ et al. (2000) NF-kappa B-dependent fractalkine induction in rat aortic endothelial cells stimulated by IL-1 beta, TNF-alpha, and LPS. J Leukoc Biol 67:577–584
Hellerbrand C, Jobin C, Licato LL, Sartor RB, Brenner DA (1998) Cytokines induce NF-kappa B in activated but not in quiescent rat hepatic stellate cells. Am J Physiol Gastrointest Liver Physiol 275:269–278
Han B, Logsdon CD (1999) Cholecystokinin induction of mob-1 chemokine expression in pancreatic acinar cells requires NF-kappa B activation. Am J Physiol Cell Physiol 277:74–82
Moon MR, Parikh AA, Pritts TA et al. (1999) Complement component C3 production in IL-1beta-stimulated human intestinal epithelial cells is blocked by NF-kappa B inhibitors and by transfection with Ser 32/36 mutant I-kappa Balpha. J Surg Res 82:48–55
Liu L, Paul A, MacKenzie CJ et al. (2001) Nuclear factor kappa B is involved in lipopolysaccharide-stimulated induction of interferon regulatory factor-1 and GAS/GAF DNA-binding in human umbilical vein endothelial cells. Br J Pharmacol 134:1629–1638
Vermeulen L, De-Wilde G, Notebaert S, Vanden-Berghe W, Haegeman G (2002) Regulation of the transcriptional activity of the nuclear factor-kappa B p65 subunit. Biochem Pharmacol 64:963–970
Cottet S, Dupraz P, Hamburger F et al. (2001) SOCS-1 protein prevents Janus kinase/STAT-dependent inhibition of beta cell insulin gene transcription and secretion in response to interferon-gamma. J Biol Chem 276:25862–25870
Flodström-Tullberg M, Yadav D, Hagerkvist R et al. (2003) Target cell expression of suppressor of cytokine signaling-1 prevents diabetes in the NOD mouse. Diabetes 52:2696–2700
Laybutt DR, Weir GC, Kaneto H et al. (2002) Overexpression of c-Myc in beta-cells of transgenic mice causes proliferation and apoptosis, downregulation of insulin gene expression, and diabetes. Diabetes 51:1793–1804
Servomaa K, Rytomaa T (1988) Suicidal death of rat chloroleukaemia cells by activation of the long interspersed repetitive DNA element (L1Rn). Cell Tissue Kinet 21:33–43
Servomaa K, Rytomaa T (1990) UV light and ionizing radiations cause programmed death of rat chloroleukaemia cells by inducing retropositions of a mobile DNA element (L1Rn). Int J Radiat Biol 57:331–343
Noguchi K, Kitanaka C, Yamana H et al. (1999) Regulation of c-Myc through phosphorylation at Ser-62 and Ser-71 by c-Jun N-terminal kinase. J Biol Chem 274:32580–32587
Noguchi K, Yamana H, Kitanaka C et al. (2000) Differential role of the JNK and p38 MAPK pathway in c-Myc- and s-Myc-mediated apoptosis. Biochem Biophys Res Commun 267:221–227
Ammendrup A, Maillard A, Nielsen K et al. (2000) The c-Jun amino-terminal kinase pathway is preferentially activated by interleukin-1 and controls apoptosis in differentiating pancreatic beta-cells. Diabetes 49:1468–1476
Bonny C, Oberson A, Negri S, Sauser C, Schorderet DF (2001) Cell-permeable peptide inhibitors of JNK. Novel blockers of beta-cell death. Diabetes 50:77–82
Furano AV, Usdin K (1995) DNA “fossils” and phylogenetic analysis. Using L1 (LINE-1, long interspersed repeated) DNA to determine the evolutionary history of mammals. J Biol Chem 270:25301–25304
Furano AV, Somerville CC, Tsichlis PN, D’Ambrosio E (1986) Target sites for the transposition of rat long interspersed repeated DNA elements (LINEs) are not random. Nucleic Acids Res 14:3717–3727
Rieneck K, Bovin L, Josefsen K et al. (2000) Massive parallel gene expression profiling of RINm5F pancreatic islet β-cells stimulated with interleukin-1β. APMIS 108:855–872
Eizirik DL, Kutlu B, Rasschaert J, Darville M, Cardozo AK (2003) Use of microarray analysis to unveil transcription factor and gene networks contributing to β cell dysfunction and apoptosis. Ann NY Acad Sci 1005:55–74
Riachy R, Vandewalle B, Conte JK et al. (2002) 1,25-Dihydroxyvitamin D3 protects RINm5F and human islet cells against cytokine-induced apoptosis: implication of the antiapoptotic protein A20. Endocrinology 143:4809–4819
Acknowledgements
We thank Susanne Munch, Anette Hellgren and Hanne Steen for excellent technical assistance. S.G. Rønn is the recipient of a fellowship from the University of Copenhagen. This work was supported in part by grants from the Danish Diabetes Association, the Juvenile Diabetes Foundation International (no. 1-2001-706) and Novo Nordisk.
Author information
Authors and Affiliations
Corresponding author
Additional information
A.E. Karlsen and P.E. Heding shared first authorship
Rights and permissions
About this article
Cite this article
Karlsen, A.E., Heding, P.E., Frobøse, H. et al. Suppressor of cytokine signalling (SOCS)-3 protects beta cells against IL-1β-mediated toxicity through inhibition of multiple nuclear factor-κB-regulated proapoptotic pathways. Diabetologia 47, 1998–2011 (2004). https://doi.org/10.1007/s00125-004-1568-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-004-1568-3