Abstract
Key message
R-BPMV is located within a recently expanded TNL cluster in the Phaseolus genus with suppressed recombination and known for resistance to multiple pathogens including potyviruses controlled by the I gene.
Abstract
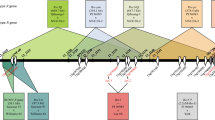
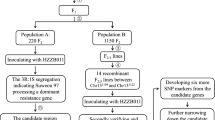
Bean pod mottle virus (BPMV) is a comovirus that infects common bean and legumes in general. BPMV is distributed throughout the world and is a major threat on soybean, a closely related species of common bean. In common bean, BAT93 was reported to carry the R-BPMV resistance gene conferring resistance to BPMV and linked with the I resistance gene. To fine map R-BPMV, 182 recombinant inbred lines (RILs) derived from the cross BAT93 × JaloEEP558 were genotyped with polymerase chain reaction (PCR)-based markers developed using genome assemblies from G19833 and BAT93, as well as BAT93 BAC clone sequences. Analysis of RILs carrying key recombination events positioned R-BPMV to a target region containing at least 16 TIR-NB-LRR (TNL) sequences in BAT93. Because the I cluster presents a suppression of recombination and a large number of repeated sequences, none of the 16 TNLs could be excluded as R-BPMV candidate gene. The evolutionary history of the TNLs for the I cluster were reconstructed using microsynteny and phylogenetic analyses within the legume family. A single I TNL was present in Medicago truncatula and lost in soybean, mirroring the absence of complete BPMV resistance in soybean. Amplification of TNLs in the I cluster predates the divergence of the Phaseolus species, in agreement with the emergence of R-BPMV before the separation of the common bean wild centers of diversity. This analysis provides PCR-based markers useful in marker-assisted selection (MAS) and laid the foundation for cloning of R-BPMV resistance gene in order to transfer the resistance into soybean.




Similar content being viewed by others
Data availability
Phenotype data generated or analyzed during this study are included in this published article (and its supplementary files). Genotypic data are available from the corresponding author on reasonable request.
References
Altschul S (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. https://doi.org/10.1093/nar/25.17.3389
Ammiraju JSS, Lu F, Sanyal A et al (2008) Dynamic evolution of oryza genomes is revealed by comparative genomic analysis of a genus-wide vertical data set. Plant Cell 20:3191–3209. https://doi.org/10.1105/tpc.108.063727
Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Report 9:415–415. https://doi.org/10.1007/BF02672016
Bailey TL, Johnson J, Grant CE, Noble WS (2015) The MEME suite. Nucleic Acids Res 43:W39–W49
Barragan AC, Weigel D (2021) Plant NLR diversity: the known unknowns of pan-NLRomes. Plant Cell 33:814–831. https://doi.org/10.1093/plcell/koaa002
Bello MH, Moghaddam SM, Massoudi M et al (2014) Application of in silico bulked segregant analysis for rapid development of markers linked to Bean common mosaic virusresistance in common bean. BMC Genomics 15:903. https://doi.org/10.1186/1471-2164-15-903
Bellucci E, Aguilar OM, Alseekh S et al (2021) The INCREASE project: intelligent collections of food-legume genetic resources for European agrofood systems. Plant J 108:646–660
Bitocchi E, Rau D, Bellucci E et al (2017) Beans (Phaseolus ssp.) as a model for understanding crop evolution. Front. Plant Sci 8:722
Broughton WJ, Hernández G, Blair M et al (2003) Beans (Phaseolus spp.)—model food legumes. Plant Soil. pp 55–128
Burset M, Guigó R (1996) Evaluation of gene structure prediction programs. Genomics 34:353–367. https://doi.org/10.1006/geno.1996.0298
Cesari S, Bernoux M, Moncuquet P et al (2014) A novel conserved mechanism for plant NLR protein pairs: the “integrated decoy” hypothesis. Front Plant Sci 5:606. https://doi.org/10.3389/fpls.2014.00606
Chalhoub B, Belcram H, Caboche M (2004) Efficient cloning of plant genomes into bacterial artificial chromosome (BAC) libraries with larger and more uniform insert size. Plant Biotechnol J 2:181–188. https://doi.org/10.1111/j.1467-7652.2004.00065.x
Chen NWG, Sévignac M, Thareau V et al (2010) Specific resistances against Pseudomonas syringae effectors AvrB and AvrRpm1 have evolved differently in common bean ( Phaseolus vulgaris ), soybean ( Glycine max ), and Arabidopsis thaliana. New Phytol 187:941–956. https://doi.org/10.1111/j.1469-8137.2010.03337.x
Chen NWG, Thareau V, Ribeiro T et al (2018) Common bean subtelomeres are hot spots of recombination and favor resistance gene evolution. Front Plant Sci 9:1185. https://doi.org/10.3389/fpls.2018.01185
Choi K, Reinhard C, Serra H et al (2016) Recombination rate heterogeneity within arabidopsis disease resistance genes. PLOS Genet 12:e1006179. https://doi.org/10.1371/journal.pgen.1006179
Creusot F, Macadré C, Cana EF et al (1999) Cloning and molecular characterization of three members of the NBS-LRR subfamily located in the vicinity of the Co-2 locus for anthracnose resistance in Phaseolus vulgaris. Genome 42:254–264. https://doi.org/10.1139/g98-134
Cronk Q, Ojeda I, Pennington RT (2006) Legume comparative genomics: progress in phylogenetics and phylogenomics. Curr Opin Plant Biol 9:99–103. https://doi.org/10.1016/j.pbi.2006.01.011
Crute IR, Pink D (1996) Genetics and utilization of pathogen resistance in plants. Plant Cell 8:1747–1755. https://doi.org/10.1105/tpc.8.10.1747
Dangl JL, Horvath DM, Staskawicz BJ (2013) Pivoting the plant immune system from dissection to deployment. Science (80- ) 341:746–751. https://doi.org/10.1126/science.1236011
David P, Chen NWG, Pedrosa-Harand A et al (2009) A nomadic subtelomeric disease resistance gene cluster in common bean. Plant Physiol 151:1048–1065. https://doi.org/10.1104/pp.109.142109
de Haan P, Kormelink R, de Oliveira RR et al (1991) Tomato spotted wilt virus L RNA encodes a putative RNA polymerase. J Gen Virol 72:2207–2216. https://doi.org/10.1099/0022-1317-72-9-2207
Delgado-Salinas A, Bibler R, Lavin M (2006) Phylogeny of the Genus Phaseolus (Leguminosae): a recent diversification in an ancient landscape. Syst Bot 31:779–791
Dodds PN, Rathjen JP (2010) Plant immunity: towards an integrated view of plant–pathogen interactions. Nat Rev Genet 11:539–548. https://doi.org/10.1038/nrg2812
Doyle J, Doyle J (1991) DNA isolation from small amount of plant tissue. Phytochem Bull. https://doi.org/10.2307/4119796
Drijfhout E (1978) Genetic interaction between Phaseolus vulgaris and bean common mosaic virus with implications for strain identification and breeding for resistance. Versl van Landbouwkd Onderz 1–89
Ferreira JJ, Campa A, Pérez-Vega E et al (2012) Introgression and pyramiding into common bean market class fabada of genes conferring resistance to anthracnose and potyvirus. Theor Appl Genet 124:777–788. https://doi.org/10.1007/s00122-011-1746-x
Ferrier-Cana E, Geffroy V, Macadré C et al (2003) Characterization of expressed NBS-LRR resistance gene candidates from common bean. Theor Appl Genet 106:251–261. https://doi.org/10.1007/s00122-002-1032-z
Ferrier-Cana E, Macadré C, Sévignac M et al (2005) Distinct post-transcriptional modifications result into seven alternative transcripts of the CC-NBS-LRR gene JA1tr of Phaseolus vulgaris. Theor Appl Genet 110:895–905. https://doi.org/10.1007/s00122-004-1908-1
Fisher ML, Kyle MM (1994) Inheritance of resistance to potyviruses in Phaseolus vulgaris L. III. Cosegregation of phenotypically similar dominant responses to nine potyviruses. Theor Appl Genet 89–89:818–823. https://doi.org/10.1007/BF00224503
Fisher ML, Kyle MM (1996) Inheritance of resistance to potyviruses in Phaseolus vulgaris L. IV. Inheritance, linkage relations, and environmental effects on systemic resistance to four potyviruses. Theor Appl Genet 92:204–212. https://doi.org/10.1007/BF00223377
Fisher RA (1937) The design of experiments. 2nd edition. Oliver & Boyd. Edinburgh, London, p 260
Flor HH (1956) The complementary genic systems in flax and flax rust. Adv Genet, pp pp 29–54
Foyer CH, Lam H-M, Nguyen HT et al (2016) Neglecting legumes has compromised human health and sustainable food production. Nat Plants 2:16112. https://doi.org/10.1038/nplants.2016.112
Freyre R, Skroch PW, Geffroy V et al (1998) Towards an integrated linkage map of common bean. 4. Development of a core linkage map and alignment of RFLP maps. Theor Appl Genet 97. https://doi.org/10.1007/s001220050964
Garcia T, Duitama J, Zullo SS et al (2021) Comprehensive genomic resources related to domestication and crop improvement traits in Lima bean. Nat Commun 12. https://doi.org/10.1038/s41467-021-20921-1
Geffroy V, Creusot F, Falquet J et al (1998) A family of LRR sequences in the vicinity of the Co-2 locus for anthracnose resistance in Phaseolus vulgaris and its potential use in marker-assisted selection. Theor Appl Genet 96:494–502. https://doi.org/10.1007/s001220050766
Geffroy V, Sévignac M, Billant P et al (2008) Resistance to Colletotrichum lindemuthianum in Phaseolus vulgaris: a case study for mapping two independent genes. Theor Appl Genet 116:407–415. https://doi.org/10.1007/s00122-007-0678-y
Geffroy V, Macadré C, David P et al (2009) Molecular analysis of a large subtelomeric nucleotide-binding-site–leucine-rich-repeat family in two representative genotypes of the major gene pools of Phaseolus vulgaris. Genetics 181:405–419. https://doi.org/10.1534/genetics.108.093583
Gepts P, Bliss FA (1985) F1 hybrid weakness in the common bean. J Hered 76:447–450. https://doi.org/10.1093/oxfordjournals.jhered.a110142
Ghabrial SA (1982) Radioimmunosorbent assay for detection of lettuce mosaic virus in lettuce seed. Plant Dis 66:1037. https://doi.org/10.1094/PD-66-1037
Giesler LJ, Ghabrial SA, Hunt TE, Hill JH (2002) Bean pod mottle virus: a threat to U.S. soybean production. Plant Dis 86:1280–1289. https://doi.org/10.1094/PDIS.2002.86.12.1280
Gonthier L, Bellec A, Blassiau C et al (2010) Construction and characterization of two BAC libraries representing a deep-coverage of the genome of chicory (Cichorium intybus L., Asteraceae). BMC Res Notes 3:. https://doi.org/10.1186/1756-0500-3-225
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. https://doi.org/10.1080/10635150390235520
Hart JP, Griffiths PD (2015) Genotyping‐by‐sequencing enabled mapping and marker development for the By‐2 Potyvirus resistance allele in common bean. Plant Genome 8:plantgenome2014–09. https://doi.org/10.3835/plantgenome2014.09.0058
Hill JH, Whitham SA (2014) Control of virus diseases in soybeans. In: Advances in virus research. United States, pp 355–390
Innes RW, Ameline-Torregrosa C, Ashfield T et al (2008) Differential accumulation of retroelements and diversification of NB-LRR disease resistance genes in duplicated regions following polyploidy in the ancestor of soybean. Plant Physiol 148:1740–1759. https://doi.org/10.1104/pp.108.127902
Jones JDG, Witek K, Verweij W et al (2014) Elevating crop disease resistance with cloned genes. Philos Trans R Soc B Biol Sci 369:20130087. https://doi.org/10.1098/rstb.2013.0087
Katoh K (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518. https://doi.org/10.1093/nar/gki198
Kawashima CG, Guimarães GA, Nogueira SR et al (2016) A pigeonpea gene confers resistance to Asian soybean rust in soybean. Nat Biotechnol 34:661–665. https://doi.org/10.1038/nbt.3554
Kelly JD, Bornowski N (2018) Biotechnologies of crop improvement, vol 3. Springer International Publishing, Cham
Koinange EMK, Gepts P (1992) Hybrid weakness in Wild Phaseolus vulgaris L. J Hered 83:135–139. https://doi.org/10.1093/oxfordjournals.jhered.a111173
Kosambi DD (1943) The estimation of map distances from recombination values. Ann Eugen 12:172–175. https://doi.org/10.1111/j.1469-1809.1943.tb02321.x
Kourelis J, van der Hoorn RAL (2018) Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30:285–299. https://doi.org/10.1105/tpc.17.00579
Lander ES, Green P, Abrahamson J et al (1987) MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181. https://doi.org/10.1016/0888-7543(87)90010-3
Lavin M, Herendeen PS, Wojciechowski MF (2005) Evolutionary rates analysis of leguminosae implicates a rapid diversification of lineages during the tertiary. Syst Biol 54:575–594. https://doi.org/10.1080/10635150590947131
Lukashin A (1998) GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res 26:1107–1115. https://doi.org/10.1093/nar/26.4.1107
Märkle H, Saur IML, Stam R (2022) Evolution of resistance (R) gene specificity. Essays Biochem 66:551–560. https://doi.org/10.1042/EBC20210077
McClean PE, Mamidi S, McConnell M et al (2010) Synteny mapping between common bean and soybean reveals extensive blocks of shared loci. BMC Genomics 11:184. https://doi.org/10.1186/1471-2164-11-184
Meyers BC, Dickerman AW, Michelmore RW et al (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20:317–332. https://doi.org/10.1046/j.1365-313X.1999.00606.x
Meyers BC, Kozik A, Griego A et al (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15:809–834. https://doi.org/10.1105/tpc.009308
Meziadi C, Richard MMS, Derquennes A et al (2016) Development of molecular markers linked to disease resistance genes in common bean based on whole genome sequence. Plant Sci 242:351–357. https://doi.org/10.1016/j.plantsci.2015.09.006
Meziadi C, Blanchet S, Geffroy V, Pflieger S (2017) Genetic resistance against viruses in Phaseolus vulgaris L.: State of the art and future prospects. Plant Sci 265:39–50. https://doi.org/10.3390/v13071239
Meziadi C, Lintz J, Naderpour M, et al (2021) R-BPMV-mediated resistance to bean pod mottle virus in Phaseolus vulgaris L. is heat-stable but elevated temperatures boost viral infection in susceptible genotypes. Viruses 13:1239. https://doi.org/10.3390/v13071239
Michelmore RW, Christopoulou M, Caldwell KS (2013) Impacts of resistance gene genetics, function, and evolution on a durable future. Annu Rev Phytopathol 51:291–319. https://doi.org/10.1146/annurev-phyto-082712-102334
Miklas PN, Kelly JD, Beebe SE, Blair MW (2006) Common bean breeding for resistance against biotic and abiotic stresses: from classical to MAS breeding. Euphytica 147:105–131. https://doi.org/10.1007/s10681-006-4600-5
Moghaddam SM, Oladzad A, Koh C et al (2021) The tepary bean genome provides insight into evolution and domestication under heat stress. Nat Commun 12:2638. https://doi.org/10.1038/s41467-021-22858-x
Morales FJ, Singh SP (1997) Inheritance of the mosaic and necroses reactions induced by bean severe mosaic comoviruses in Phaseolus vulgaris L. Euphytica 93:223–226. https://doi.org/10.1023/A:1002902207395
Mundt CC (2014) Durable resistance: a key to sustainable management of pathogens and pests. Infect Genet Evol 27:446–455. https://doi.org/10.1016/j.meegid.2014.01.011
Ngou BPM, Ding P, Jones JDG (2022) Thirty years of resistance: zig-zag through the plant immune system. Plant Cell 34:1447–1478. https://doi.org/10.1093/plcell/koac041
Peterson DG, Tomkins JP, Frisch DA et al (2000) Construction of Plant Bacterial Artificial Chromosome (BAC) libraries: an illustrated guide. J Agric Genomics, 5.
Pflieger S, Blanchet S, Meziadi C et al (2014) The “one-step” Bean pod mottle virus (BPMV)-derived vector is a functional genomics tool for efficient overexpression of heterologous protein, virus-induced gene silencing and genetic mapping of BPMV R-gene in common bean (Phaseolus vulgaris L.). BMC Plant Biol 14. https://doi.org/10.1186/s12870-014-0232-4
Pflieger S, Blanchet S, Meziadi C et al (2015) Bean pod mottle virus (BPMV) viral inoculation procedure in common bean (Phaseolus vulgaris L.). BIO-PROTOCOL 5:e1524–e1524. https://doi.org/10.21769/BioProtoc.1524
Pierce WH (1934) Viruses of the bean. Phytopathology 24:87–115
Porch TG, Blair MW, Lariguet P et al (2009) Generation of a mutant population for TILLING common bean genotype BAT 93. J Am Soc Hortic Sci 134:348–355. https://doi.org/10.21273/JASHS.134.3.348
Richard MMS, Chen NWG, Thareau V et al (2013) The Subtelomeric khipu Satellite Repeat from Phaseolus vulgaris: lessons learned from the genome analysis of the andean genotype G19833. Front Plant Sci 4:109. https://doi.org/10.3389/fpls.2013.00109
Richard MMS, Pflieger S, Sévignac M et al (2014) Fine mapping of Co-x, an anthracnose resistance gene to a highly virulent strain of Colletotrichum lindemuthianum in common bean. Theor Appl Genet 127:1653–1666. https://doi.org/10.1007/s00122-014-2328-5
Richard MMS, Gratias A, Alvarez Diaz JC et al (2021) A common bean truncated CRINKLY4 kinase controls gene-for-gene resistance to the fungus Colletotrichum lindemuthianum. J Exp Bot 72:3569–3581. https://doi.org/10.1093/jxb/erab082
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386. https://doi.org/10.1385/1-59259-192-2:365
Rutherford K, Parkhill J, Crook J et al (2000) Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. https://doi.org/10.1093/bioinformatics/16.10.944
Schmutz J, Cannon SB, Schlueter J et al (2010) Genome sequence of the palaeopolyploid soybean. Nature 463:178–183. https://doi.org/10.1038/nature08670
Schmutz J, McClean PE, Mamidi S et al (2014) A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet 46:707–713. https://doi.org/10.1038/ng.3008
Shakiba E, Chen P, Gergerich R et al (2012) Reactions of commercial soybean cultivars from the mid south to soybean mosaic virus. Crop Sci 52:1990–1997. https://doi.org/10.2135/cropsci2011.04.0221
Shao Z-Q, Zhang Y-M, Hang Y-Y et al (2014) Long-term evolution of nucleotide-binding site-leucine-rich repeat genes: understanding gained from and beyond the legume family. Plant Physiol 166:217–234. https://doi.org/10.1104/pp.114.243626
Shii CT, Mok MC, Temple SR, Mok DWS (1980) Expression of developmental abnormalities in hybrids of Phaseolus vulgaris L. J Hered 71:218–222. https://doi.org/10.1093/oxfordjournals.jhered.a109353
Shoemaker RC, Schlueter J, Doyle JJ (2006) Paleopolyploidy and gene duplication in soybean and other legumes. Curr Opin Plant Biol 9:104–109. https://doi.org/10.1016/j.pbi.2006.01.007
Soderlund C, Bomhoff M, Nelson WM (2011) SyMAP v3.4: a turnkey synteny system with application to plant genomes. Nucleic Acids Res 39:e68–e68. https://doi.org/10.1093/nar/gkr123
Stefanović S, Pfeil BE, Palmer JD, Doyle JJ (2009) Relationships among phaseoloid legumes based on sequences from eight chloroplast regions. Syst Bot 34:115–128. https://doi.org/10.1600/036364409787602221
Steuernagel B, Witek K, Krattinger SG et al (2020) The NLR-annotator tool enables annotation of the intracellular immune receptor repertoire. Plant Physiol 183:468–482. https://doi.org/10.1104/pp.19.01273
Tai TH, Dahlbeck D, Clark ET et al (1999) Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc Natl Acad Sci 96:14153–14158. https://doi.org/10.1073/pnas.96.24.14153
Tamborski J, Krasileva KV (2020) Evolution of plant NLRs: from natural history to precise modifications. Annu Rev Plant Biol 71:355–378
Thareau V, Dehais P, Serizet C et al (2003) Automatic design of gene-specific sequence tags for genome-wide functional studies. Bioinformatics 19:2191–2198. https://doi.org/10.1093/bioinformatics/btg286
Tock AJ, Fourie D, Walley PG et al (2017) Genome-wide linkage and association mapping of halo blight resistance in common bean to Race 6 of the globally important bacterial pathogen. Front Plant Sci 8. https://doi.org/10.3389/fpls.2017.01170
Toro O, Tohme J, Debouck D (1990) Wild bean (Phaseolus vulgaris L.): description and distribution. CIAT, p 106
Vallejos CE, Astua-Monge G, Jones V et al (2006) Genetic and molecular characterization of the I locus of Phaseolus vulgaris. Genetics 172:1229–1242. https://doi.org/10.1534/genetics.105.050815
Vlasova A, Capella-Gutiérrez S, Rendón-Anaya M et al (2016) Genome and transcriptome analysis of the Mesoamerican common bean and the role of gene duplications in establishing tissue and temporal specialization of genes. Genome Biol 17:1–18. https://doi.org/10.1186/s13059-016-0883-6
Wang Y, Hobbs HA, Hill CB et al (2005) Evaluation of ancestral lines of U.S. Soybean cultivars for resistance to four soybean viruses. Crop Sci 45:639–644. https://doi.org/10.2135/cropsci2005.0639
Wenger AM, Peluso P, Rowell WJ et al (2019) Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat Biotechnol 37:1155–1162. https://doi.org/10.1038/s41587-019-0217-9
Whitham S, Dinesh-Kumar SP, Choi D et al (1994) The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell 78:1101–1115
Wisser RJ, Oppenheim SJ, Ernest EG et al (2021) Genome assembly of a Mesoamerican derived variety of lima bean: a foundational cultivar in the Mid-Atlantic USA. G3 (Bethesda) 11. https://doi.org/10.1093/g3journal/jkab207
Yang S, Li J, Zhang X et al (2013) Rapidly evolving R genes in diverse grass species confer resistance to rice blast disease. Proc Natl Acad Sci 110:18572–18577. https://doi.org/10.1073/pnas.1318211110
Young ND, Debellé F, Oldroyd GED et al (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480:520–524. https://doi.org/10.1038/nature10625
Zhang J, Li W, Xiang T et al (2010) Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7:290–301. https://doi.org/10.1016/j.chom.2010.03.007
Acknowledgements
We are grateful to Mireille Sévignac and Manon Richard for DNA isolation and RILs genotyping and Sophie Blanchet for her contribution to virus disease scoring. We thank Hélène Bergès and Arnaud Bellec at CNRGV (Toulouse, France) for their help with the BAC library construction. CM was supported by fellowships from the French Research Ministry. JCAD was supported by a fellowship from the Colombian Institute of Education support ICETEX.
Funding
This work was supported by INRAe and IDEEV fundings and ANR-22-CE20-0022. IPS2 benefits from the support of Saclay Plant Sciences-SPS (ANR-17-EUR-0007).
Author information
Authors and Affiliations
Contributions
CM, YCAD and VT developed molecular markers and performed bioinformatic analysis. CM and SP conducted virus resistance evaluation and genetic analysis and were actively involved in the writing of the manuscript. WM constructed and screened the BAC library. AGW, YCAD, SP, ASG and PNM participated in the data analysis, discussion and revised the manuscript. VG designed and supervised the research, discussed the results, coordinated the group work with the help of SP and wrote the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interests
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Ethical standards
On behalf of all authors, the corresponding author states that the experiments comply with the current laws of the country in which they were performed.
Additional information
Communicated by Matthew N. Nelson.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Meziadi, C., Alvarez-Diaz, JC., Thareau, V. et al. Fine-mapping and evolutionary history of R-BPMV, a dominant resistance gene to Bean pod mottle virus in Phaseolus vulgaris L.. Theor Appl Genet 137, 8 (2024). https://doi.org/10.1007/s00122-023-04513-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00122-023-04513-9