Abstract
Key message
The wheat transcription factor bZIPC1 interacts with FT2 and affects spikelet and grain number per spike. We identified a natural allele with positive effects on these two economically important traits.
Abstract
Loss-of-function mutations and natural variation in the gene FLOWERING LOCUS T2 (FT2) in wheat have previously been shown to affect spikelet number per spike (SNS). However, while other FT-like wheat proteins interact with bZIP-containing transcription factors from the A-group, FT2 does not interact with any of them. In this study, we used a yeast-two-hybrid screen with FT2 as bait and identified a grass-specific bZIP-containing transcription factor from the C-group, designated here as bZIPC1. Within the C-group, we identified four clades including wheat proteins that show Y2H interactions with different sets of FT-like and CEN-like encoded proteins. bZIPC1 and FT2 expression partially overlap in the developing spike, including the inflorescence meristem. Combined loss-of-function mutations in bZIPC-A1 and bZIPC-B1 (bzipc1) in tetraploid wheat resulted in a drastic reduction in SNS with a limited effect on heading date. Analysis of natural variation in the bZIPC-B1 (TraesCS5B02G444100) region revealed three major haplotypes (H1–H3), with the H1 haplotype showing significantly higher SNS, grain number per spike and grain weight per spike than both the H2 and H3 haplotypes. The favorable effect of the H1 haplotype was also supported by its increased frequency from the ancestral cultivated tetraploids to the modern tetraploid and hexaploid wheat varieties. We developed markers for the two non-synonymous SNPs that differentiate the bZIPC-B1b allele in the H1 haplotype from the ancestral bZIPC-B1a allele present in all other haplotypes. These diagnostic markers are useful tools to accelerate the deployment of the favorable bZIPC-B1b allele in pasta and bread wheat breeding programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat contributes to almost 20% of the world caloric intake, with approximately 770 million tons produced annually across the globe (fao.org, marketing year of 2020/2021). Current rates of yield increase (0.9%) are insufficient to match the projected growth of the human population (Ray et al. 2013), which has generated a renewed interest in understanding and improving this trait. However, yield is difficult to study because it is determined by multiple genes with complex epistatic interactions and interactions with the environment (Reynolds et al. 2012). To facilitate its genetic dissection, total grain yield can be divided into several yield components, including number of spikes per surface unit, grains per spike (GNS) and thousand kernel weight (TKW). The number of grains per spike can be further divided into spikelet number per spike (SNS) and grains per spikelet (also known as fertility).
Of the different yield components, SNS has the highest heritability (Zhang et al. 2018) which facilitates its genetic characterization. The number of spikelets per spike is determined when the inflorescence meristem (IM) stops producing lateral meristems and transitions into a terminal spikelet. This transition occurs early after the initiation of the reproductive phase, limiting the influence of the environment later in the growing season and resulting in a higher heritability than other grain yield components that are affected by the environment throughout the growing season. Increases in SNS have been associated with increases in grain yield in productive genotypes grown in optimum environments (Boden et al. 2015; Dobrovolskaya et al. 2014; Hai et al. 2008; Kuzay et al. 2019; Wolde et al. 2019). However, in suboptimal environments or in source-limited genotypes the increases in SNS are no longer associated with yield increases due to reductions in fertility and/or kernel weight (Kuzay et al. 2019).
In wheat, SNS has been shown to be correlated with flowering time, with genotypes flowering earlier usually having lower SNS than those flowering later (Shaw et al. 2013). Flowering time in wheat is mainly regulated by the vernalization (long exposures to low temperatures providing the competency to flower) and photoperiod pathways (requirement for long days), which converge on the regulation of FT1 (Distelfeld et al. 2009), the wheat homolog of Arabidopsis thaliana (Arabidopsis) FLOWRING LOCUS T (FT) and Oryza sativa (rice) HEADING DATE 3 (Hd3) (Corbesier et al. 2007; Distelfeld et al. 2009; Tamaki et al. 2007). The FT-encoded protein, also referred to as florigen, has been shown to be a mobile signal transported through the plant phloem from the leaves to the shoot apical meristem (SAM) in both Arabidopsis and rice (Corbesier et al. 2007; Tamaki et al. 2007).
In wheat, FT1 is also a strong promoter of flowering that is expressed in the leaves and not in the SAM, so it is also assumed to be a mobile signal (Lv et al. 2014; Yan et al. 2006). In the absence of FT1 expression (e.g. under short days in photoperiod sensitive wheats), the SAM transitions to the reproductive phase but spike development is arrested, and spikes fail to emerge from the leaf sheaths or head extremely late (Shaw et al. 2019; Gauley and Boden 2021). In rice, it has been shown that Hd3 activates the expression of MADS-box meristem identity genes by forming complexes with 14-3-3 and FD-like proteins (Florigen Activation Complex), which bind to the MADS-box gene promoters (Taoka et al. 2011). In wheat, FT1 has also been shown to interact with the bZIP transcription factor FDL2 and a variety of 14-3-3 proteins which are thought to act as a bridge facilitating the interaction between FT1 and FD-like proteins (Li et al. 2015; Li and Dubcovsky 2008). Once FT1 arrives at the SAM, the floral activation complex plays a critical role in accelerating the transition from the vegetative to the reproductive phase by activating meristem identity genes such as VERNALIZATION1 (VRN1) (Li et al. 2015).
The expansion of the FT-like gene family in monocot and eudicot species has led to the diversification of FT functions (Ballerini and Kramer 2011; Chardon and Damerval 2005; Higgins et al. 2010; Pin et al. 2010; Woods et al. 2019). In wheat, twelve different FT-like genes have been identified (Halliwell et al. 2016; Lv et al. 2014) and evidence of sub-functionalization has been found. For example, FT3 and FT5 have a more critical role in the induction of flowering under SD than other LD expressed FT-like genes (Faure et al. 2007; Kikuchi et al. 2009; Lv et al. 2014; Zikhali et al. 2017). In addition, FT2 was shown to have a more predominant influence on SNS than on heading time (Glenn et al. 2022; Shaw et al. 2019).
The functional differences in FT2 are particularly interesting, because this gene is the closest paralog of FT1 (78% identical at the protein level) and the only FT-like gene in wheat that has been shown to be transcribed directly in the SAM and developing spike (Shaw et al. 2019). Loss-of-function mutations in both homoeologs of FT2 in tetraploid wheat (henceforth ft2) result in small differences in heading time but significant increases in SNS. However, the ft2 mutant also shows reduced fertility precluding its utilization in breeding applications (Shaw et al. 2019). Fortunately, a natural variant of FT-A2, resulting in an Aspartic Acid (D) change to Alanine (A) at position 10 (henceforth, D10A), has been recently associated with increases in SNS, GNS and grain weight per spike (GWS) indicating no negative impacts on fertility (Glenn et al. 2022). The beneficial A10 allele was almost absent in tetraploid wheat but was present in more than half of the analyzed hexaploid (or common wheat) varieties suggesting that this allele has been favored by selection for improved grain yield in common wheat (Glenn et al. 2022).
The functional differentiation of FT1 and FT2 was paralleled by differences in their protein–protein interactions. Whereas FT1 was able to interact in yeast-two-hybrid (Y2H) assays with five of the six tested 14-3-3 proteins, FT2 showed no interactions with any of them (Li et al. 2015). The two proteins also differed in their interactions with FDL proteins. Specifically, FT1 showed positive Y2H interactions with FDL2 and FDL6, in contrast FT2 showed a positive interaction with FDL13 (Li and Dubcovsky 2008). However, later it was found that FDL13 was an alternative splice form of FDL15 with a retained intron and a premature stop codon (Li et al. 2015), and the complete FDL15 did not interact with FT2. In summary, no protein interactors of FT2 were identified before this study.
Here, we explored the proteins with which FT2 can interact by performing a Y2H screen and identified a bZIP-containing transcription factor from the grass-specific C-group that we designated as bZIPC1. We show that the proteins encoded by bZIPC1 and three other members of the C-group can interact with FT- and CEN-like wheat proteins, a function that was previously shown only for bZIP members of the A-group (FDL2, FDL6 and FDL15) (Li et al. 2015). Functional characterization of bZIPC1 mutants revealed that this gene impacts SNS but has limited effect on heading time. Finally, we characterized natural variation in this gene and identified variants associated with differences in SNS that may be of value for wheat breeding applications.
Materials and methods
Plant materials and growth conditions
We identified the bZIPC1 orthologs in the durum wheat cultivar Kronos using the sequences from Chinese Spring bZIPC-A1 (TraesCS5A02G440400) and bZIPC-B1 (TraesCS5B02g444100) and identified loss-of-function mutations in the sequenced Kronos mutant population (Krasileva et al. 2017). We then combined mutations in the A and B homoeologs from each gene to generate three loss-of-function lines designated as bzipc1-1, bzipc1-2 and bzipc1-3.
bZIPC1 mutant lines were initially grown in growth chambers at 16 h long-day with temperatures oscillating between 22 and 18 °C during the day and night, respectively. After heading and phenotyping, plants were moved to a greenhouse with supplemental lighting during the winter for drying and grain multiplication.
Phylogenetic and statistical analyses
Phylogenetic analyses of bZIP C-group genes were performed using BdbZIPC1, AtbZIP9, AtbZIP10, AtbZIP25 and AtbZIP63, as seed sequences for BLAST searches using Phytozome and NCBI as described previously (Woods et al. 2011). Amino acid sequences were aligned using MUSCLE (Edgar 2004) before a manual alignment of amino acid sequences in Mesquite (Maddison and Maddison 2007). Unalignable regions were pruned from the analysis to minimize noise in the phylogenetic signal, and only 83 contiguous amino acids including the basic region and the leucine zipper domain were used in the final analysis. The Neighbor-Joining (Saitou and Nei 1987) and the Maximum Likelihood methods were used to infer the evolutionary history of the bZIP C-group across flowering plant diversification using five Arabidopsis bZIP proteins from the S-group as outgroups. For the Maximum Likelihood analysis, we used the Le and Gascuel (2008) model and obtained the initial tree(s) for the heuristic search automatically by applying the Neighbor-Joining algorithm to a matrix of pairwise distances estimated using the Jones-Taylor-Thornton model (Jones et al. 1992), and then selecting the topology with superior log likelihood value. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches (Felsenstein 1985). The evolutionary distances were computed using the Poisson correction method and are presented in number of amino acid substitutions per site. Evolutionary analyses were done using MEGA X (Kumar et al. 2018).
Analysis of Variance was conducted with the “Anova” function in R package “car” (Fox and Weisberg 2019) with type 3 sum of squares and LS Means to accommodate unbalanced designs.
Yeast two-hybrid (Y2H) assays
The full-length coding region of FT2 (TraesCS3A02G143100) was cloned from Chinese Spring as described in Li et al. (2015). The FT2 coding region was recombined into pDONRzeo using Life Technologies BP Clonase following the manufacturer’s protocol. FT2 in pDONRzeo was subsequently recombined into the pDEST32 and pDEST22 yeast destination vectors using Life Technologies LR Clonase II following the manufacturer’s protocol. Clones were verified by sequencing at each cloning step to ensure sequence integrity. All direct assays and library screens were performed using the MaV203 yeast strain as described in the ProQuest manual (Invitrogen). Before screening the cDNA libraries, we confirmed that FT2 was not auto-activated when used as prey or bait by testing it against the empty vector.
We screened a cDNA “photoperiod” library previously developed in B. distachyon (Cao et al. 2011). The photoperiod library was generated from shoots of two-week-old plants collected over 24 h grown under long (20 h light) and short day (8 h light) photoperiods (Cao et al. 2011). For the screen, transformants were selected on plates with synthetic minimal media lacking leucine (L) and tryptophan (W) and were replica plated on synthetic minimal media lacking L, W and uracil (U) to identify putative FT2 interactors. Yeast colony polymerase chain reaction (PCR) was done using the Phire polymerase following manufacturer’s instructions (Thermo Fisher).
To confirm the Y2H screen results, we cloned the full-length coding sequence from the wheat homolog of Bradi1g05480. The complete bZIPC1 coding region was amplified by PCR and cloned into the pJET vector (ThermoFisher), and from there amplified using bZIPC1 BP primers (Table S1) and recombined into pDONRzeo using Life Technologies BP clonase. The pDONRzeo vector containing the desired bZIPC1 coding region was recombined into pDEST22 and pDEST32 using Life Technologies LR Clonase II. Clones were verified by sequencing at each cloning step. We also cloned the wheat bZIPC1 paralogs bZIPC3 and bZIPC4, and genes CEN4, CEN5, FT2, FT3 and FT5 using the same cloning strategy described for bZIPC1 (Table S1), whereas bZIPC2 was synthesized and cloned. bZIPC1 and all genes used in the directed Y2H assays were confirmed to not auto-activate when either used as prey or bait. FT1 and CEN2 were previously cloned (Li et al. 2015) and transferred into pDEST22 and pDEST32.
Bimolecular fluorescence complementation (BiFC)
Bimolecular fluorescence complementation (BiFC, also known as split-YFP) assays were used to validate the interaction between FT2 and bZIPC1 in wheat protoplasts using the methods described in Li et al. (2021). Briefly, the coding regions (including the stop codon) of FT-A2 (allele A10) and bZIPC-A1 were transferred from pDONRzeo to the modified Gateway-compatible vectors UBIpro:N-YFP-GW and UBIpro:C-YFP-GW by recombination reactions (UBIpro = maize UBIQUITIN promoter; N-YFP = YFP-N-terminal fragment; C-YFP = YFP-C-terminal fragment). The resulting vectors UBIpro:N-YFP:FT-A2 and UBIpro:C-YFP:bZIPC-A1 were simultaneously transfected into wheat protoplasts, that were prepared, transfected, and visualized as described in Shan et al. (2014). An interaction between FT-A2 and bZIPC1 would bring together the two non-fluorescent C- and N-fragments of the YELLOW FLUORESCENT PROTEIN (YFP) resulting in the formation of the complete fluorescent protein.
Spatial Transcriptomics
We used the Molecular Cartography™ technology from Resolve BioSciences combinatorial single-molecule fluorescence in-situ hybridization according to the manufacturer’s instructions.
Tissue preparation and hybridization Wheat shoot apical meristems were collected from the vegetative to the floret primordia stage. The samples were immediately fixed in 4% PFA after harvest, dehydrated, and embedded in paraffin following the user guide for sample preparation from Resolve BioSciences. Sections from the central plane of the developing spikes (10 µm-thick) were placed within the capture areas of Resolve BioSciences slides. The slides were dried overnight at 37 °C, followed by a 10-min bake at 50 °C to enhance section adhesion. The sections were then deparaffinized, permeabilized, and refixed according to the user guide. After complete dehydration, the sections were mounted using SlowFade-Gold Antifade reagent, covered with a thin glass coverslip, and sent to Resolve BioSciences on dry ice for analysis.
Upon arrival, tissue sections were washed twice in 1 × PBS for 2 min, followed by 1-min washing in 50% and 70% ethanol at room temperature. Ethanol was removed by aspiration and DST1 buffer was added. This was followed by tissue priming for 30 min at 37 °C followed by a 48-h hybridization using probes specific for the target genes. Samples were washed to remove excess probes and fluorescently tagged in a two-step color development process. After imaging, fluorescent signals were removed during decolorization. Color development, imaging, and decolorization were repeated for multiple cycles to generate a unique combinatorial code for each target gene.
Probe design Probes were designed using Resolve BioSciences’ proprietary design algorithm and gene annotations from Chinese Spring RefSeq v1.1. To identify potential off-target sites, searches were confined to the coding regions. Each target sequence underwent a single scan for all k-mers, favoring regions with rare k-mers as seeds for full probe design. A probe candidate was generated by extending a seed sequence until reaching a certain target stability. After these initial screens, probes were aligned with the background transcriptome, and probes with stable off-target hits were discarded. From the pool of accepted probes, the final set was composed by selecting the highest scoring probes.
Specifically, for each of the selected tetraploid wheat genes, we choose the homoeolog expressed at higher levels in a Kronos transcriptome including different stages of spike development (VanGessel et al. 2022): bZIPC-A1 (TraesCS5A02G440400), bZIPC-B3 (TraesCS5B02G059200), bZIPC-A4 (TraesCS6A02G154600) and FT-A2 (TraesCS3A02G143100). bZIPC2 (TraesCS1A02G329900 and TraesCS1B02G343500) was excluded from this study because it is primarily expressed during grain development (Choulet et al. 2014) and was not detected in the Kronos spike transcriptome study (VanGessel et al. 2022).
We also provided Resolve BioSciences the sequences of the homoeologs of the selected genes, and they excluded them in their quality control for specificity performed against all the coding sequences of the wheat genome (Ref Seq v1.1). Therefore, probes are not genome-specific and can detect both homoeologs for each gene. While probe design was successful for bZIPC1, bZIPC3, and bZIPC4 (Catalogue Nos. PP4XE, PQ4XF, and PR4XG, respectively) specific probes for FT2 could not be designed due to its close similarity with FT1. However, following the masking of FT-A1 and FT-B1 sequences from the reference genome, Resolve BioSciences generated specific probes that differentiate FT2 and FT1 from other wheat genes (Catalogue No. PGGXX, all these probes are part of kit number K7128). Although these probes can potentially detect both genes, the absence of FT1 expression in the Kronos developing spike (VanGessel et al. 2022) ensures that the signals presented for this tissue are specific to FT2. Based on a comparison with the spike RNAseq data the FT-A2 probes seem to be less efficient than the bZIPC1 probes.
Imaging Samples were imaged by Resolve BioSciences using a Zeiss Celldiscoverer 7 with a 50 × Plan Apochromat water immersion objective having a numerical aperture (NA) of 1.2 and a 0.5 × magnification changer, resulting in a final magnification of 25 × . Standard CD7 LED excitation light sources, filters, and dichroic mirrors were used in conjunction with customized emission filters optimized for detecting specific signals. The excitation time per image was fixed at 1000 ms for each channel and 1 ms for calcofluor white. A z-stack was captured at each region with a z-slice spacing according to the Nyquist-Shannon sampling theorem. A custom CD7 CMOS camera (Zeiss Axiocam Mono 712, 3.45 μm pixel size) was employed. For each region, a z-stack was acquired for each fluorescent color (two colors) in each imaging round. A total of 8 imaging rounds were performed for each position, resulting in 32 z-stacks per region. The entirely automated imaging process per round, which included water immersion generation and precise relocation of regions for imaging in all three dimensions, was implemented through a custom Python script utilizing the scripting API of the Zeiss ZEN software (Open application development). Imaging for the cell wall-specific stain, Calcofluor White, was conducted after the primary 8-round imaging.
In the initial image processing steps, all images underwent background fluorescence correction. An iterative filtering process adjusted thresholds for factors like absolute brightness, local background, and background of the periphery, all based on 3D image volume. For signal segmentation and decoding, the raw data images were aligned using transformation matrices derived from the feature point clouds. Final image analysis was performed in ImageJ using the Polylux tool plugin to examine specific Molecular Cartography™ signals.
Haplotype analysis and marker development
We performed a haplotype analysis using the exome capture data from 55 tetraploid and hexaploid wheat accessions from the genotyping project “2017_WheatCAP_UCD” (https://wheat.triticeaetoolbox.org/downloads/download-vcf.pl) available in the Wheat T3 database (Blake et al. 2016). Using the 242 SNPs detected in a 1.3 Mb region on chromosome 5B flanking bZIPC-B1 (CS RefSeq v1.1 615,695,210 to 617,038,639), we performed a cluster analysis using R functions ‘dist’ and ‘hclust’ (method = ‘ward.D2’) (R Core Team 2021).
We used the available exome capture sequence from Berkut and Patwin-515HP (Wheat/T3) to develop KASP markers for the bZIPC-B1 SNPs. We also created KASP markers for the induced mutant alleles (Table S1).
Results
Identification of FT2 interactors by Y2H screening
To identify proteins that interact with FT2, we performed Y2H screens using the wheat Chinese Spring FT-A2 protein (TraesCS3A02G143100) as bait and a B. distachyon photoperiod cDNA library developed by harvesting whole plants across a diurnal cycle in long and short photoperiods (Cao et al. 2011) as prey. In the three screens performed against this library (transformation efficiencies of 4.5 × 105, 4.6 × 106 and 4.1 × 106), we detected 121 positive colonies corresponding to 26 different genes (Table S2). Among them, the gene represented by the largest number of colonies (67) was Bradi1g35230 (annotated as a kinectin-related protein with motor activity). The second most abundant gene was Bradi1g05480, which was detected in 29 colonies. This gene was annotated as an uncharacterized bZIP transcription factor. We decided to prioritize this bZIP for functional analyses because it was the only interactor detected in all three Y2H screens and also because of the known protein–protein interactions between FT1 and other bZIP proteins (Li et al. 2015). The interactors detected in the Y2H screens and their functional annotation are presented in Table S2.
Phylogenetic analyses of bZIPC-like genes
The large family of bZIP domain transcription factors includes 13 subfamilies (A-L and S) (Guedes Corrêa et al. 2008; Peviani et al. 2016). Reciprocal BLAST searches of Bradi1g05480 between B. distachyon and Arabidopsis did not reveal a clear one-to-one orthologous relationship but were sufficient to place Bradi1g05480 within the C-group (Fig. 1), which includes Arabidopsis genes bZIP9 (At5g24800), bZIP10 (At4g02640), bZIP25 (At3g54620), and bZIP63 (At5g28770). To reflect Bradi1g05480’s phylogenetic relationship within the C-group of this large and diverse family of bZIP transcription factors, we designated this gene as bZIPC1 (Table 1).
Phylogenetic analysis of C-group and S-group bZIP proteins. The tree was constructed using the Neighbor Joining method using an alignment of 83 amino acids (Fig. S1). Bootstrap values are indicated at the branching points. Bar indicates substitutions per site. Genes discussed in the manuscript are labeled in bold font. At = Arabidopsis thaliana, Os = Oryza sativa, Zm = Zea mays, Cl = Chasmanthium laxum, Sb = Sorghum bicolor, Si = Setaria italica, Hv = Hordeum vulgare, Ta = Triticum aestivum, Bd = Brachypodium distachyon, Pl = Pharus latifolius, Ja = Joinvillea ascendens
To understand the evolutionary history within the bZIP C-group and to identify the closest proteins to Bradi1g05480 in other species, we selected 55 bZIP proteins from the C-group spanning flowering plant diversification and five Arabidopsis bZIP proteins from the close S-group as an outgroup (Fig. 1). Relationships among these C- and S-class bZIP proteins were estimated using both Neighbor-Joining (Fig. 1) and Maximum Likelihood (Fig. S1) methods on an alignment of 83 contiguous amino acids including the conserved basic region and leucine zipper (Fig. S1).
The bZIPC1 clade included grass species and the near grass relative Joinvillea (Fig. 1). The closest clade to bZIPC1, which we designate here as bZIPC2, also included grass species from the major grass subfamilies (Fig. 1). These results suggest that bZIPC1 and bZIPC2 genes are likely the result of a grass specific duplication, which is consistent with the known whole genome duplication event at the base of the grasses (Paterson et al. 2004). These two clades, together with the eudicot clade including the duplicated Arabidopsis bZIP10, bZIP25 and bZIP63 proteins, correspond to the possible orthologous group C1 in Guedes Corrêa et al. (2008).
Two additional clades within the bZIP-C group, designated here as bZIPC3 and bZIPC4, were more distantly related to the bZIPC1/bZIPC2 cluster (Fig. 1). The Arabidopsis C-group protein bZIP9 was included in the same clade as bZIPC4 from grasses, forming a group similar to the proposed orthologous group C3 in Guedes Corrêa et al. (2008). This result suggests that some of the diversification of the bZIP C-group occurred before the monocot—eudicot divide. The Neighbor-Joining (Fig. 1) and Maximum Likelihood (Fig. S1) trees were similar with the exception of the position of the bZIPC3 and bZIPC4 clades relative to the bZIPC1-bZIPC2 group (Figs. 1, S1). This is not an unexpected result since the nodes among these three groups were supported by low bootstrap values (< 30).
To generate a nomenclature for the wheat bZIP transcription factors that reflects their evolutionary relationships, we propose to incorporate the group name to the bZIP gene name and designate the wheat bZIP genes from the C-group as bZIPC1, bZIPC2, bZIPC3, and bZIPC4 (Table 1).
Interactions between bZIPC-like and FT-like proteins
The B. distachyon clones identified in the Y2H library were not full-length genes based on BLASTN and BLASTP comparisons. To validate the interaction with full-length bZIPC1, we conducted direct Y2H assays using the full-length sequence of wheat bZIPC1 protein (TraesCS5A02G440400, 392 amino acids). Since there are two different natural variants in FT-A2 known to impact SNS (D10 and A10) (Glenn et al. 2022), we tested both protein variants in the direct Y2H assays. We found that bZIPC1 can interact with both FT-A2 protein variants in yeast when used as either baits or preys, confirming the initial Y2H screen results (Table 2; Fig. 2a). The interaction between FT2 and bZIPC1 was validated using bimolecular fluorescence complementation (BiFC) in wheat protoplasts (Fig. 2e–g). We detected fluorescent signals in the nucleus and cytoplasm of the protoplasts transfected simultaneously with UBIpro:N-YFP:FT-A2 − UBIpro:C-YFP:bZIPC-A1 (Fig. 2e). No fluorescent signal was detected in the protoplasts transfected with the negative controls UBIpro:N-YFP:FT-A2 − UBIpro:C-YFP (Fig. 2f) and UBIpro:N-YFP − UBIpro:C-YFP:bZIPC-A1 (Fig. 2g).
Interactions between bZIPC-like and FT/CEN-like proteins. Yeast two-hybrid assays between a bZIPC1, b bZIPC2, c bZIPC3, d bZIPC4 and FT1, FT2-D10, FT2-A10 (bZIPC1 only), FT3, FT5, CEN2, CEN4, CEN5 and empty pDEST32 (auto-activation test). Left panel: SD medium lacking Leucine and Tryptophan (–L–W) to select for yeast transformants containing both bait and prey. Right panel: interaction on –L–W–U medium (no L, W, and Uracil). Dilution factors = 1, 1:10 and 1:100. e–g FT2 and bZIPC1 interactions using bimolecular fluorescence complementation (BiFC) in wheat protoplast. e Fluorescent signals detected in the nucleus and cytoplasm for UBIpro:N-YFP:FT-A2 − UBIpro:C-YFP:bZIPC-A1, f, g Only background noise signal was detected in negative controls UBIpro:N-YFP:FT-A2 − UBIpro:C-YFP (f) and UBIpro:N-YFP − UBIpro:C-YFP:bZIPC-A1 (g)
To explore if bZIPC1 can interact with other FT-like proteins, we performed pairwise Y2H assays between bZIPC1 and FT-A1 (TraesCS7A02G115400), FT-B3 (TraesCS1B02G351100), FT-B5 (TraesCS4B02G379100), CEN-B2 (TraesCS2B02G310700), CEN-A4 (TraesCS4A02G409200) and CEN-A5 (TraesCS5A02G128600). We found that in addition to FT2, bZIPC1 can interact with FT3 and weakly with FT5 (Table 2; Fig. 2).
We also tested if the bZIPC1 paralogs were able to interact with the same set of FT-like proteins in a set of Y2H pairwise tests (Fig. 2). bZIPC2 interacted strongly with FT5 and CEN2 and weakly with the other genes, excluding CEN5, which showed no detectable interaction (Fig. 2b). bZIPC3 only interacted with FT3 and CEN5 and weakly with FT5, whereas the bZIPC4 protein only interacted weakly with FT1 and CEN4 (Table 2; Fig. 2).
Expression of bZIPC1 and FT2 overlap in the developing spikelet
We took advantage of published RNAseq data in Chinese Spring from five different tissues at three developmental stages (Choulet et al. 2014) to evaluate the expression levels of the four different bZIPC genes. In CS, bZIPC1 transcripts for the three homoeologs (bZIPC-A1, bZIPC-B1, and bZIPC-D1) were detected across all tissues with the highest expression in the roots (Fig. S2a). The spike tissues, all collected after the initiation of the elongation phase (Zadoks scale Z32, Z39 and Z65 (Zadoks et al. 1974)), showed high and consistent expression levels of all three bZIPC1 homoeologs. bZIPC2 transcripts were detected only in the developing grains (Fig. S2b), whereas bZIPC3 transcript levels were low in the developing grains, but abundant in other tissues (Fig. S2c). bZIPC4 transcripts were detected in all tissues, with higher levels in the spike (particularly at the latest stage Z65) and stems (Fig. S2d).
To study the expression of bZIPC1 earlier in spike development, we used an RNAseq study in tetraploid wheat Kronos including samples collected at the vegetative (W1.0), double ridge (W2.0), glume primordia (W3.0), lemma primordia (W3.25), and floret primordia (W3.5) stages (VanGessel et al. 2022), with W values based on the Waddington spike developmental scale (Waddington et al. 1983). Overall expression was similar between the A and B genomes, with expression levels increasing from W1.0 to W3.0 and remaining constant through W3.5 (Fig. 3a). FT2 expression levels in the same RNAseq samples were extremely low in W1.0 but then increased significantly in W2.0 to W3.0 before stabilizing, with FT-A2 expressed at higher levels than FT-B2 (Fig. 3b). Thus, bZIPC1 and FT2 expression overlaps during the early stages of wheat spike development.
Overlapping expression of bZIPC1 and FT2 in the developing wheat spike. a Expression of bZIPC-A1 (TraesCS5A02G44044) and bZIPC-B1 (TraesCS5B02G444100) in tetraploid wheat Kronos across the early stages of spike development (n = 4). b Expression of FT-A2 and FT-B2 in tetraploid wheat Kronos across the early stages of spike development (n = 4). Waddington spike developmental scale (Waddington et al. 1983): W1.0 = vegetative stage, W2.0 = early double ridge stage, W3.0 = glume primordium present, W3.25 = lemma primordium present, W3.50 = floret primordium present
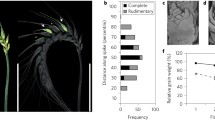
To explore if the bZIPC1 and FT2 genes were co-expressed in the same regions of the developing spike, we performed a spatial transcriptomics experiment. Figure 4a shows the distribution of bZIPC1, bZIPC3, and bZIPC4 transcripts in a developing spike of tetraploid wheat Kronos at the lemma primordia stage (W3.25). At this stage, bZIPC1 showed a strong and uniform distribution across the developing spike (Fig. 4a, b). bZIPC3 also showed a relative strong signal and a similar distribution, except for a reduced presence in the distal region of the spikelet meristems (Fig. 4a). Finally, bZIPC4 showed a lower signal localized in the central part of the spike (Fig. 4a). The different distribution of the three bZIPC genes suggests good specificity of the hybridization probes.
Spatial distribution of bZIPC and FT2 genes in the wheat developing spike. Kronos spike at the lemma primordia stage (W3.25). a bZIPC1 (red), bZIPC3 (yellow) and bZIPC4 (green). b bZIPC1 (dark green) and FT2 (yellow). c Detail of (b) showing co-localization of bZIPC1 and FT2 in the IM. Cell walls stained with calcofluor are shown in gray in (a, b) and in red in (c). IM = inflorescence meristem, SM = spikelet meristem, GP = glume primordium, LP = lemma primordium. Scale = 100 μm (color figure online)
The signal of FT2 was localized mainly at the base and along the central part of the developing spike reaching the IM (Fig. 4b). Therefore, FT2 transcripts overlapped with bZIPC1 across its distribution, and the two genes were co-localized in the IM (Fig. 4c), where the transition to the terminal spikelet is close to occurring (usually at W3.5). These results support the hypothesis that the physical interaction between bZIPC1 and FT2 may be biologically relevant.
Loss-of-function mutations in bZIPC1 are associated with reduced SNS
To determine the function of bZIPC1, we combined loss-of-function mutations in the two homoeologs of this gene in tetraploid wheat Kronos. We identified one Kronos mutant line with a loss-of-function mutation in the A genome homoeolog bZIPC-A1 (TraesCS5A02G440400) and three in the B genome homoeolog bZIPC-B1 (TraesCS5B02G444100). The mutation in bZIPC-A1 (CS RefSeq v1.1, chromosome 5A position 621,763,602) in the Kronos mutant K3308 results in a premature stop codon at position 97 (Q97*) in the first exon. In bZIPC-B1, we identified two independent mutants with identical mutations (K2991 and K3532). This mutation is located on chromosome 5B position 616,654,824 at the acceptor splicing site at the end of intron one (Fig. 5a). Sequencing of the bZIPC-B1 mRNA products in mutants K2991 and K3532 revealed that the second exon (94 bp) was spliced out, generating a shift in the reading frame and a premature stop codon (Fig. S3). We also identified a truncation mutation in Kronos mutant line K2038 at chromosome 5B position 616,655,991 that results in a premature stop codon (W117*).
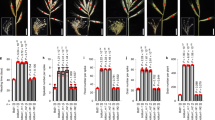
Characterization of F2 bZIPC1 mutants for SNS, and flowering time in a growth chamber. a Selected mutations for bZIPC-A1 and bZIPC-B1 in Kronos. b Spike morphology of WT and mutant sister lines for bzipc1-1, bzipc1-2 and bzipc1-3. Scale bar = 2 cm c Spikelet number (No.)/spike (SNS). d Days to heading. e Leaf number at heading time. c–e Different letters above bars indicate significant differences in Tukey tests (P < 0.05). n = 5–9
We designed KASP markers for each of the EMS-induced mutations (Table S1) to trace them during subsequent experiments. We made crosses between the plants carrying the A genome mutation with those carrying the three independent B genome mutants. In the three F2 progenies, we selected plants homozygous for the different mutant combinations using the KASP markers (Table S1). We designated the three combined mutants as bzipc1-1 (K3308 + K3532), bzipc1-2 (K3308 + K2991), and bzipc1-3 (K3308 + K2038, Fig. 5a).
All three double mutants showed highly significant reductions in SNS (P < 0.001) compared to their wildtype sister lines (average reduction 6.7 spikelets / spike across the three mutants, Table S3), but the individual spikelets showed no obvious differences (Fig. 5b, c). Significant reductions were also observed in a greenhouse experiment using F2 plants (average 4.4 spikelets / spike, Table S3) and in a growth chamber experiment using more advanced F3 plants (average 6.9 spikelets / spike, Fig. S4, Table S4). These last two experiments included only the bzipc1-1 and bzipc1-2 combined mutants and sister wildtype lines due to seed limitations. Among the single mutants, bzipc-A1-1, bzipc-B1-1 and bzipc-B1-2 showed modest but significant reductions in SNS relative to the wildtype (Fig. 5c).
In the same three experiments, we found no consistent significant differences in days to heading (DTH) or leaf number at heading (LN) between the mutants and their corresponding wildtype sister lines. These results suggest that bZIPC1 has a limited effect on the timing of the transition between the vegetative and reproductive phase stages and the duration of the elongation phase (Fig. 5d, e).
Natural variation in bZIPC1
The significant differences in SNS observed in the bzipc1 mutants motivated us to look at natural variation in both bZIPC1 homoeologs in a set of 6 tetraploid and 49 hexaploid wheat sequenced by exome capture (Fig. 6). We found two non-synonymous SNPs in bZIPC-B1 (TraesCS5B02G444100) but none in bZIPC-A1 (TraesCS5A02G440400) or bZIPC-D1 (TraesCS5D02G447500) in hexaploid wheat, so we focused our haplotype analysis in a 1.3 Mb region on chromosome 5B flanking bZIPC-B1 (CS RefSeq v1.1 615,695,210 to 617,038,639).
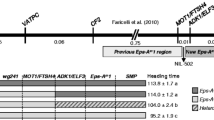
Haplotypes in the bZIPC1 region. Cluster analysis based on 242 SNPs detected in chromosome 5B (CS RefSeq v1.1 615,695,210 to 617,038,639 bp) in exome capture data extracted from T3/Wheat (https://wheat.triticeaetoolbox.org/). Blue arrows indicate spring lines from haplotypes H2 and H3 used as parental lines in crosses with Berkut (H1). Red arrows indicate winter lines used as parental lines in the soft red winter mapping population (DeWitt et al. 2021). Clustering scale calculated from method “ward D2” (color figure online)
A cluster analysis based on the 242 SNPs detected in this region (Table S5) revealed four haplotypes designated here as H1 to H4 (Fig. 6). The H1 haplotype was the most frequent (72.7%) among these accessions and was more closely related to H2 than to the other two haplotypes. Haplotypes H3 and H4 were related to each other, but since H4 included only a single tetraploid accession (280–1-Yr15) used for the introgression of Yr15 from T. turgidum subsp. dicoccoides (Yaniv et al. 2015), we did not characterize it further. We identified three independent historical recombination events among haplotypes within this 1.34 Mb region in varieties 26R61, LA95135, Kronos and Gredho (summarized in Table S5).
We then compared the four haplotypes for polymorphisms within the bZIPC-B1 coding region and identified one synonymous SNP at position 616,652,860 that differentiates H3 from all other three haplotypes, and two missense SNPs at positions 616,654,272 (N151K) and 616,654,229 (V166M) that differentiate the H1 haplotype (Table S6). The bZIPC-B1 linked amino acids K151 and M166 (KM henceforth) were found only in the H1 haplotypes, whereas the amino acid combination N151 and V166 (NV henceforth) were found in the other three haplotypes, bZIPC-A1, bZIPC-D1, and Hordeum vulgare, suggesting that NV is the ancestral state and KM the derived state. In addition, while we observed variation at position 166 (Table S6), the N151 amino acid was conserved in B. distachyon, rice, Setaria, and Panicum supporting the previous hypothesis.
The H1 haplotype was detected in the genomes of tetraploid wheat Svevo (Maccaferri et al. 2019) and Kronos (https://opendata.earlham.ac.uk/opendata/data/Triticum_turgidum/EI/v1/) and PanGenome hexaploid varieties Chinese Spring, Lancer, SY Mattis, Stanley, Jagger, and CDC Landmark (Walkowiak et al. 2020). A comparison of the bZIPC-B1 promoter region including 1500 bp upstream of the start codon revealed no polymorphisms among these diverse accessions, suggesting that H1 may be a relatively recent haplotype. A comparison of the bZIPC-B1 promoter region (1500 bp) between the H1 and H2 haplotypes (ArinaLrFor, Julius, Cadenza, Paragon, and Clair) revealed 9 SNPs and 4 indels, whereas a comparison between the H1 and H3 haplotypes (Norin61, Spelt, Mace, and Rubigus) revealed 19 SNPs and two indels, confirming the closer relationship between the H1 and H2 haplotypes relative to H3.
To explore the variation of the bZIPC-B1 haplotypes during the evolution of the polyploid wheat species, we screened a collection of 63 T. turgidum subsp. dicoccoides, 77 T. turgidum subsp. dicoccon, and 303 T. turgidum subsp. durum with KASP markers for the N151K and V166M SNPs (primers bZIPC1-4229 and bZIPC1-4272, Table S1). We found that 35% of the T. turgidum subsp. dicoccoides accessions and 4% of the T. turgidum subsp. dicoccon accessions have the derived KM allele (Tables 3, S7). For the T. turgidum subsp. durum, we split the group into old landraces and more modern cultivars and breeding lines. Of the 175 landraces, we found that 37% had the derived KM allele while of the 128 cultivars and breeding lines, 65% carried the KM allele (Tables 3, S7), indicating an increased frequency of the derived allele in the modern cultivated durum wheats.
A similar analysis of exome capture data from 482 T. aestivum subsp. aestivum accessions (He et al. 2019) revealed that 71% of the accessions have the derived bZIPC-B1 KM allele and only 29% the ancestral haplotype (Tables 3, S7). Taken together, these results suggest positive selection for the derived KM allele in modern wheat breeding programs. To explore this hypothesis further, we tested the effect of the bZIPC-B1 KM allele in four segregating populations.
Effect of bZIPC1 alleles on grain yield components
The haplotype analysis revealed that the group of varieties with the H1 haplotype included the spring wheat variety Berkut, which is the central parent in the spring wheat Nested Association Mapping (NAM) population (Blake et al. 2019). We previously characterized populations generated from crosses between Berkut (H1) and spring common wheat accessions Patwin-515HP (H3, Table S8), RSI5 (H3, Table S9), and Dharwar Dry (H2, Table S10) for SNS and other grain yield components in multiples locations and under two watering regimes (Zhang et al. 2018).
These populations were previously genotyped with the Illumina 90 K SNP chip (Jordan et al. 2015) and among these SNPs, we identified IWB56221 (RefSeq v1.1 5B 616,652,623) as the closest diagnostic marker for the H1 haplotype (H1 = T, H2, H3 and H4 = C). Using this marker, we performed t-tests between the two alleles for the different traits in the different environments (Table 4).
The comparison between the H1 and H3 haplotypes in the crosses Berkut × Patwin-515HP and Berkut × RSI5 showed that the H1 haplotype (= KM bZIPC-B1 allele) was associated with significant increases in the number of total and fertile spikelets per spike (3.1–6.1%), grain number per spike (11.0–11.5% increase), and grain weight per spike (10.6–11.9%, Table 5) relative to H3 (NV) in multiple locations. In the cross Berkut × Dharwar Dry, the H1 haplotype was also associated with a significant increase in grain number per spike (7.3%) and grain weight per spike (10.6%, Table 4) relative to the H2 haplotype (NV). Although the number of total and fertile spikelets per spike was not significantly different in this population, it showed the same trend as in the other two populations, with higher values in H1 (1.1–1.7%) relative to H2.
We also explored the effect of the H1 haplotype relative to H3 in the winter wheat population LA95135 (H1) × SS-MPV57 (H3) evaluated in five different environments in North Carolina, USA (DeWitt et al. 2021). In addition to bZIPC-B1, this population segregates for another four genes affecting SNS including PPD-D1, RHT-D1, FT-A2 (Glenn et al. 2022), and WAPO-A1 (Kuzay et al. 2019). We inferred the bZIPC-B1 genotype from the consensus of two SNPs flanking this gene at 616,367,516 and 616,955,782, and used this information together with the genotypes of the other four genes in a factorial ANOVA including the five genes as factors, all gene interactions, and environments as blocks (Table 5, raw data in Table S11).
The combined ANOVA across the five environments explained 44% of the variation in SNS and revealed highly significant differences in SNS for all five genes including bZIPC-B1 (P < 0.0001, Table 5). PPD-D1 and WAPO-A1 showed the strongest effects, whereas bZIPC-B1 had the weakest effect, with the Berkut allele showing on average a 1.4% increase in SNS (Table 5). The differences in SNS for bZIPC-B1 were significant in three out of the five environments when the ANOVAs were performed separately by environment. None of the two-way interactions involving bZIPC-B1 with the other four genes were significant, but the interaction between bZIPC-B1 and FT-A2 was the closest to significance (P = 0.0916, Table 5). When the same population was analyzed only for bZIPC-B1 and FT-A2, the interaction was highly significant (P = 0.0004, Fig. S5a, b), with the FT-A2 A10—bZIPC-B1 H1 allele combination showing the highest SNS.
We also analyzed the effect of bZIPC-B1 on DTH, fertility, GNS, and GWS in the same LA95135 × SS-MPV57 population. We found non-significant differences for DTH or fertility, but we observed significant increases of 3.07% in GNS (P = 0.0203) and 3.15% in GWS (P = 0.0094) associated with the H1 haplotype (LA95135) relative to the H3 haplotype (SS-MPV57, Table 5). The highest GWS was observed among the RILs combining the bZIPC-B1 H1 and FT-A2 A10 alleles (Fig. S5c).
Taken together, these results indicate that the derived bZIPC-B1 KM allele (H1 haplotype) is associated with beneficial effects on SNS, GNS, and GWS relative to the ancestral NV allele, providing a possible explanation for the rapid frequency increase of the derived KM allele during wheat domestication and improvement (Table 3).
Discussion
bZIP transcription factors are a large and functionally diversified family
The basic leucine zipper (bZIP) transcription factors (TF) are a large and diverse family present in eukaryotes from Saccharomyces cerevisiae to Homo sapiens (Deppmann et al. 2006). They are characterized by two conserved domains. The first one is a DNA-binding domain rich in basic amino acids, which is followed by a leucine zipper consisting of several heptad repeats of hydrophobic amino acids. This last region favors the formation of homodimers or heterodimers, resulting in multiple combinations with unique effects on transcriptional regulation (Deppmann et al. 2006). In plants, the bZIP transcription factors have a variety of roles ranging from hormonal responses, light signaling, nitrogen/carbon metabolism, biotic and abiotic stress, flower induction, development, and seed storage and maturation (Agarwal et al. 2019; Alonso et al. 2009; Alves et al. 2013; Baena-González et al. 2007; Choi et al. 2000; Fujita et al. 2005; Frank et al. 2018; Jakoby et al. 2002).
The plant bZIP transcription factors have diversified into 13 subfamilies (A-L and S) (Guedes Corrêa et al. 2008). The bZIP proteins from the C-group, which is the focus of this study, preferentially form heterodimers with other members of the C-group as well as with members of the S1-group (Ehlert et al. 2006). Known as the “C/S1 network”, this network has been shown to be involved in amino acid metabolism, stress response, and energy homeostasis (Alonso et al. 2009; Dröge-Laser and Weiste 2018; Feng et al. 2021). The C-group bZIP genes in Arabidopsis (bZIP9, bZIP10, bZIP25 and bZIP63) participate in a wide range of functions. For example, bZIP63 has been shown to regulate the starvation response (Mair et al. 2015) and to play an important role in fine-tuning of ABA-mediated, sugar-dependent abiotic stress responses (Matiolli et al. 2011), whereas bZIP10 was found to modulate basal defense and cell death (Kaminaka et al. 2006) and to work with glutathione to induce various heat shock proteins (Kumar and Chattopadhyay 2018).
Evolutionary and functional characterization of bZIPC genes in grasses
In grasses, the bZIP proteins of the C-group form a family of four paralogous clades, with groups bZIPC1 and bZIPC2 more closely related to each other than to the bZIPC3 and bZIPC4 groups (Fig. 1). The bZIPC1 clade includes previously studied proteins such as wheat SPA HETERODIMERIZING PROTEIN (TaSHP) (Boudet et al. 2019), barley BLZ1 (Vicente-Carbajosa et al. 1998) and maize OHP1 (Pysh et al. 1993), whereas the bZIPC2 clade includes the wheat STORAGE PROTEIN ACTIVATOR (TaSPA = bZIPC2) (Albani et al. 1997), barley BLZ2 (Oñate et al. 1999), and maize OPAQUE2 proteins (Pysh et al. 1993). The bZIPC3 and bZIPC4 subgroups have been less characterized.
TaSPA (= bZIPC2) was shown to be involved in the regulation of grain storage proteins (Albani et al. 1997) and in starch accumulation (Guo et al. 2020). This protein is orthologous to barley BLZ2, which interacts with BLZ1 (= bZIPC1) (Oñate et al. 1999). Both BLZ1 and BLZ2 genes are involved in the regulation of seed storage proteins (Vicente-Carbajosa et al. 1998). The orthologs of bZIPC2 in maize OPAQUE2 (O2) (Pysh et al. 1993) and rice RISBZ1 (OsbZIP58) (Kawakatsu et al. 2009) were initially identified as positive regulators of grain storage protein. Similarly, the orthologs of the bZIPC1 protein in barley (BLZ1) and maize (OHP1 and OHP1b) have been shown to bind to the promoters of the genes encoding storage proteins promoting their activation (Vicente-Carbajosa et al. 1998; Zhang et al. 2015). TaSHP, however, has been suggested to encode a repressor of glutenin storage protein synthesis in wheat (Boudet et al. 2019). These results suggest that bZIP proteins from the C-group can act both as transcriptional activators or repressors of storage proteins.
In addition to their role in the regulation of grass storage proteins, bZIPC1 and bZIPC2 genes have been shown to have additional functions. The rice ortholog of bZIPC1, OsbZIP33 (Os03g58250), is involved in the resistance of abiotic stresses via ABA-dependent stress signal transduction, with transgenic rice plants overexpressing OsbZIP33 showing significantly higher survival rates than their wildtype counterparts after dehydration treatment (Chen et al. 2015). The significant reduction in SNS detected in the bzipc1 wheat mutants in this study demonstrates a previously unreported role of bZIPC1 in the regulation of spike development.
The grass bZIPC3 and bZIPC4 genes, which are more distantly related to bZIPC1 and bZIPC2 (Fig. 1), have been less characterized. The three wheat bZIPC3 homoeologs have been shown to be upregulated by cold (Tian et al. 2022), whereas bZIPC-A4 (TraesCS6A02G154600 = Traes_6AS_F1CEB89EE.2) was upregulated after drought and heat treatments in wheat seedlings (Liu et al. 2015). Given the high level of expression of bZIPC3 and bZIPC4 genes in the wheat developing spike (Fig. 4), it would be interesting to determine if their mutants also show variation in spike-related traits.
In summary, the previous studies suggest some degree of sub-functionalization within the bZIP C-group clade. This sub-functionalization is reflected in their different expression profiles (Fig. 4) and in our Y2H results (Table 2), which showed that each of the four wheat bZIPC proteins can interact with different sets of proteins encoded by FT-like and CEN-like genes.
bZIPC1 interacts with FT2 and regulates SNS
Our Y2H and BiFC experiments showed that the FT2 and bZIPC1 proteins can physically interact with each other in both yeast and wheat protoplasts. Two indirect sources of evidence suggest that this interaction is biologically relevant. First, bZIPC1 and FT-A2 are both expressed in the developing spike at the same stages and are co-localized in the IM (Fig. 4b, c). However, it is important to note that if FT2 proteins are mobile as FT1, they might additionally be present outside the regions where transcripts were detected. In addition, we observed a genetic interaction between the bZIPC-B1 and FT-A2 natural variants on SNS (Fig. S5), which may be related to the observed physical interaction between the proteins encoded by these two genes.
In spite of its sequence similarity with FT1, FT2 shows some unique characteristics that set it apart from other FT-like genes. FT2 is the only FT-like gene in wheat that is expressed directly in the developing spike (Shaw et al. 2019; VanGessel et al. 2022). It is also the only one that encodes a protein that does not interact with any of the known bZIP proteins of the A-group (FDL2, FDL6, and FDL15) or the known 14-3-3 proteins that interact with the other FT-like proteins (Li et al. 2015).
These unique characteristics motivated us to look for FT2 protein interactors and led to the discovery of bZIPC1 as an FT2 interactor. We initially hypothesized that knock-out mutants in bZIPC1 would limit the ability of FT2 to accelerate the formation of the terminal spikelet and would result in an increase in SNS as in the ft2 mutants. However, all three bzipc1 mutants in tetraploid wheat showed a significant decrease in SNS (Fig. 5), negating our initial hypothesis. We discuss below three alternative hypotheses that may explain the reduced SNS in the bzipc1 mutant.
Based on the observed physical and genetic interactions between FT2 and bZIPC1 our favored hypothesis is that bZIPC1 interaction with FT2 may result in a non-functional protein complex that competes with other FT2 interactors required for FT2 function. Under this scenario, the bzipc1 mutant could reduce competition with an interactor required for FT2 function, resulting in increased activity and reduced SNS. However, since bZIPC1 can also interact with FT3 and weakly with FT5 (Fig. 2; Table 2), we cannot rule out alternative hypotheses in which the bzipc1 effect on SNS is mediated by FT3 and/or FT5. Finally, bZIPC1 may impact SNS by its effects on stress or nutrition pathways. This hypothesis is based on the known role of Arabidopsis bZIP genes from the C-group on the regulation of energy metabolism (Frank et al. 2018; Matiolli et al. 2011). Under starvation, the C/S1 network (Ehlert et al. 2006) reprograms metabolic gene expression to support survival by controlling plant growth, development and stress responses through fine tuning carbon and nitrogen responses (Dröge-Laser and Weiste 2018). Since wheat plants affected by abiotic stresses or reduced nitrogen show reduced SNS (Frank and Bauer 1982; Frank et al. 1987; Maas and Grieve 1990), it will be interesting to investigate if wheat bZIPC genes are involved in these responses.
Natural variation in bZIPC1 and potential applications to plant breeding
Analysis of natural variation in the bZIPC-B1 chromosome region revealed three major haplotypes (H1-H3), with the H1 haplotype associated with increases in SNS or GNS in four wheat RIL populations evaluated in multiple environments (Tables 4, 5). The frequency of the H1 haplotype was higher in the durum landraces (37%) than in the ancestral domesticated Emmer (4%), and increased again in cultivated durum (65%) and common wheat (71%, Table 3). The historical increase in the H1 haplotype frequency and its favorable effect on SNS suggest that this haplotype has been favored by positive selection in wheat breeding programs. The current frequencies also indicate that there is still a significant number of common and durum wheat varieties that can potentially benefit from the incorporation of the H1 haplotype.
The bZIPC-B1 protein in the H1 haplotype differs from other haplotypes by two missense SNPs that result in two linked amino acid changes (KM in H1 and NV in the other haplotypes). Since loss-of-function mutations in this gene are associated with variation in SNS, we hypothesize that these amino acid changes can be the cause of the observed natural differences in SNS. However, a more precise genetic map of this trait will be required to rule out the possibility that other genes linked to bZIPC-B1 contributed to the differences in SNS and the changes in allele frequency.
The two amino acid changes in the bZIPC-B1 protein may also affect its interactions with other proteins, and may explain the significant genetic interaction for SNS detected between bZIPC-B1 and FT-A2 in the RIL population LA95135 × SS-MPV57 (Fig. S5). In this population, the effect of bZIPC-B1 on SNS was significant in the presence of the FT-A2 A10 allele but not in the presence of the D10 allele. Similarly, the effect of FT2 on SNS was significant in the presence of the bZIPC-B1 H1 haplotype but not in the presence of the NV allele (Fig. S5). This epistatic interaction may be associated with the simultaneous and rapid increase of the FT2 A10 (60–80%, Glenn et al. 2022) and bZIPC-B1 H1 haplotype frequencies in modern common wheat cultivars, and suggests that simultaneous selection for these two alleles may be a useful breeding strategy to increase SNS.
In addition to its favorable effect on SNS, the H1 haplotype showed significantly higher GNS and GWS than the H2 and H3 haplotypes in multiple locations and in both spring and winter wheat. This is an encouraging result because it suggests that increases in grain number were not offset by a decrease in kernel weight. The interaction between bZIPC-B1 and FT-A2 for GWS was not significant but the trend was the same as for SNS, with the largest increase in GWS detected in the RILs combining the FT-A2 A10 and bZIPC-B1 H1 alleles (Fig. S5c). The magnitude of the bZIPC-B1 H1 haplotype effect on GWS and its interactions with FT-A2 varied across environments, similar to what has been reported previously for other wheat genes affecting SNS such as FT-A2 (Glenn et al. 2022) and WAPO-A1 (Kuzay et al. 2019, 2022). Therefore, before deployment of these alleles into breeding programs, it would be prudent to test their effects in local genetic backgrounds and environments.
Fortunately, the favorable H1 haplotype in bZIPC-B1 and the A10 allele in FT-A2 are frequent in the wheat germplasm, which together with the haplotype analysis presented here, will facilitate the evaluation of the effect of these two alleles in breeding programs that have access to genotypic information. The diagnostic molecular markers developed in this study for the beneficial bZIPC-B1 allele together with the markers previously developed for the FT-A2 alleles (Glenn et al. 2022) are useful tools to evaluate the distribution and the effect of these alleles in a breeding program, and to accelerate their deployment. The sequences of these markers are publicly available to facilitate the deployment of these potentially beneficial alleles.
Data availability
All data generated or analyzed during this study are included in the manuscript and supporting files. Kronos mutants are available from the authors upon request without any restrictions for use and from the Germplasm Resources Unit (GRU) at the John Innes Centre. Images for the spike sections used in the Spatial Transcriptomics study and the hybridization coordinates for the four genes presented in this study are available at https://dubcovskylab.ucdavis.edu/content/spatial-transcriptomic.
References
Agarwal P, Baranwal VK, Khurana P (2019) Genome-wide analysis of bZIP transcription factors in wheat and functional characterization of a TabZIP under abiotic stress. Sci Rep 9:1–18. https://doi.org/10.1038/s41598-019-40659-7
Albani D, Hammond-Kosack MCU, Smith C et al (1997) The wheat transcriptional activator SPA: a seed-specific bZIP protein that recognizes the GCN4-like motif in the bifactorial endosperm box of prolamin genes. Plant Cell 9:171–184. https://doi.org/10.1105/tpc.9.2.171
Alonso R, Oñate-Sánchez L, Weltmeier F et al (2009) A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 21:1747–1761. https://doi.org/10.1105/tpc.108.062968
Alves MS, Dadalto SP, Gonçalves AB et al (2013) Plant bZIP transcription factors responsive to pathogens: a review. Int J Mol Sci 14:7815–7828. https://doi.org/10.3390/ijms14047815
Baena-González E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448:938–942. https://doi.org/10.1038/nature06069
Ballerini ES, Kramer EM (2011) In the light of evolution: a reevaluation of conservation in the CO-FT regulon and its role in photoperiodic regulation of flowering time. Front Plant Sci 2:81. https://doi.org/10.3389/fpls.2011.00081
Blake V, Birkett C, Matthews DE et al (2016) The Triticeae Toolbox: combining phenotype and genotype data to advance small-grains breeding. Plant Genome 9:plantgenome2014.12.0099. https://doi.org/10.3835/PLANTGENOME2014.12.0099
Blake NK, Pumphrey M, Glover K et al (2019) Registration of the Triticeae-CAP spring wheat nested association mapping population. J Plant Regist 13:294–297. https://doi.org/10.3198/jpr2018.07.0052crmp
Boden SA, Cavanagh C, Cullis BR et al (2015) Ppd-1 is a key regulator of inflorescence architecture and paired spikelet development in wheat. Nat Plants 1:14016. https://doi.org/10.1038/nplants.2014.16
Boudet J, Merlino M, Plessis A et al (2019) The bZIP transcription factor SPA Heterodimerizing Protein represses glutenin synthesis in Triticum aestivum. Plant J 97:858–871. https://doi.org/10.1111/tpj.14163
Cao S, Siriwardana CL, Kumimoto RW, Holt BF (2011) Construction of high quality Gateway™ entry libraries and their application to yeast two-hybrid for the monocot model plant Brachypodium distachyon. BMC Biotechnol 11:53. https://doi.org/10.1186/1472-6750-11-53
Chardon F, Damerval C (2005) Phylogenomic analysis of the PEBP gene family in cereals. J Mol Evol 61:579–590. https://doi.org/10.1007/s00239-004-0179-4
Chen H, Dai XJ, Gu ZY (2015) OsbZIP33 is an ABA-dependent enhancer of drought tolerance in rice. Crop Sci 55:1673–1685. https://doi.org/10.2135/CROPSCI2014.10.0697
Choi HI, Hong JH, Ha JO et al (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275:1723–1730. https://doi.org/10.1074/JBC.275.3.1723
Choulet F, Alberti A, Theil S et al (2014) Structural and functional partitioning of bread wheat chromosome 3B. Science 345:1249721. https://doi.org/10.1126/science.1249721
Corbesier L, Vincent C, Jang S et al (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316:1030–1033. https://doi.org/10.1126/SCIENCE.1141752
Deppmann CD, Alvania RS, Taparowsky EJ (2006) Cross-Species annotation of basic leucine zipper factor interactions: Insight into the evolution of closed interaction networks. Mol Biol Evol 23:1480–1492. https://doi.org/10.1093/molbev/msl022
DeWitt N, Guedira M, Lauer E et al (2021) Characterizing the oligogenic architecture of plant growth phenotypes informs genomic selection approaches in a common wheat population. BMC Genom 22:402. https://doi.org/10.1186/s12864-021-07574-6
Distelfeld A, Li C, Dubcovsky J (2009) Regulation of flowering in temperate cereals. Curr Opin Plant Biol 12:178–184. https://doi.org/10.1016/j.pbi.2008.12.010
Dobrovolskaya O, Pont C, Sibout R et al (2014) FRIZZY PANICLE drives supernumerary spikelets in bread wheat. Plant Physiol 167:189–199. https://doi.org/10.1104/PP.114.250043
Dröge-Laser W, Weiste C (2018) The C/S1 bZIP network: a regulatory hub orchestrating plant energy homeostasis. Trends Plant Sci 23:422–433. https://doi.org/10.1016/J.TPLANTS.2018.02.003
Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform 5:113. https://doi.org/10.1186/1471-2105-5-113
Ehlert A, Weltmeier F, Wang X et al (2006) Two-hybrid protein-protein interaction analysis in Arabidopsis protoplasts: establishment of a heterodimerization map of group C and group S bZIP transcription factors. Plant J 46:890–900. https://doi.org/10.1111/j.1365-313X.2006.02731.x
Faure S, Higgins J, Turner A, Laurie DA (2007) The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare). Genetics 176:599. https://doi.org/10.1534/GENETICS.106.069500
Felsenstein J (1985) Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39:783–791. https://doi.org/10.2307/2408678
Feng Y, Wang Y, Zhang G et al (2021) Group-C/S1 bZIP heterodimers regulate MdIPT5b to negatively modulate drought tolerance in apple species. Plant J 107:399–417. https://doi.org/10.1111/tpj.15296
Fox J, Weisberg S (2019) An R companion to applied regression, 3rd edn. Sage, Thousand Oaks
Frank AB, Bauer A (1982) Effect of temperature and fertilizer N on apex development in spring wheat. Agron J 74:504–509. https://doi.org/10.2134/agronj1982.00021962007400030024x
Frank AB, Bauer A, Black AL (1987) Effects of air temperature and water stress on apex development in spring wheat. Crop Sci 27:113–116. https://doi.org/10.2135/cropsci1987.0011183x002700010028x
Frank A, Matiolli CC, Viana AJC et al (2018) Circadian entrainment in Arabidopsis by the sugar-responsive transcription factor bZIP63. Curr Biol 28:2597–2606. https://doi.org/10.1016/J.CUB.2018.05.092
Fujita Y, Fujita M, Satoh R et al (2005) AREB1 is a transcription activator of novel ABRE-Dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17:3470–3488. https://doi.org/10.1105/tpc.105.035659
Gauley A, Boden SC (2021) Stepwise increases in FT1 expression regulate seasonal progression of flowering in wheat (Triticum aestivum). New Phytol 229:1163–1176. https://doi.org/10.1111/nph.16910
Glenn P, Zhang J, Brown-Guedira G et al (2022) Identification and characterization of a natural polymorphism in FT-A2 associated with increased number of grains per spike in wheat. Theor Appl Genet 135:679–692. https://doi.org/10.1007/s00122-021-03992-y
Guedes Corrêa LG, Riaño-Pachón DM, Guerra Schrago C et al (2008) The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PLoS ONE 3:e2944. https://doi.org/10.1371/journal.pone.0002944
Guo D, Hou Q, Zhang R et al (2020) Over-expressing TaSPA-B reduces prolamin and starch accumulation in wheat (Triticum aestivum L.) grains. Int J Mol Sci 21:3257. https://doi.org/10.3390/IJMS21093257
Hai L, Guo H, Wagner C et al (2008) Genomic regions for yield and yield parameters in Chinese winter wheat (Triticum aestivum L.) genotypes tested under varying environments correspond to QTL in widely different wheat materials. Plant Sci 175:226–232. https://doi.org/10.1016/J.PLANTSCI.2008.03.006
Halliwell J, Borrill P, Gordon A et al (2016) Systematic Investigation of FLOWERING LOCUS T-like Poaceae gene families identifies the short-day expressed flowering pathway gene TaFT3 in wheat (Triticum aestivum L.). Front Plant Sci 7:857. https://doi.org/10.3389/fpls.2016.00857
He F, Pasam R, Shi F et al (2019) Exome sequencing highlights the role of wild-relative introgression in shaping the adaptive landscape of the wheat genome. Nat Genet 51:896–904. https://doi.org/10.1038/s41588-019-0382-2
Higgins JA, Bailey PC, Da L (2010) Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS ONE 5:e10065. https://doi.org/10.1371/journal.pone.0010065
Jakoby M, Weisshaar B, Dröge-Laser W et al (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7:106–111. https://doi.org/10.1016/S1360-1385(01)02223-3
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. https://doi.org/10.1093/bioinformatics/8.3.275
Jordan KW, Wang S, Lun Y et al (2015) A haplotype map of allohexaploid wheat reveals distinct patterns of selection on homoeologous genomes. Genome Biol 16:48. https://doi.org/10.1186/s13059-015-0606-4
Kaminaka H, Näke C, Epple P et al (2006) bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J 25:4400–4411. https://doi.org/10.1038/sj.emboj.7601312
Kawakatsu T, Yamamoto MP, Touno SM et al (2009) Compensation and interaction between RISBZ1 and RPBF during grain filling in rice. Plant J 59:908–920. https://doi.org/10.1111/j.1365-313X.2009.03925.x
Kikuchi R, Kawahigashi H, Ando T et al (2009) Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiol 149:1341–1353. https://doi.org/10.1104/PP.108.132134
Krasileva KV, Vasquez-Gross HA, Howell T et al (2017) Uncovering hidden variation in polyploid wheat. Proc Natl Acad Sci USA 114:E913–E921. https://doi.org/10.1073/pnas.1619268114
Kumar D, Chattopadhyay S (2018) Glutathione modulates the expression of heat shock proteins via the transcription factors BZIP10 and MYB21 in Arabidopsis. J Exp Bot 69:3729–3743. https://doi.org/10.1093/jxb/ery166
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Kuzay S, Xu Y, Zhang J et al (2019) Identification of a candidate gene for a QTL for spikelet number per spike on wheat chromosome arm 7AL by high-resolution genetic mapping. Theor Appl Genet 132:2689–2705. https://doi.org/10.1007/s00122-019-03382-5
Kuzay S, Lin H, Li C et al (2022) WAPO-A1 is the causal gene of the 7AL QTL for spikelet number per spike in wheat. PLoS Genet 18:e1009747. https://doi.org/10.1371/JOURNAL.PGEN.1009747
Le SQ, Gascuel O (2008) An improved general amino acid replacement matrix. Mol Biol Evol 25:1307–1320. https://doi.org/10.1093/molbev/msn067
Li C, Dubcovsky J (2008) Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J 55:543–554. https://doi.org/10.1111/J.1365-313X.2008.03526.X
Li C, Lin H, Dubcovsky J (2015) Factorial combinations of protein interactions generate a multiplicity of florigen activation complexes in wheat and barley. Plant J 84:70. https://doi.org/10.1111/TPJ.12960
Li K, Debernardi JM, Li C, Lin H, Zhang C, Dubcovsky J (2021) Interactions between SQUAMOSA and SVP MADS-box proteins regulate meristem transitions during wheat spike development. Plant Cell 33:3621–3644. https://doi.org/10.1093/plcell/koab243
Liu C, Mao B, Ou S et al (2014) OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol Biol 84:19–36. https://doi.org/10.1007/S11103-013-0115-3
Liu Z, Xin M, Qin J et al (2015) Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol 15:152. https://doi.org/10.1186/S12870-015-0511-8
Lv B, Nitcher R, Han X et al (2014) Characterization of Flowering Locus T1 (FT1) gene in Brachypodium and wheat. PLoS ONE 9:e494171. https://doi.org/10.1371/JOURNAL.PONE.0094171
Maas EV, Grieve CM (1990) Spike and leaf development of sal-stressed wheat. Crop Sci 30:1309–1313. https://doi.org/10.2135/cropsci1990.0011183x003000060031x
Maccaferri M, Harris NS, Twardziok SO et al (2019) Durum wheat genome highlights past domestication signatures and future improvement targets. Nat Genet 51:885–895. https://doi.org/10.1038/s41588-019-0381-3
Maddison WP (2007) Mesquite: a modular system for evolutionary analysis. Version 2.0: http://mesquiteproject.org
Mair A, Pedrotti L, Wurzinger B et al (2015) SnRK1-triggered switch of bZIP63 dimerization mediates the low-energy response in plants. Elife 4:e05828. https://doi.org/10.7554/eLife.05828.001
Matiolli CC, Tomaz JP, Duarte GT et al (2011) The Arabidopsis bZIP gene AtbZIP63 is a sensitive integrator of transient abscisic acid and glucose signals. Plant Physiol 157:692–705. https://doi.org/10.1104/pp.111.181743
Oñate L, Vicente-Carbajosa J, Lara P et al (1999) Barley BLZ2, a seed-specific bZIP protein that interacts with BLZ1 in vivo and activates transcription from the GCN4-like motif of B-hordein promoters in barley endosperm. J Biol Chem 274:9175–9182. https://doi.org/10.1074/jbc.274.14.9175
Paterson AH, Bowers JE, Chapman BA (2004) Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA 101:9903–9908. https://doi.org/10.1073/pnas.0307901101
Peviani A, Lastdrager J, Hanson J, Snel B (2016) The phylogeny of C/S1 bZIP transcription factors reveals a shared algal ancestry and the pre-angiosperm translational regulation of S1 transcripts. Sci Rep 6:30444. https://doi.org/10.1038/srep30444
Pin PA, Benlloch R, Bonnet D et al (2010) An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330:1397–1400. https://doi.org/10.1126/SCIENCE.1197004
Pysh LD, Aukerman MJ, Schmidt’ RJ, (1993) OHPI: A maize basic domain/leucine zipper protein that interacts with Opaque2. Plant Cell 5:227–236. https://doi.org/10.1105/tpc.5.2.227
R Core Team (2021) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/
Ray DK, Mueller ND, West PC, Foley JA (2013) Yield trends are insufficient to double global crop production by 2050. PLoS ONE 8:66428. https://doi.org/10.1371/journal.pone.0066428
Reynolds M, Foulkes J, Furbank R et al (2012) Achieving yield gains in wheat. Plant Cell Environ 35:1799–1823. https://doi.org/10.1111/j.1365-3040.2012.02588.x
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Shan Q, Wang Y, Li J, Gao C (2014) Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc 9:2395–2410. https://doi.org/10.1038/nprot.2014.157
Shaw LM, Turner AS, Herry L et al (2013) Mutant alleles of Photoperiod-1 in Wheat (Triticum aestivum L.) that confer a late flowering phenotype in long days. PLoS ONE 8:e79459. https://doi.org/10.1371/JOURNAL.PONE.0079459
Shaw LM, Lyu B, Turner R et al (2019) FLOWERING LOCUS T2 regulates spike development and fertility in temperate cereals. J Exp Bot 70:193–204. https://doi.org/10.1093/jxb/ery350
Tamaki S, Matsuo S, Hann LW et al (2007) Hd3a protein is a mobile flowering signal in rice. Science 316:1033–1036. https://doi.org/10.1126/SCIENCE.1141753
Taoka KI, Ohki I, Tsuji H et al (2011) 14–3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476:332–335. https://doi.org/10.1038/NATURE10272
Tian Y, Peng K, Lou G et al (2022) Transcriptome analysis of the winter wheat Dn1 in response to cold stress. BMC Plant Biol 22:277. https://doi.org/10.1186/S12870-022-03654-1
VanGessel C, Hamilton J, Tabbita F et al (2022) Transcriptional signatures of wheat inflorescence development. Sci Rep-Uk 12:17224. https://doi.org/10.1038/s41598-022-21571-z
Vicente-Carbajosa J, Oñ L, Lara P et al (1998) Barley BLZ1: a bZIP transcriptional activator that interacts with endosperm-specific gene promoters. Plant J 13:629–640. https://doi.org/10.1111/j.1365-313X.1998.00068.x
Waddington SR, Cartwright PM, Wall PC (1983) A quantitative scale of spike initial and pistil development in barley and wheat. Ann Bot 51:119–130. https://doi.org/10.1093/oxfordjournals.aob.a086434
Walkowiak S, Gao L, Monat C et al (2020) Multiple wheat genomes reveal global variation in modern breeding. Nature 588:277–283. https://doi.org/10.1038/s41586-020-2961-x
Wolde GM, Mascher M, Schnurbusch T (2019) Genetic modification of spikelet arrangement in wheat increases grain number without significantly affecting grain weight. Mol Genet Genom 294:457–468. https://doi.org/10.1007/S00438-018-1523-5
Woods D, Hope CL, Malcomber ST (2011) Phylogenomic analyses of the BARREN STALK1/LAX PANICLE1 (BA1/LAX1) genes and evidence for their roles during axillary meristem development. Mol Biol Evol 28:2147–2159. https://doi.org/10.1093/MOLBEV/MSR036
Woods D, Dong Y, Bouche F et al (2019) A florigen paralog is required for short-day vernalization in a pooid grass. Elife 8:e42153. https://doi.org/10.7554/ELIFE.42153
Yan L, Fu D, Li C et al (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Nat Acad Sci USA 103:19581–19586. https://doi.org/10.1073/pnas.0607142103
Yaniv E, Raats D, Ronin Y et al (2015) Evaluation of marker-assisted selection for the stripe rust resistance gene Yr15, introgressed from wild emmer wheat. Mol Breed 35:43. https://doi.org/10.1007/s11032-015-0238-0
Zadoks JC, Chang TT, Konzak CF (1974) A decimal code for the growth stages of cereals. Weed Res 14:415–421. https://doi.org/10.1111/J.1365-3180.1974.TB01084.X
Zhang Z, Yang J, Wu Y (2015) Transcriptional regulation of Zein gene expression in maize through the additive and synergistic action of opaque2, prolamine-box binding factor, and O2 heterodimerizing proteins. Plant Cell 27:1162–1172. https://doi.org/10.1105/tpc.15.00035
Zhang J, Gizaw SA, Eligio B et al (2018) Identification and validation of QTL for grain yield and plant water status under contrasting water treatments in fall-sown spring wheats. Theor Appl Genet 131:1741–1759. https://doi.org/10.1007/s00122-018-3111-9
Zikhali M, Wingen LU, Leverington-Waite M et al (2017) The identification of new candidate genes Triticum aestivum FLOWERING LOCUS T3–B1 (TaFT3-B1) and TARGET OF EAT1 (TaTOE1-B1) controlling the short-day photoperiod response in bread wheat. Plant Cell Environ 40:2678–2690. https://doi.org/10.1111/pce.13018
Funding
This project was supported by the Agriculture and Food Research Initiative Competitive Grant 2022-68013-36439 (WheatCAP) from the USDA National Institute of Food and Agriculture and by the Howard Hughes Medical Institute. DPW is Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation.
Author information
Authors and Affiliations
Contributions
PG collected and analyzed most of the experimental data. DPW performed the Y2H screen, identified the bZIPC1 interactor, contributed to data analyses and supervised PG and NO. NO contributed to the Y2H screen, JZ contributed natural variation data and statistical analyses and the bimolecular fluorescent complementation experiment. GG provided valuable intellectual and writing contributions. JD and DPW initiated and coordinated the project. JD obtained funding and administered the project, contributed data analyses, supervised JZ, PG and DPW, and was responsible for the final manuscript. All authors reviewed the manuscript and provided suggestions.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Communicated by Beat Keller.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Glenn, P., Woods, D.P., Zhang, J. et al. Wheat bZIPC1 interacts with FT2 and contributes to the regulation of spikelet number per spike. Theor Appl Genet 136, 237 (2023). https://doi.org/10.1007/s00122-023-04484-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00122-023-04484-x