Abstract
Aims/hypothesis
It has been suggested that uncoupling protein 3 (UCP3) can increase energy expenditure, thereby regulating body weight. Although studies on UCP3 knock-out mice suggest that lack of UCP3 function does not cause obesity or Type 2 diabetes, it is possible that up-regulation of UCP3 function improves these disorders or their clinical sequelae. A 10- to 20-fold increase of UCP3 gene expression is achievable through physiological or pharmacological stimuli. We examined the phenotype of transgenic mice with approximately 18-fold overexpression of mouse UCP3 mRNA in skeletal muscle.
Methods
We generated transgenic mice with approximately 18-fold overexpression of mouse UCP3 mRNA in skeletal muscle under control of the skeletal muscle-specific muscle creatine kinase gene promoter. The phenotype of these mice was analysed either on a standard diet or on a 4-week high-fat diet.
Results
In mice on standard chow, there was no difference in body weight, oxygen consumption and mitochondrial protonmotive force between transgenic mice and non-transgenic littermates. However, transgenic mice tended to have lower body weight, increased oxygen consumption and decreased mitochondrial protonmotive force than the control mice. Transgenic mice on a 4-week high-fat diet consumed much more oxygen and had noticeably less weight gain and less epididymal fat, as well as better glucose tolerance than non-transgenic littermates.
Conclusions/interpretation
Our study shows that 18-fold overexpression of UCP3 mRNA in the skeletal muscle reduced diet-induced obesity. An 18-fold increase of UCP3 mRNA can be attained by physiological or pharmacological stimuli, suggesting that UCP3 has therapeutic potential in the treatment of obesity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Uncoupling protein 3 (UCP3), which has been identified by several groups [1, 2, 3, 4], is reported to be involved in energy metabolism by uncoupling electron transport from ATP synthesis in mitochondria and is expressed at high levels in the skeletal muscle [3], an important organ in glucose and lipid metabolism [5]. Uncoupling protein 3 mRNA is expressed at much higher levels than uncoupling protein 2 (UCP2) mRNA in the skeletal muscle in vivo although accurate comparison is difficult [1, 2, 3, 4]. As uncoupling protein 1 (UCP1) gene expression is almost undetectable in the skeletal muscle [6], UCP3 is considered to be the most relevant UCP in the skeletal muscle. Several studies have reported that UCP3 gene expression is up-regulated by triiodothyronine, catecholamines, fatty acids and peroxisome-proliferator-activated receptor (PPAR) agonists [4, 7, 8, 9].
It has been suggested that UCP3 is involved in energy expenditure, possibly leading to regulation of body weight. In two studies on UCP3 knock-out mice [10, 11] there was no difference between UCP3 knock-out and wild-type mice with regard to obesity, body temperature and serum concentrations of insulin, glucose, triglycerides and fatty acids. No obvious phenotype of UCP3 knock-out mice was observed when comparing their responses to fasting, exposure to cold, a high-fat diet and treatment with thyroid hormones with those of the control mice. This suggests that lack of UCP3 is not a major determinant of energy expenditure and obesity.
Although loss of the UCP3 function does not cause obesity or Type 2 diabetes, it is still possible that up-regulation of the UCP3 function improves these disorders. One study reported that glucose transport and GLUT4 translocation to the cell surface were increased in L6 myotubes in which UCP3 was overexpressed in vitro by adenovirus-mediated gene transfer [12]. Another study reported that transgenic mice overexpressing human UCP3 in the skeletal muscle weighed approximately 30% less than their wild-type littermates and that these transgenic mice were insulin-sensitive, with lower fasting plasma glucose and insulin concentrations [13]. These data suggest that drugs increasing UCP3 could have anti-obesity and/or antidiabetic effects. However, the levels of UCP3 overexpression in this study were very high (approximately 66-fold at the mRNA level), and no known stimulus will increase UCP3 gene expression by that amount. Several reports noted that UCP3 gene expression increased by a maximum of 10- to 20-fold in response to physiological or pharmacological stimuli in vivo [14]. We have reported that an agonist for PPARδ increased UCP3 gene expression by approximately 20-fold using L6 myotubes in vitro [15]. In this context we produced transgenic mice that overexpress mouse UCP3 mRNA in the skeletal muscle by approximately 18-fold. Furthermore, to explain the effect of the increased UCP3 on diet-induced obesity, we examined the phenotype of transgenic mice on high-fat diet.
Materials and methods
Generation of transgenic mice overexpressing UCP3
Based on the non-coding sequence of rat UCP3 cDNA [3], sense and antisense primers (sense: 5′-CAA AGG AAC CAG GCC ATC CTC CGG AAC C-3′; antisense: 5′-AA AGT ACC AAG CGG CCT GCT TGC CTT GT-3′) were prepared. Reverse transcription-PCR was done using Superscript (Invitrogen, Carlsbad, Calif., USA) with 10 µg of total RNA from mouse gastrocnemius muscle [16]. The PCR products were subcloned and sequenced (Accession ID: AB008216) by the dideoxy method [3]. A fusion gene comprising the mouse muscle creatine kinase (MCK) promoter and mouse UCP3 cDNA coding sequences was designed to enable UCP3 expression to be targeted mainly to the skeletal muscle (Fig. 1a) [17]. The purified Hind III-Sac I fragment (10 µg/ml) was microinjected into the pronucleus of fertilised BDF 1 (DBA2 X C57/B6) mouse eggs (Japan SLC, Hamamatsu, Japan). The viable eggs were transferred into the oviducts of pseudopregnant female ICR mice (Japan CLEA, Osaka, Japan) using standard techniques [18]. Transgenic founder mice were identified by Southern blot analysis of tail DNAs using the mouse UCP3 cDNA fragment as a probe [18]. Transgenic mice were used as heterozygotes. Transgenic mice were back-crossed with C57/B6 mice. Mice of F4 generation or later were used in this study. Animals were housed in a temperature-, humidity-, and light-controlled room (temperature: 22±0.5°C, humidity: 50%, 12-h light, 12-h darkness) with free access to water and food. All experimental procedures were approved by the Kyoto University Graduate School of Medicine Committee on Animal Research and carried out in accordance with the Declaration of Helsinki as revised in 2000. The principles of laboratory animal care (NIH publication no. 85-23, revised 1985); were followed, as well as any specific national laws.
Generation of transgenic mice overexpressing UCP3. Schematic representation (a) of the mouse creatine kinase promoter and mouse UCP3 fusion gene. The coding region of mouse UCP3 cDNA is denoted by the closed box. Northern blot analysis (b) of transgene expression in the gastrocnemius muscle from 8-week-old transgenic mice of Line A and Line B, and UCP3 mRNA expression in the gastrocnemius muscle from 8-week-old non-transgenic littermates. Total RNA analysed: 30 µg. Tg, transgenic mice; non Tg, non-transgenic littermates. The bar graph (c) shows quantitative analysis of UCP3 mRNA expression. Levels of UCP3 mRNA expression increased approximately 18-fold in transgenic mice Line A in comparison with the non-transgenic littermates. Data are expressed as means ± SE (n=5). Western blot analysis (d) using antirat UCP3 antibody. Amounts analysed were: 100 µg of total protein and 600 ng of protein from mitochondrial fraction. No added peptide or inhibitory peptide (to which the antibody was raised) was included during the first antibody incubation. Western blot analysis (e) using human UCP3 antibody. Amounts analysed were: 100 µg of total protein and 600 ng of protein from mitochondrial fraction
RNA extraction and northern blot analysis
Total RNA was extracted from tissues using Trizol reagent (Invitrogen) as described previously [19]. Filters containing 30 µg of total RNA were prepared. Northern blot analyses were done using cDNA probes of mouse UCP3 [9]. The density of 18S rRNA stained with ethidium bromide was used to monitor the amount of total RNA in each sample.
Western blot analysis
The gastrocnemius muscle was removed and homogenised in ice-cold buffer (50 mmol/l Tris (pH 7.4), 150 mmol/l NaCl, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% SDS) containing 1 mmol/l phenylmethlysulfonyl fluoride, 0.01 mmol/l leupeptin and 5 µg/ml aprotinin. Homogenates were centrifuged and supernatants were used for analysis of total protein. Mitochondrial fraction was obtained from gastrocnemius muscle as described previously [20]. Antibody for rat UCP3, its inhibitory peptide and antibody for human UCP3 were purchased from Alpha Diagnostic International (San Antonio, Tex., USA). Total and mitochondrial protein were used for SDS-PAGE and western blot analyses using antibodies for rat UCP3 or human UCP3 [21].
Isolation of mitochondria
Mitochondria were isolated from gastrocnemius skeletal muscle (n=7) as described previously [22, 23]. Mice were decapitated before removal of skeletal muscle. The skeletal muscle was homogenised in buffer containing 250 mmol/l sucrose, 1 mmol/l HEPES and 0.2 mmol/l EDTA (pH 7.2 with KOH). Fractionation of the homogenate was achieved by spinning for 10 min at 1500 g and at 4°C. The supernatant was then poured through a 250 µmol/l Nitex screen (Dynamic Aqua-Supply, British Columbia, Canada) and re-spun for 14 min at 16 000 g and at 4 C to obtain a mitochondrial pellet. The pellet was re-suspended on ice in 175 µl of suspension medium containing 120 mmol/l KCl, 20 mmol/l sucrose, 3 mmol/l HEPES, 2 mmol/l MgCl2, 2 mmol/l EGTA and 0.5% BSA (pH 7.2 with KOH). Stock 9% BSA was defatted by the Chen method [24] and dialysed against 153 mmol/l NaCl and 11 mmol/l KCl. The protein concentration of the mitochondrial suspension was assayed by the modified Lowry method using BSA as the reference standard.
Measurement of mitochondrial oxygen consumption and mitochondrial protonmotive force (Δp)
Mitochondrial suspensions (0.5 mg protein/ml) were placed in the incubation chamber (incubation temperature: 37°C) and magnetically stirred. Succinate-driven respiration was assessed by using 5 µmol/l rotenone to block complex 1 and 0.4 µg nigericin to assess total protonmotive force through measurements of membrane potential. State 4 respiration (maximum non-phosphorylating or leak-dependent respiration) was measured in the presence of saturating amounts of the ATP synthase inhibitor, oligomycin (10 µg/mg protein). Mitochondrial proton leak kinetics were assessed by adding malonate in increments (0.3 mmol/l to 3.6 mmol/l). The respiration rate of muscle mitochondria was measured using a Clark-type oxygen electrode (Hansatech, Norfolk, UK) [22]. Protonmotive force was measured using a methyltriphenylphosphonium-sensitive electrode as described previously [22].
Measurements of body weight and cumulative food intake
Body weight was measured daily, beginning at 4 weeks of age. Food intake was measured daily over a 1-week period at the age of 26 weeks. It was measured in male mice, which were maintained in individual metabolic cages.
Measurement of oxygen consumption
The gas analyser used to assess the metabolic rate consisted of six acrylic metabolic chambers, CO2 and O2 analysers (RL-600, AlcoSystem, Tokyo, Japan), and a switching system (ANI6-A-S, AlcoSystem) to sample gas from each metabolic chamber. Each metabolic chamber had 125.4 cm2 floor space and was 6.5 cm in height. Oxygen consumption was calculated by measuring the O2 and CO2 concentrations of each chamber [25].
Glucose and insulin tolerance tests
For the glucose tolerance test 20-week-old transgenic mice and their non-transgenic littermates were treated with an intraperitoneal injection of 2.0 mg/g glucose after an overnight fast [18]. For the insulin tolerance test, mice were injected intraperitoneally with 0.5 mU/g human regular insulin (Novolin R, Novo Nordisk, Bagsvard, Denmark) [18]. Blood samples were taken from the mouse tail vein before and 30, 60 and 90 min after the injection. Plasma glucose concentrations were measured by the glucose oxidase method with a reflectance glucometer (One Touch II, Lifescan, Milpitas, Calif., USA).
High-fat diet study
To examine the effect of UCP3 overexpression on diet-induced obesity, UCP3 transgenic mice and non-transgenic littermates were fed a high-fat diet for 4 weeks from 24 weeks of age. The high-fat diet was produced by Oriental Bioservice (Kyoto, Japan) [26]. Its fat content was 23%, whereas that of standard chow was 7%.
Histology
We did histological examinations of skeletal muscle, white and brown adipose tissues, heart, liver, and pancreas. This was done by light microscopy, using 5-µm-thick sections from 10% buffered, formalin-fixed, paraffin-embedded specimens which were stained with hematoxylin and eosin [18].
Measurement of plasma concentrations of glucose, triglyceride, fatty acids, cholesterol and insulin
Blood samples were taken from the retro-orbital sinus of mice at 9:00 hours after overnight fasting or at 14:00 hours. Plasma glucose, triglyceride, and insulin concentrations were measured by the glucose oxidase method with a reflectance glucometer, the enzymatic kit (Wako Pure Chemical, Osaka, Japan) and by RIA with mouse insulin standards (Morinaga BioLab, Yokohama, Japan), respectively [18].
Statistical analysis
Data were expressed as means ± SE. Statistical significance was tested by one-way ANOVA. If F was found to be significant, the Student’s t test was used to test individual differences. A p value of less than 0.05 was considered statistically significant.
Results
Generation of transgenic mice overexpressing UCP3
A fusion gene comprising the mouse MCK promoter and mouse UCP3 cDNA coding sequence was designed to enable UCP3 expression to be targeted to the skeletal muscle (Fig. 1a). Several transgenic lines with different copy numbers of the transgene were obtained. With the mouse UCP3 cDNA probe, northern blot analysis showed that an mRNA species of 2.4 kb in size, which was expected from the fusion gene of MCK promoter and mouse UCP3 cDNA, was expressed abundantly in the skeletal muscle from several lines of the transgenic mice (Fig. 1b). The highest UCP3 mRNA levels were obtained in Line A of the transgenic lines and were approximately 18-fold higher than in non-transgenic littermates (Fig. 1b,c). The second highest mRNA levels were approximately 15-fold higher (Line B; Fig. 1b).
Western blot analysis of protein expression in the skeletal muscle of these two lines detected multiple bands, separated by electrophoresis, of total protein extracted from the skeletal muscle (Fig. 1d). When protein of mitochondrial fraction was electrophoresed, antirat UCP3 antibody detected, as expected from the molecular mass of mouse UCP3, a single band near 34 Mr (Fig. 1d). This band at 34 Mr disappeared when inhibitory peptide was simultaneously incubated (Fig. 1d). Similar results were obtained using antihuman UCP3 antibody (Fig. 1e).
To quantify UCP3 concentrations in the skeletal muscle of transgenic mice, we examined transgenic mice of Line A. Western blot analyses were done using various amounts of protein from skeletal muscle mitochondria. Figure 2a is a representative result. It shows that the density of bands of 600 ng of protein from a UCP3 transgenic mouse was nearly equivalent to that of 10 µg of protein from a non-transgenic littermate. UCP3 protein was approximately 15 times higher in transgenic mice than in nontrasgenic littermates (n=5; Fig. 2b). As the MCK promoter also induces gene expression in the cardiac muscle, we examined the UCP3 gene expression there, finding that UCP3 mRNA levels were slightly higher in the cardiac muscle of Line A and Line B than in those of non-transgenic littermates. In Line A and Line B, UCP3 mRNA levels in the cardiac muscle were less than 1% of those in the skeletal muscle. UCP3 protein levels were not detected in the cardiac muscle either of transgenic mice or of non-transgenic littermates. UCP3 mRNA levels in the brown adipose tissue of transgenic mice were not significantly different from those of non-transgenic littermates (107±12% in Line A and 112±9% in Line B, p>0.05). Northern blot analysis with 30 µg of total RNA failed to detect any band of UCP3 mRNA in tissues of transgenic mice other than skeletal muscle, cardiac muscle and brown adipose tissue. The transgenic mice of both lines were fertile, and viable throughout adulthood with no appreciable complications. All these results were observed in transgenic mice of each sex.
Quantification of UCP3 protein in mitochondria from skeletal muscle of transgenic mice. Representative result of western blot analysis (a) using antirat UCP3 antibody. Line A transgenic mice were examined. Various amounts of protein from mitochondria were analysed. The density of bands of 600 ng of protein from UCP3 transgenic mice was nearly equal to that of 10 µg of protein from non-transgenic littermates. The bar graph (b) shows quantitative analysis of UCP3 protein, which increased approximately 15-fold in transgenic mice as compared with non-transgenic littermates. Data are expressed as means ± SE (n=5)
Mitochondrial proton leak
The kinetic response curve for the proton leak reactions showed trends towards increased mitochondrial oxygen consumption over a range of values for Δp (Fig. 3). The absolute values for oxygen consumption and Δp at State 4 (the point on each curve at the furthermost point on the right) were not statistically different between groups (Fig. 3). However, UCP3 transgenic mice tended to have higher State 4 oxygen consumption than the non-transgenic littermates (202.9±10.1 nmol O2 mg−1 min−1 vs 193.1±7.1 nmol O2 mg−1 min−1, p=0.3). UCP3 transgenic mice also tended to have a lower State 4 Δp value (196.5±2.2 mV for transgenic mice vs 201.3±4.1 mV for non-transgenic littermates, p=0.2).
Kinetic response of the proton leak to protonmotive force in mitochondria isolated from hindlimb muscle of UCP3 transgenic mice (▲) and non-transgenic littermates (■) (n=7, each). State 4 respiration was established using a saturating concentration of oligomycin (10 µg/mg protein). Subsequent additions of malonate (0.3 mmol/l to 3.6 mmol/l) provided the overall kinetics of the proton leak reactions. Data are expressed as means ± SE. Statistical significance was determined by Student’s t test. A p value of less than 0.05 was considered statistically significant
Phenotype of UCP3 transgenic mice on standard chow diet
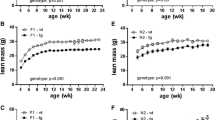
In transgenic mice and non-transgenic littermates on a standard chow diet no significant difference in body weight was observed, although the transgenic mice tended to be lighter (27.6±0.6 g vs 29.9±0.6 g at 21 weeks, p=0.07; Fig. 4). No differences in food intake (2.8±0.1 g/day vs 2.7±0.1 g/day, p>0.05), oxygen consumption (46.1±2.6 ml kg−1 min−1 vs 41.3±1.1 ml kg−1 min−1, p>0.05), rectal temperature (36.1±0.6 C vs 35.9±0.5°C, p>0.05) and histological analysis were observed between transgenic and non-transgenic littermates. No significant differences were noted in serum concentrations of glucose, insulin, cholesterol, triglyceride and fatty acids, although these tended to be lower in transgenic mice than in non-transgenic littermates (Table 1). In glucose and insulin tolerance tests on 28-week-old transgenic mice and non-transgenic littermates no significant differences were noted (Fig. 5a, Fig. 6a). As the phenotype described above was noted in transgenic mice of Line A and Line B, we used Line A, the line expressing the highest levels of UCP3, for further examination.
Changes in body weight of male transgenic mice and non-transgenic littermates on standard chow and on high-fat diets. UCP3 transgenic mice and non-transgenic littermates were fed a standard chow diet for the first 24 weeks (n=22 to 25). For the following 4 weeks, some were fed a high-fat diet (n=7 to 9), while others continued on the standard chow diet (n=16 to 17). Data are expressed as means ± SE. *p<0.05 vs non-transgenic littermates on high-fat diet
Glucose tolerance test in (a) male 28-week-old transgenic mice and non-transgenic littermates on a standard chow diet and (b) in male 28-week-old transgenic mice and non-transgenic littermates after 4 weeks on a high-fat diet. Data are expressed as means ± SE (n=7 to 9). *p<0.05 vs non-transgenic littermates
Insulin tolerance test in (a) male 28-week-old transgenic mice and non-transgenic littermates on a standard chow diet and (b) in male 28-week-old transgenic mice and non-transgenic littermates after 4 weeks on a high-fat diet. Serum glucose concentrations are expressed as percentages of the initial values of transgenic mice and non-transgenic littermates. Data are expressed as means ± SE (n=7 to 9). *p<0.05 vs non-transgenic littermates
Body weight of transgenic mice on high-fat diet
One week after beginning the high-fat diet, the transgenic mice were approximately 10% less obese than the non-transgenic littermates (28.3±0.6 g vs 31.6±0.8 g, p<0.05; Fig. 4). After 4 weeks of a high-fat diet, the transgenic mice were approximately 14% less obese than the non-transgenic littermates (32.4±1.3 g vs 37.5±1.3 g, p<0.05; Fig. 4). Body weight gain for this 4-week period was nearly 49% less in transgenic mice than in non-transgenic littermates (4.4±1.1 g vs 8.6±0.8 g, p<0.05). The weight of the epidydimal fat pad was approximately 20% lower in transgenic mice than in non-transgenic littermates after the 4-week high-fat diet (1693±131 mg vs 2110±17 mg, p<0.05). No difference was observed in the weight of livers or other organs. Histological analyses did not show any obvious difference between transgenic mice and non-transgenic littermates after the 4-week high-fat diet.
Food intake and oxygen consumption of transgenic mice on high-fat diet
No difference in food intake and rectal temperature was noted between transgenic mice and non-transgenic littermates (food intake: 2.5±0.15 g/day vs 2.3±0.08 g/day, p>0.05; rectal temperature: 35.8±0.5°C vs 35.6±0.7°C, p>0.05). However, total oxygen consumption for 24 h was higher in transgenic mice than in non-transgenic littermates (54.1±4.5 ml kg−1 min−1 vs 43.1±1.6 ml kg−1 min−1, p<0.05).
Blood analysis of transgenic mice on high-fat diet
No significant differences in serum concentrations of glucose, insulin, cholesterol, triglyceride and fatty acids were found between transgenic mice and non-transgenic littermates. However, the relevant values tended to be lower in transgenic mice (Table 1).
Glucose homeostasis in transgenic mice on high-fat diet
In the glucose tolerance test, increases in serum glucose were less marked in transgenic mice than in non-transgenic littermates (n=5 to 7, p<0.05; Fig. 5b). In the insulin tolerance test, hypoglycaemic responses 60 and 90 min after the injection were more pronounced in transgenic mice than in non-transgenic littermates (n=5 to 7, p<0.05; Fig. 6b).
Discussion
We generated transgenic mice that overexpressed mouse UCP3 in the skeletal muscle by using MCK promoter. In these mice UCP3 overexpression was targeted mainly to the skeletal muscle. Levels of UCP3 mRNA and protein were approximately 18-fold and 15-fold higher in the lines with the highest expression. Northern blot analyses detected a slight increase of UCP3 mRNA in the cardiac muscle. UCP3 mRNA levels in the cardiac muscle of transgenic mice were less than 1% of those in their skeletal muscle. Western blot analyses failed to detect the band of UCP3 in the cardiac muscle in our study. UCP3 mRNA levels in the brown adipose tissue of transgenic mice were not significantly different from those of non-transgenic littermates. No band of UCP3 mRNA was detected in tissues of transgenic mice other than skeletal muscle, cardiac muscle and brown adipose tissue. The phenotype described here therefore results primarily from the expression of the transgene in the skeletal muscle.
Expression of the UCP3 gene has been reported to increase 10- to 20-fold in response to physiological stimuli such as fasting [14]. An approximately 18-fold increase of UCP3 mRNA, as observed in transgenic mice in our study, seems to be feasible by physiological or pharmacological stimuli in vivo.
Our findings for mice on a standard chow diet, with no significant differences across a wide range of factors studied, indicate that an approximately 18-fold overexpression of UCP3 mRNA does not strongly affect energy expenditure on a standard chow diet. However, after a 4-week high-fat diet, transgenic mice were approximately 14% less obese than non-transgenic littermates. This finding is consistent with the tendency of transgenic mice to weigh less on a standard chow diet than their non-transgenic littermates. The increase in body weight of transgenic mice for this 4-week period was nearly 50% less than that of non-transgenic littermates. Moreover, the mean weight of the epididymal fat pad was approximately 20% lower in transgenic mice than in non-transgenic littermates after a 4-week high-fat diet. As no significant difference in food intake was noted between transgenic mice and non-transgenic littermates, the reduction of body weight and epididymal fat pad can be explained by the significantly increased oxygen consumption of transgenic mice on a high-fat diet. The reason why oxygen consumption was significantly increased on a high-fat diet but not on a standard chow diet is not clear. It is also not clear whether the improved glucose tolerance and insulin sensitivity seen in transgenic mice as compared with non-transgenic littermates can be explained by the reduction of obesity or by any other mechanisms.
In one study on transgenic mice which overexpressed human UCP3 mRNA by approximately 66-fold in the skeletal muscle [13] transgenic mice on standard chow were leaner and had decreased adipose tissue mass, improved glucose metabolism and increased metabolic rates (oxygen consumption). The phenotype of transgenic mice on a standard chow diet in this study was consistent with that of our transgenic mice on a high-fat diet. The discrepancy between their and our phenotypes on a standard chow diet could be explained by the difference in mRNA levels of overexpressed UCP3 (66-fold vs 18-fold). Recently the same authors reported that the increase in protein levels of overexpressed UCP3 in their transgenic mice was approximately 23-fold [27], which was also larger than in our transgenic mice (23-fold vs 15-fold). Although this difference in the UCP3 protein levels is not so large, it is possible that a small difference of UCP3 protein concentration in this range is critical for the discrepancy between phenotypes on a standard chow diet. Alternatively, the discrepancy could be due to the difference in the species of overexpressed transgene (human UCP3 vs. mouse UCP3). Taken together with these other findings, our study suggests that overexpression of UCP3 can cause increased energy expenditure, resulting in less adipose-tissue mass and improved glucose metabolism.
In conclusion, our transgenic mice with approximately 18-fold overexpression of UCP3 mRNA did not show any obvious phenotype on a standard chow diet, but were less obese and had improved glucose tolerance on a high-fat diet. As an 18-fold increase of UCP3 mRNA could be attained by physiological or pharmacological stimuli, our results suggest that drugs which increase UCP3 could be effective against obesity and Type 2 diabetes, with few adverse effects.
Abbreviations
- UCP3:
-
uncoupling protein 3
- UCP2:
-
uncoupling protein 2
- UCP1:
-
uncoupling protein 1
- PPAR:
-
peroxisome-proliferator-activated receptor
- MCK:
-
muscle creatine kinase
- Δp:
-
mitochondrial protonmotive force
References
Larkin S, Mull E, Miao W et al. (1997) Regulation of the third member of the uncoupling protein family, UCP3, by cold and thyroid hormone. Biochem Biophys Res Commun 240:222–227
Boss O, Samec S, Paoloni-Giacobino A et al. (1997) Uncoupling protein-3: a new member of the mitochondrial carrier family with tissue-specific expression. FEBS Lett 408:39–42
Matsuda J, Hosoda K, Itoh H et al. (1997) Cloning of rat uncoupling protein-3 and uncoupling protein-2 cDNAs: their gene expression in rats fed high-fat diet. FEBS Lett 418:200–204
Gong DW, He Y, Karas M, Reitman M (1997) Uncoupling protein-3 is a mediator of thermogenesis regulated by thyroid hormone, beta3-adrenergic agonists, and leptin. J Biol Chem 272:24129–24132
Zurlo F, Larson K, Bogardus C, Ravussin E (1990) Skeletal muscle metabolism is a major determinant of resting energy expenditure. J Clin Invest 86:1423–1427
Klaus S, Casteilla L, Bouillaud F, Ricquier D (1991) The uncoupling protein UCP: a membraneous mitochondrial ion carrier exclusively expressed in brown adipose tissue. Int J Biochem 23:791–801
Weigle DS, Selfridge LE, Schwartz MW et al. (1998) Elevated free fatty acids induce uncoupling protein 3 expression in muscle: a potential explanation for the effect of fasting. Diabetes 47:298–302
Nagase I, Yoshida T, Saito M (2001) Up-regulation of uncoupling proteins by beta-adrenergic stimulation in L6 myotubes. FEBS Lett 494:175–180
Matsuda J, Hosoda K, Itoh H et al. (1998) Increased adipose expression of the uncoupling protein-3 gene by thiazolidinediones in Wistar fatty rats and in cultured adipocytes. Diabetes 47:1809–1814
Vidal-Puig AJ, Grujic D, Zhang CY et al. (2000) Energy metabolism in uncoupling protein 3 gene knockout mice. J Biol Chem 275:16258–16266
Gong DW, Monemdjou S, Gavrilova O et al. (2000) Lack of obesity and normal response to fasting and thyroid hormone in mice lacking uncoupling protein-3. J Biol Chem 275:16251–16257
Huppertz C, Fischer BM, Kim YB et al. (2001) Uncoupling protein 3 (UCP3) stimulates glucose uptake in muscle cells through a phosphoinositide 3-kinase-dependent mechanism. J Biol Chem 276:12520–12529
Clapham JC, Arch JR, Chapman H et al. (2000) Mice overexpressing human uncoupling protein-3 in skeletal muscle are hyperphagic and lean. Nature 406:415–418
Boss O, Hagen T, Lowell BB (2000) Uncoupling proteins 2 and 3: potential regulators of mitochondrial energy metabolism. Diabetes 49:143–156
Son C, Hosoda K, Matsuda J et al. (2001) Up-regulation of uncoupling protein 3 gene expression by fatty acids and agonists for PPARs in L6 myotubes. Endocrinology 142:4189–4194
Shintani M, Nishimura H, Akamizu T et al. (1999) Thyrotropin decreases leptin production in rat adipocytes. Metabolism 48:1570–1574
Johnson JE, Wold BJ, Hauschka SD (1989) Muscle creatine kinase sequence elements regulating skeletal and cardiac muscle expression in transgenic mice. Mol Cell Biol 9:3393–3399
Ogawa Y, Masuzaki H, Hosoda K et al. (1999) Increased glucose metabolism and insulin sensitivity in transgenic skinny mice overexpressing leptin. Diabetes 48:1822–1829
Ogawa Y, Masuzaki H, Isse N et al. (1995) Molecular cloning of rat obese cDNA and augmented gene expression in genetically obese Zucker fatty (fa/fa) rats. J Clin Invest 96:1647–1652
Sivitz WI, Fink BD, Donohoue PA (1999) Fasting and leptin modulate adipose and muscle uncoupling protein: divergent effects between messenger ribonucleic acid and protein expression. Endocrinology 140:1511–1519
Yamamoto Y, Yoshimasa Y, Koh M et al. (2000) Constitutively active mitogen-activated protein kinase kinase increases GLUT1 expression and recruits both GLUT1 and GLUT4 at the cell surface in 3T3-L1 adipocytes. Diabetes 49:332–339
Monemdjou S, Kozak LP, Harper ME (1999) Mitochondrial proton leak in brown adipose tissue mitochondria of Ucp1-deficient mice is GDP insensitive. Am J Physiol 276:E1073–E1082
Monemdjou S, Hofmann WE, Kozak LP, Harper ME (2000) Increased mitochondrial proton leak in skeletal muscle mitochondria of UCP1-deficient mice. Am J Physiol Endocrinol Metab 279:E941–E946
Chen RF (1967) Removal of fatty acids from serum albumin by charcoal treatment. J Biol Chem 242:173–181
Ishihara K, Oyaizu S, Onuki K, Lim K, Fushiki T (2000) Chronic (-)-hydroxycitrate administration spares carbohydrate utilization and promotes lipid oxidation during exercise in mice. J Nutr 130:2990–2995
Masuzaki H, Ogawa Y, Hosoda K, Kawada T, Fushiki T, Nakao K (1995) Augmented expression of the obese gene in the adipose tissue from rats fed high-fat diet. Biochem Biophys Res Commun 216:355–358
Cadenas S, Echtay KS, Harper JA et al. (2002) The basal proton conductance of skeletal muscle mitochondria from transgenic mice overexpressing or lacking uncoupling protein-3. J Biol Chem 277:2773–2778
Acknowledgements
This work was supported in part by research grants from the Japanese Ministry of Education, Culture, Sports, Science and Technology; the Japanese Ministry of Health, Labour and Welfare; Ono Medical Research Foundation; Japan Diabetes Foundation; Tanabe Medical Frontier Conference; and in Canada from the Natural Sciences and Engineering Research Council and the Canadian Institutes of Health Research. We thank Professor S.D. Hauschka for providing p3300MCKCAT, which contains MCK promoter. We also gratefully acknowledge the assistance of Dr S. Monemdjou in the studies of muscle mitochondrial proton leak kinetics, and thank Dr M. Hirode and Dr Y. Sakura for the histological analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Son, C., Hosoda, K., Ishihara, K. et al. Reduction of diet-induced obesity in transgenic mice overexpressing uncoupling protein 3 in skeletal muscle. Diabetologia 47, 47–54 (2004). https://doi.org/10.1007/s00125-003-1272-8
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-003-1272-8