Abstract
Key message
Sedimentation values and falling number in the last decades have helped maintain high baking quality despite rigorous selection for grain yield in wheat. Allelic combinations of major loci sustained the bread-making quality while improving grain yield. Glu-D1, Pinb-D1, and non-gluten proteins are associated with sedimentation values and falling number in European wheat.
Abstract
Zeleny sedimentation values (ZSV) and Hagberg-Perten falling number (HFN) are among the most important parameters that help determine the baking quality classes of wheat and, thus, influence the monetary benefits for growers. We used a published data set of 372 European wheat varieties evaluated in replicated field trials in multiple environments. ZSV and HFN traits hold a wide and significant genotypic variation and high broad-sense heritability. The genetic correlations revealed positive and significant associations of ZSV and HFN with each other, grain protein content (GPC) and grain hardness; however, they were all significantly negatively correlated with grain yield. Besides, GPC appeared to be the major predictor for ZSV and HFN. Our genome-wide association analyses based on high-quality SSR, SNP, and candidate gene markers revealed a strong quantitative genetic nature of ZSV and HFN by explaining their total genotypic variance as 41.49% and 38.06%, respectively. The association of known Glutenin (Glu-1) and Puroindoline (Pin-1) with ZSV provided positive analytic proof of our studies. We report novel candidate loci associated with globulins and albumins—the non-gluten monomeric proteins in wheat. In addition, predictive breeding analyses for ZSV and HFN suggest using genomic selection in the early stages of breeding programs with an average prediction accuracy of 81 and 59%, respectively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat producers and downstream value chains place a high premium on grain’s baking quality because it affects the end-user value and yields high monetary profits. Baking quality, therefore, becomes an important selection criterion in wheat breeding programs across the globe. In Germany, wheat varieties are released according to different quality classes, and several traits form the basis for determining a given class (Sortenliste 2021). Zeleny sedimentation values (ZSV) and Hagberg-Perten falling number (HFN) are among the most important parameters that help determine the quality class of a given wheat line. During registration trials, the lines are evaluated at two management intensity levels, i.e., without (level-1) and with (level-2) the application of plant growth regulators and fungicides. At level-1, measurements are conducted mainly for the agronomic and disease traits. On the other hand, data gathered at level-2 primarily form the basis for describing quality characteristics. Grain yield (GY) is measured at both intensity levels (Sortenliste 2021). It is known that nitrogen fertilizers strongly impact both grain yield and quality in wheat. However, given the Nitrates Directive (91/676/EEC)—a core legislation to reduce agriculture-based nitrate emissions into water bodies in the European Union—German Fertilizer Ordinance (Düngeverordnung) regulates nitrogen and phosphorous emissions into water bodies (Justiz 2017). Thus, the limited application of organic and inorganic fertilizers to reduce the risks of nutrient emissions warrant the exploitation of genetics and breeding approaches to enhance the genetic gain for grain quality traits for climate-smart agricultural practices. Wheat grain quality is primarily determined by its texture, protein content, and quality. Historically, wheat proteins are divided into four major fractions: glutenin, gliadin, globulins, and albumins (Osborne 1907). Glutenin and gliadin together form “gluten”—the largest natural macropolymer in wheat that affects wheat dough’s viscoelastic properties and the appearance and structure of flour-based products (Lukow et al. 1989; Payne 1987). Globulins and albumins, on the other hand, form the “non-gluten” fraction of wheat proteins, and, although present in minor fraction, their ratio and quality were reported to affect dough quality and flour processing (Gupta et al. 1992; Pence et al. 1954; Zhang et al. 2021).
The Zeleny sedimentation test determines the wheat gluten content and quality whereby the flour is mixed in a lactic acid solution that causes the gluten to expand and sediment. Larger ZSV (slower sedimentation) represents high gluten content, strength, and quality—a means for predicting the bread-making quality of flour (Zeleny 1962). Major high-molecular-weight (HMW) glutenin (Glu-1) loci, i.e., Glu-A1, -B1, and -D1, are present on the long arm of group-1 wheat chromosomes (Payne and Lawrence 1983). The low molecular weight (LMW) glutenin loci (Glu-3), on the other hand, are Glu-A3, -B3, and -D3 that are present on the short arm of group-1 homoeologous chromosomes (Singh and Shepherd 1988). The Glu-3 loci, consisting of several alleles, were reported to be linked with gliadin (Gli-1; Gli-A1, -B1, and -D1) loci (Brown and Flavell 1981). Variations in the haplotypes of gluten loci yield varying gluten content and qualities, consequently determining the overall grain quality. Würschum et al. (2016) recently reported gluten loci’s association with sedimentation values. From the major known loci, the puroindoline (Pin-1) genes—present at the Hardness locus—which mainly affects the grain texture (Morris 2002), were also associated with ZSV (Kristensen et al. 2018; Mohler et al. 2012; Würschum et al. 2016). In addition to Glu and Pin loci, peroxidases (PODs)—that are widely distributed in cereals—have also been associated with improved rheological properties of flour doughs, loaf volume, crumb structure, and overall bread making (Geng et al. 2019; van Oort et al. 2000; Zhou et al. 2021).
Hagberg-Perten falling number test indicates starch degrading enzyme α-amylase’s activity in wheat flour (Perten 1964). Higher α-amylase activity quickly breaks down the grain starch particles into glucose and maltose reducing the viscosity of the slurry (mix of wheat flour in distilled water). Consequently, a viscometer (stirrer or plunger) falls rapidly (time measured in seconds) through lesser viscous slurry which results in a lower HFN (shorter time). However, if the starch particles are intact, the viscometer falls slowly through the thick slurry resulting in high HFN. Low HFN (high α-amylase activity) is associated with pre-harvest spouting that imposes a significant negative impact on the grain quality resulting in flour that produces sticky and weaker doughs as well as smaller and deformed bread loaves.
Most grain quality traits harbor quantitative genetic control, i.e., their total genetic variance is controlled by the concerted action of large- as well as small-effect loci. In such a scenario, prediction of the total genetic value based on a large number of marker genotypes conferring both large and small effects on the trait becomes a method-of-choice (Meuwissen et al. 2001). Wheat grain quality traits, e.g., ZSV, HFN, flour water retention, and flour yield, are usually evaluated at the later stages of the breeding programs because, on the one hand, the kernel availability is usually scarce in earlier generations, and, on the other hand, most analyses are usually time- and cost-intensive and, thus, analyzing a large number of candidates becomes a virtual impossibility. Hence, quality data are gathered at the later breeding stages on the lines that show promising agronomic trait values and good disease resistance packages. Genome-wide prediction, by using phenotypic data collected at later stages on a relatively larger set of environments as a “training set,” can help select the promising lines for quality traits in early breeding cycles by predicting their genetic values. In addition, in line and hybrid breeding programs, genome-wide prediction of traits can help design the parental crosses with better genetic gain. Recent studies have shown the promise of genome-wide prediction of quality traits for genomic selection (Kristensen et al. 2018; Liu et al. 2016; Muqaddasi et al. 2020; Sandhu et al. 2021).
Gogna et al (2022) published several agronomic, disease, and quality traits’ phenotypic and genotypic data in the GABI panel of 372 registered European wheat varieties. In this study, we took advantage of the published data on ZSV and HFN plus further traits and analyzed them in more detail. We show the relationships and genetic trends of grain quality traits, especially with grain yield (GY). Moreover, the value of glutenin, puroindoline, and plant height loci is illustrated via constructing haplotypes to breed for sustainable or better GY. Our association analyses revealed the quantitative genetic nature of ZSV and HFN and identified known and novel putative loci annotated as globulins, albumins, and peroxide. These genes affect the quality profiles of wheat grain. We also performed genome-wide predictions for ZSV and HFN to evaluate the prospects of predictive breeding: the results point to a promising use of genomic selection for these traits to increase the genetic gain per unit of time and perhaps cost.
Material and methods
The phenotypic and genomic datasets used in this study were previously published (Gogna et al. 2022). Therefore, in the following, we have limited ourselves to introducing only the essential information on phenotyping and genotyping.
Field trials and phenotypic data collection
A European wheat panel (GABI) comprising 372 varieties (358 winter type; 14 spring type) was evaluated for two major grain quality traits viz. Zeleny sedimentation values (ZSV; ml) and Hagberg-Perten falling number (HFN; s). The phenotypic data of these two traits were gathered from eight and four environments, respectively. Each environment was considered as a location-by-year combination. The field trials were conducted using an alpha lattice design with two replications per environment. We did not observe conditions conducive to pre-harvest sprouting (e.g., heterogeneous plots, uneven ripening, and lodging) that can strongly influence HFN. A major reason is that data were gathered by keeping in view the German registration authority’s (Bundessortenamt) practices to obtain quality traits’ data from field trials where standard crop protection and plant growth regulator applications (intensity level-2) are executed: this reduces lodging and consequently impacts quality profiles, including HFN. A further detailed description of field trials, agronomic practices, climatic conditions, and calculation of the adjusted entry means per environment was given previously (Gogna et al. 2022; Zanke et al. 2014). Briefly, the phenotypic measurements for ZSV and HFN were carried out by the collaborating seed companies KWS Lochow GmbH and Syngenta Seeds GmbH by using sample volumes of 400 g grains per harvested field plot based on methods described by the International Association for Cereal Science and Technology (ICC) standard number 116/1 and 107/1, respectively.
Phenotypic data analyses
The consistency or stability among the environments for both traits was investigated by drawing environment-specific adjusted entry mean values as box-and-whisker plots. To check the general performance of a given trait across environments, we calculated the average correlation by performing Fisher’s \(z\) transformation, as described previously (Muqaddasi et al. 2020). Across-environment individual variance components of the genotype, environment, and corresponding residuals were computed based on a restricted maximum likelihood (REML) approach by employing a random model:
where \(y_{ij}\) is the phenotypic value (adjusted entry mean) of the \(i^{th}\) genotype in the \(j^{th}\) environment, \(\mu\) is the common intercept term, \(g_{i}\) is the effect of the \(i^{th}\) genotype, \(e_{j}\) is the effect of the \(j^{th}\) environment, and \(\varepsilon_{ij}\) is the corresponding residual term as \(\varepsilon \sim N\left( {0,I\sigma_{\varepsilon }^{2} } \right)\) with \(I\) and \(\sigma_{\varepsilon }^{2}\) being the identity matrix and residual variance, respectively. The broad-sense heritability (\(H^{2}\)) across environments was calculated as follows:
where \(\sigma_{g}^{2}\) and \(\sigma_{\varepsilon }^{2}\) denote the variance components of the genotype and residuals, respectively, and \(nE\) represents the number of environments. The best linear unbiased estimations (BLUEs) were computed accordingly by setting the genotype as the fixed effect and all other effects as random in Eq. 1.
Correlations, genetic trend, and determination of best-fit path analysis model
Since grain protein content (GPC; %), grain hardness (GH; %), and grain yield (GY; dt ha−1) are considered during the variety registration trials, we retrieved their phenotypic data (i.e., genotypic values or BLUEs) from a previously conducted study based on multiple environments (Muqaddasi et al. 2020). We calculated the genetic correlations among all five traits based on their across-environments BLUEs, as described previously (Muqaddasi et al. 2020).
To test the genetic trend or progress \((\updelta )\), we performed linear regression based on BLUEs calculated from Eq. 1 and the year-of-registration of only winter wheat varieties (n = 325) covering three decades starting from the year 1980 as \(\updelta =\mathrm{b}/{y}_{0}\), where \(\mathrm{b}\) and \({y}_{0}\) denote the regression slope and the mean of varieties released in 1980, respectively. In addition, we highlighted Rht-D1 alleles (i.e., tall, and short) in scatterplots to show their distribution across the years of registration.
Path analysis—an extension of multiple regression—allows determining the best-fit hypothesis to explain the relationships between the dependent and independent traits by comparing different models (Streiner 2005). We set GY, ZSV, and HFN as dependent traits and examined two different models as follows:
In model 1 (Eq. 3), all the possible relationships among the investigated traits were exploited. For example, GY was predicted by GPC, GH, ZSV, and HFN; ZSV by GPC, GH, and HFN; and HFN by GPC and GH. The indices, namely χ2 test, comparative fit index (CFI), Tucker–Lewis’ index (TLI), and standard root mean square residual (SRMR) were used to analyze the goodness of fit of the model as χ2 \(\left( P \right) > 0.05\), CFI ≥ 0.90, TLI ≥ 0.95, and SRMR ≤ 0.08 (Suhr 2008). In model 1, some trait relationships were observed to be non-significant which led us to develop model 2 (Eq. 4), in which only significant relationships from model 1 were fitted.
To calculate the effect of the independent traits on the dependent, let \(p\) be the path from one trait to another and T be the trait number. In the first step of model 2 (Eq. 4), GY was only affected by GPC; therefore, the direct (or total) effect for GY was calculated as \(GY_{{T_{5} }} \sim GPC_{{[p_{51} T_{1} ]}}\); whereas, ZSV and HFN were predicted by both GPC and GH. Based on the best-fit indices, model 2 was selected to calculate the effects of each independent trait on the dependent traits (for more details, see the Results section).
Genotyping and molecular data analyses
The whole wheat panel (n = 372) was first genotyped with 732 simple sequence repeat (SSR) markers that resulted in scorable 770 loci and 3176 alleles (Kollers et al. 2013). In addition, two state-of-the-art high-density single nucleotide polymorphic (SNP) marker arrays, namely, 90,000 Illumina iSelect (Wang et al. 2014) and 35,000 Axiom Affymetrix Breeders Array (Allen et al. 2017) were employed. The genetic (cM) for SSRs were retrieved from Sorrells et al. (2011) while the physical positions (bp) for SNP arrays were retrieved from Sun et al. (2020).
Since high-molecular-weight glutenin subunits (HMW-GS; Glu-1) and grain hardness puroindoline (Pin-1) genes are reported to have a significant impact on wheat end-use quality, we sequenced the whole panel to observe their allelic frequency and impact in European wheat. Each Glu-1 locus on chromosomes 1AL (Glu-A1), 1BL (Glu-B1), and 1DL (Glu-D1) consists of two tightly linked genes encoding x- and y-type HMW-GS. PCR-based candidate/functional markers were applied to distinguish the wheat varieties for null-, x-, and y-type glutenin subunits. For Glu-A1, as described in Liu et al. (2008), the functional marker UMN19 was used to distinguish between Ax-null and Ax2* subunits. For Glu-B1, two markers were used: one distinguished between Bx6 and Bx7 or Bx17 subunits (Schwarz et al. 2004), and the second distinguished between Bx7NE (normally expressed) and Bx7OE (overexpressed) subunits (Butow et al. 2003). For Glu-D1, UMN25 and UNM26 functional markers helped discriminate Dx2 + Dy12 and Dx5 + Dy10 subunits (Liu et al. 2008).
For Pin-1 loci, i.e., Pina-D1 and Pinb-D1, pyrosequencing technology was employed to genotype the whole panel for the frequently present alleles in European wheat, viz., Pina-D1a, Pina-D1b, Pinb-D1a, Pinb-D1b, Pinb-D1c, and Pinb-D1d. The details about pyrosequencing were given previously by Huang and Röder (2005). As Morris (2002) described, the presence to both Pin wild type alleles, i.e., Pina-D1a and Pinb-D1a, leads to soft endosperm texture, whereas loss-of-function mutations in any of the two loci lead to wheats with hard endosperm. We divided the whole panel based on soft and hard wheats as boxplots and analyzed the difference between the means of both categories via Welch’s two-sided t-test by assuming unequal variances at the confidence interval of 0.95.
Introduction of reduced height (Rht) genes resulted in significant improvement in GY in wheat (Hedden 2003) and, given the negative relationship between GY and quality traits, the whole panel was genotyped with Rht-D1 to observe its allelic effect on GY and grain quality traits (Muqaddasi et al. 2017).
For all candidate markers (i.e., Glu-D1, Rht-D1, and Pin-1), boxplots were drawn for the complete set of varieties (BLUEs) and those harboring reference and variant alleles. We used Welch’s two-sided t-test to observe if there existed significant differences between the means of varieties harboring different alleles.
Genome-wide association studies and candidate gene identification
We performed genome-wide association studies (GWAS) based on the BLUEs calculated across environments and molecular markers that passed the quality criteria of < 5% missing values and > 5% minor allele frequency. We combined the SSRs, SNP arrays, and functional-gene markers for the molecular markers set. Following Yu et al. (2006), a standard linear mixed-effect model was used to perform GWAS as:
where y is the column vector of BLUEs of each genotype calculated across environments, \(\mu\) is the common intercept, \(\beta ,{ }v,{ }u\) and \(\varepsilon\) are the vectors of marker, population structure (principal components), polygenic background, and residual error effects, respectively; X, P and Z are the corresponding design matrices. \(\mu ,\tau ,{ }\beta ,\) and v were assumed to be fixed while u and \(\varepsilon\) as random with \(u\sim N\left( {0,G\sigma_{u}^{2} } \right)\) and \(\varepsilon \sim N\left( {0,I\sigma_{\varepsilon }^{2} } \right)\). The variance-covariance genomic relationship matrix (G) was calculated based on the second solution, as described by VanRaden (2008).
To declare the marker-trait associations (MTA), we employed the Bonferroni correction \(\left( {\upalpha } \right)\) criterion to account for multiple testing as \({\upalpha } = - \log_{10} \left( {\frac{0.1}{p}} \right)\) where p is the number of quality markers used in the GWAS. Since Bonferroni’s correction amounted to \(- \log_{10} \left( P \right) = 5.34\), we used an exploratory threshold of \(- \log_{10} \left( P \right) = 3.5\) to identify the MTA. As described by Utz et al. (2000), the genotypic variance (\(p_{G}\)) explained by all the QTL was determined as \(p_{G} = \left( {\frac{{R_{{{\text{adj}}}}^{2} }}{{H^{2} }}} \right) \times 100\) where \(R_{adj}^{2}\) was calculated as \(R_{{{\text{adj}}}}^{2} = R^{2} - \left( {\frac{{z^{\prime}}}{{N - z^{\prime} - 1}}} \right)\left( {1 - R^{2} } \right)\) by fitting the MTA \(\left( {z^{\prime}} \right)\) in the order of their descending P values in a multiple linear regression model; R2, N and H2 denote the regression coefficient, number of observations, and the broad-sense heritability calculated in Eq. 2. The \(p_{G}\) explained by the individual MTA was accordingly calculated from their sum of squares.
The genetic gain per unit time and cost become co-extensive with the genotypic variance explained by the trait-associated markers. For example, if the quantitative trait loci (QTL) underlying a given trait explain a small proportion of genotypic variance, marker-assisted selection for these QTL becomes cost and time intensive. Hence, we selected the markers that explained \({p}_{G}\) of ≥ ~ 10% and designated as the QTL for the respective trait. We considered the markers with the highest \(-{\mathrm{log}}_{10}\left(P\right)\) value and \({p}_{G}\) from each QTL as “representative markers” for that QTL. The QTL were named based on accepted QTL naming conventions in wheat literature (Boden et al. 2023). The sequences harboring each SNP (MTA) were taken from the genotyping arrays and BLASTed onto the wheat’s reference sequence to determine their corresponding gene identifiers and human-readable descriptions. Also, the gene-start, -stop, and the SNP position (bp) were retrieved from Sun et al. (2020).
Genome-wide predictions
We evaluated the genome-wide prediction (GP) accuracies of ZSV and HFN by using five genomic selection (GS) models with different marker variance assumptions. The GS models were genomic best linear unbiased prediction (GBLUP), BayesA, BayesB, BayesC, and reproducing kernel Hilbert space regression (RKHSR) (Gianola and Van Kaam 2008; Habier et al. 2007; Meuwissen et al. 2001; Pérez and Los Campos 2014; VanRaden 2008). The implementation of GBLUP and RKHSR on the same panel has been reported previously (Muqaddasi et al. 2019). For Bayesian implementation (BayesA–C), we followed Pérez and Los Campos 2014 and implemented the same prior densities and default hyperparameters mentioned in the BGLR package.
We evaluated the accuracy \(\left({r}_{GP}\right)\) of all prediction models by using a fivefold cross-validation scenario. The varieties were randomly divided into five subsets: four of them were used as the training set to estimate the genetic values of the remaining test set. The accuracy of prediction was defined as Pearson’s product-moment correlation between the observed \(\left(y\right)\) and predicted \(\left(\widehat{y}\right)\) genetic values standardized by the square root of the broad-sense heritability, as \({r}_{GP}=\frac{cor\left(y, \widehat{y}\right)}{H}\). Since the cross-validation runs were repeated for 50 cycles, the mean and standard deviation values were calculated to show the performance of the individual GS model. Unless stated otherwise, all calculations were performed in software R (Team 2013) mainly by using packages lme4 (Bates et al. 2015), lavaan (Rosseel 2012), and rrBLUP (Endelman 2011).
Results
Phenotypic data analyses reveal consistent performance, significant genotypic variation, and stable genetic trend for sedimentation values and falling number
Grain quality traits, viz. Zeleny sedimentation values (ZSV) and Hagberg-Perten falling number (HFN) were assessed on a panel of 372 wheat varieties registered in European markets (Table S1). The field trials to collect data were performed in replicated trials in eight and four environments, respectively. Boxplots based on adjusted entry means drawn for individual environments showed consistent performance of both traits yielding positive average Pearson’s product-moment correlation \(({\overline{r} }_{\left(\mathrm{ZSV}\right)}=0.78;{\overline{r} }_{\left(\mathrm{HFN}\right)}=0.44)\) across all the environments (Fig. S1a,b). Grain protein content (GPC) and grain hardness (GH) are high-throughput traits that can be relatively easily scored non-destructively (e.g., via near-infrared reflectance; NIR) especially in an early generation where the kernel availability is usually scarce. We retrieved the genotypic values (BLUEs) for all 372 varieties from a previous study to evaluate if there existed any genetic correlation and trend. Grain yield (GY) is the principal parameter to select in breeding programs. Like GPC and GH, we retrieved the GY genotypic values calculated based on eight environments from a previous study to find its association with quality traits (Muqaddasi et al. 2020).
Our across years individual variance component analyses revealed wide genotypic variation that was significantly (P < 0.001) larger than zero for both ZSV and HFN, with their BLUEs approximating normal distribution (Table 1; Fig. 1a–e). Despite a significant environmental variance, the large genotypic variance translated into high broad-sense heritability estimates of 0.96 and 0.74 for ZSV and HFN, respectively (Table 1). Large genotypic variance coupled with high broad-sense heritability estimates point to, on one hand, a high selection advantage; and, on the other hand, a reasonable underlying allelic diversity that should favor reliable downstream analyses to evaluate promises of genomic approaches.
Phenotypic distribution and correlation of the investigated traits in a panel of 372 European wheat varieties. Distribution of (a) Zeleny sedimentation value (ZSV), (b) Hagberg-Perten falling number (HFN), (c) grain protein content (GPC), (d) grain hardness (GH), and (e) grain yield (GY); (f) Pearson’s product-moment correlation \(\left(r\right)\) among the investigated traits where *** and ** denote the significance of respective correlation at probability (P) of < 0.001 and < 0.01, respectively
Our regression analyses to evaluate the effect of year-of-registration on varieties revealed noteworthy results (Fig. 2a–e). For example, the genetic trend for GPC significantly (P < 0.001) decreased by 0.17% per annum (Fig. 2c) whereas GY significantly (P < 0.001) increased (0.36%; 0.321 dt ha−1y−1; Fig. 2e). Statistically non-significant (P > 0.05) genetic trends were observed for ZSV, HFN, and GH (Fig. 2a,b,d). In addition to the regression on the absolute trait-BLUEs, we performed the regression on the ratio calculated from ZSV and GPC: these values are indicative of the protein (gluten) quality and content simultaneously. For ZSV/GPC ratio—in contrast to GPC where a negative selection trend was observed—we observed a positive although non-significant genetic trend (Fig. 2e). This suggests that albeit the protein content has decreased, the breeders were able to develop varieties with good protein quality that consistently met baking requirements during registration trials.
Scatterplot of (a) Zeleny sedimentation value (ZSV), (b) Hagberg-Perten falling number, (c) grain protein content (GPC), (d) grain hardness, (e) grain yield, and (f) ratio of ZSV/GPC as a function of year-of-registration of 325 European winter wheat varieties covering three decades (1980–2009). Regression estimations, coefficient of determination (R2), significance (P) values, and increase per annum (δ) for traits are given in each sub-figure. Black and red dots represent the Rht-D1a and -D1b alleles, respectively, while the gray solid line represents the linear regression line
Correlation and path analysis uncover genetic relationships among quality traits and grain yield
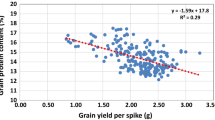
The genetic correlation among all five investigated traits based on their BLUEs revealed that all quality traits, viz. ZSV, HFN, GPC, and GH showed a negative correlation with GY. The most pronounced significant negative (− 0.76, P < 0.001) correlation was observed between GPC and GY (Fig. 1f). Among the grain quality traits, ZSV showed a higher negative and significant correlation with GPC and GH in comparison with HFN (Fig. 1f). HFN showed significant but lower correlations with all quality as well as GY—this points to a possible different genetic underpinning of ZSV and HFN. Besides, we observed genetic associations among the traits by regressing the quality trait BLUEs on GY: this also showed that (1) the most variance in GY is explained by GPC (R2 = 0.57) followed by ZSV (R2 = 0.26), and (2) there, nevertheless, exist varieties with above average values for each combination (Fig. S2a–d).
Our path analysis revealed underlying genetic relationships among the grain quality parameters and grain yield (Table S2, Fig. 3) with the most pronounced negative effect of GPC on GY, i.e., every unit increase in GPC resulted in 6.82 units decrease in GY (Fig. 3). In contrast to GY, GPC showed a direct positive effect on ZSV and HFN, i.e., every unit increase in the GPC increased the ZSV and HFN by 6.78 and 14.6 units, respectively. GH displayed positive effects on ZSV and HFN but with overall lower values than GPC. Interestingly, ZSV, HFN, and GH did not significantly affect GY. Interaction between ZSV and HFN was also non-significant (Table S2). These results further cement (1) the possibility of different genetic architecture and interaction of ZSV and HFN, especially concerning GPC, GH, and GY (Fig. 3), and (2) the interpretation of the above-mentioned genetic trend analyses where although the GY increased, except GPC, none of the other quality traits significantly changed.
Path diagram elucidating all possible relationships among the investigated traits, viz. Zeleny sedimentation value (ZSV), Hagberg-Perten falling number (HFN), grain protein content (GPC), grain hardness (GH), and grain yield (GY). Single-headed and dashed double-headed curved arrows indicate a trait’s direct effect on another and covariance between the GPC and GH, respectively. The numbers on the single-headed arrows denote the path-coefficients as positive and negative effect of one trait on another. T1–T5 and ns denote the trait numbers and non-significant relationships, respectively
GWAS identifies large-effect loci and putative candidate genes for sedimentation values and falling number
We elucidated the genetic control of ZSV and HFN based on 372 wheat varieties and genomic markers (Fig. 4). For ZSV, in total, we identified 59 marker-trait associations (MTA) representing four QTL, viz., QZsv.ipk-1A, QZsv.ipk-1B, QZsv.ipk-1D, and QZsv.ipk-5D (Tables 2, S3, Figs. 4, S3–S7). However, only seven MTA were identified for HFN, each on separate chromosomes, which signified two QTL with ≥ 10% of genotypic variance as QHfn.ipk-1A and QHfn.ipk-5B (Tables 2, S3, Figs. 4, S8, S9). Each QTL from ZSV and HFN was represented by the most significant MTA, explaining the largest genotypic variance. The total genotypic variance explained by all the MTA for ZSV and HFN amounted to 41.49% and 38.06%, respectively.
Summary of genome-wide association studies (GWAS) of Zeleny sedimentation values and Hagberg-Perten falling number in a panel of European wheat varieties. (a) Manhattan plots show the distribution of marker loci’s significance \({-\mathrm{log}}_{10}(P-\mathrm{value})\) along 21 wheat chromosomes. The dashed gray line indicates the significance threshold based on Bonferroni correction, while red continuous lines represent an exploratory significance threshold. Red dots represent the main-effect QTL’s representative markers for the respective traits. (b) Quantile-quantile plots show the distribution of observed versus expected (red dashed line) \({-\mathrm{log}}_{10}(P-\mathrm{value})\). n and p denote the number of varieties and the quality markers used in the GWAS analyses
The MTA BLASTed onto the wheat reference sequence resulted in identifying high-confidence genes. The functional annotations (human-readable descriptions) yielded highly plausible candidate genes. For example, a SNP AX-95236907 representing the ZSV-QTL QZsv.ipk-1A corresponded to the gene TraesCS1A02G317900 with functional annotation of Peroxidase—an enzyme that is known to influence the wheat quality (Table S3, Fig. S3). For QZsv.ipk-1D, the functional marker for the glutenin gene Glu-D1 (UMN25 and UMN26) that distinguished between Dx5 + DY10 and Dx2 + Dy12 subunits was significantly associated with the ZSV and explained 25.56% of the genotypic variance (Fig. S5). The QTL QZsv.ipk-1D was also represented by the marker BS00022768_51 (gene ID: TraesCS1D02G317300) that explained 26.05% of the genotypic variance with functional annotation of globulin—a non-gluten wheat protein that is associated with wheat quality (Table S3, Fig. S6). QZsv.ipk-5D, represented by BS00000020_51 (TraesCS5D02G004300), explained 11.39% of the genotypic variance and corresponded to Puroindoline-b (Pinb-D1)—a known gene at the Hardness locus responsible for controlling wheat grain texture (Morris 2002). For HFN, SNP AX-94430348 (TraesCS1A02G200400) representing the QHfn.ipk-1A explained 12.74% of the genotypic variance and corresponded to Albumin-2 protein—another non-gluten protein in wheat’s endosperm (Tables 2, S3, Figs. 4,S8).
Impact of Glu-1, Rht-D1, and Pin-1 loci on wheat quality traits and grain yield
The entire wheat panel was genotyped for major loci associated with grain quality and yield (Glu-1, Pin-1, and Rht-D1; Table S1, Fig. 5a–e), and several most abundant haplotypes were studied (Fig. 6).
Boxplots showing the allele-wise effect of major loci on (a) Zeleny sedimentation value, (b) Hagberg-Perten falling number, (c) grain protein content, (d) grain hardness, and (e) grain yield. The header and x-axis represent the gene names and corresponding alleles (subunits), respectively. The first boxplot shows the distribution of best linear unbiased estimations (BLUEs) of each trait. n denote the number of varieties in which the corresponding allele was observed. ***, **, *, and – denote the significance values of Welch's two-sided t-test at probability (P) values of < 0.001, < 0.01, < 0.05, and > 0.05 , respectively
Ten major haplotypes (allelic combinations) based on major loci for glutenins (Glu), plant height (Rht), and puroindolines (Pin), the number of varieties harboring the corresponding haplotype, and trait values based on best linear unbiased estimations (BLUEs) of varieties. The x- and y-type subunits (alleles) are colored for haplotype construction. Welch's two-sided t-test was used to observe if there existed significant differences between the means of haplotype-based trait values and total population (n = 372). α, β, γ, and – denote the significance value of the t-test at the probability (P) levels of < 0.001, < 0.01, < 0.05, and > 0.05, respectively. The trait columns (ZSV = Zeleny sedimentation value, HFN = Hagberg-Perten falling number, GPC = grain protein content, GH = grain hardness, and GY = grain yield) are colored as black = values significantly larger than the population mean, gray = values larger but insignificant than the population mean, and white = values lower than the population mean
For Glu-A1, we used the functional marker UMN19, which separated wheat varieties based on the presence of Ax-null (90.24%) and Ax2* (10.56%) subunits. Welch’s two-sided t-test revealed that the presence of Ax-null was associated with significantly (P < 0.05) higher values for HFN and GY (Fig. 5b,e); however, for ZSV, GPC, and GH, no significant differences existed (Fig. 5a,c,d).
Glu-B1, genotyped with two functional markers that distinguished between (1) Bx6 and Bx7 or Bx17, and (2) Bx7NE and Bx7OE subunits, revealed their frequencies as 29.91, 70.08, 93.82, and 6.18%, respectively. For the first marker, the presence of Bx7 or Bx17 subunit was associated with significantly higher values for ZSV and GH; however, the effect was negative for GY where the varieties harboring Bx7 or Bx17 subunit was associated with decreased (P < 0.05) values (Fig. 5e). For HFN and GPC, the difference between the means of varieties harboring these two subunits was non-significant (Fig. 5b,c). The difference between the means of varieties harboring Bx7NE and Bx7OE subunits was not significant for HFN, GPC, and GY (Fig. 5b,c,e); however, the presence of Bx7OE was associated with significantly higher values for ZSV and GH (Fig. 5a,d).
Glu-D1—genotyped with the functional markers UMN25 and UMN26—was used to differentiate varieties harboring Dx2 + Dy12 and Dx5 + Dy10 subunits that were present at the frequencies of 46.26 and 53.74%, respectively. Glu-D1 was observed as one of the most revealing loci: the presence of different subunits showed a highly significant impact on all quality traits as well as GY. For example, the presence of Dx5 + Dy10 subunits, which are known to impact quality traits positively, was associated with significantly (P < 0.001) higher values for all quality traits while (Fig. 5a–d). However—as could be deduced from the negative relationship between grain quality and GY—Dx5 + Dy10, in comparison with Dx2 + Dy12, was associated with significantly (P < 0.001) lower values for GY (Fig. 5e). The frequencies of these subunits in European varieties, nevertheless, points to the likelihood that breeders generally have been fine-tuning the grain quality traits and GY based on Glu-D1 in combination with other large-effect genes.
Rht-D1—a green revolution gene—is frequently used in European wheat breeding programs to tailor the plant height and improve lodging resistance and GY (Flintham et al. 1997). Thus, from the known negative correlation between GY and grain quality traits, it is expedient to study the effect of tall (Rht-D1a) and short (Rht-D1b) alleles on quality and GY. The difference between the means of varieties harboring tall (42.01%) and short (57.99%) alleles were non-significant for HFN and GH (Fig. 5b,d) but highly significant (P < 0.001) for ZSV, GPC, and GY (Fig. 5a,c,e): Rht-D1b presence was associated with reduced values for ZSV and GPC while increased GY.
Pina-D1—a gene present at the Hardness locus—was represented by wild-type Pina-D1a (92.43%) and null Pina-D1b (7.57%) alleles in the investigated varieties. However, the allelic influence on varieties was non-significant (Fig. 5). From allelic frequencies, it can also be safely inferred that the breeders in Europe have largely been adjusting the grain texture by exploiting the loci other than Pina-D1.
Pinb-D1 gene was genotyped for four widely present alleles in European varieties, viz., Pinb-D1a (13.35%), -D1b (42.90%), -D1c (11.08%), and -D1d (32.67%). Besides Glu-D1, Pinb-D1’s allelic influence was observed as most pronounced: the use of Pinb-D1b and -D1d to alter the grain texture in wheat varieties was evident from their frequencies (Fig. 5). The presence of Pinb-D1a (wild type) allele was associated with a significant decrease in all investigated quality traits whereas—except for HFN—the higher values were observed for mutant Pinb-D1c. As expected, wild-type Pinb-D1a and -D1c were associated with significantly higher and lower GY values, respectively. Moreover, the Pinb-D1b’s presence, compared to Pinb-D1a, was associated with significantly higher values for all quality traits while decreasing GY (Fig. 5e): This was less pronounced for Pinb-D1d where although the difference between varieties was significant for all quality traits but non-significant for GY (Fig. 6).
Of a total of 372, 350 varieties had complete information for the Pin-1 loci: 41 harbored soft while 349 hard Pin profiles (Table S1). Welch’s two-sided t-test revealed that hard wheats harbored significantly (P < 0.001) higher values for all the quality traits (Fig. 7a–d) whereas, as expected, we observed a significantly lower values for GY (Fig. 7e).
Impact of soft and hard wheats divided according to puroindoline (Pin) profiles on (a) Zeleny sedimentation value, (b) Hagberg-Perten falling number, (c) grain protein content, (d) grain hardness, and (e) grain yield. Soft wheats represent varieties harboring wild-type Pin alleles (Pina-D1a and Pinb-D1a) whereas hard wheats harbor Pin allelic variants for one or both Pina-D1 and Pinb-D1. For comparison, the first boxplot within every sub-figure represents the best linear unbiased estimations (BLUEs) across soft and hard wheats. n denote the number of varieties present in each category. P values denote the significance values of Welch's two-sided t-test
Genome-wide prediction accuracy suggests the efficient use of genome-wide selection in wheat breeding programs
We performed genome-wide predictions to assess the potential of genomics for predictive breeding measured as the correlation between the predicted and observed trait values standardized by the square root of heritability. The mean prediction accuracies resulting from the fivefold cross-validation settings of quality traits produced similar results across all five tested model scenarios, i.e., the GBLUP, Bayes-A, -B, -C, and RKHSR (Fig. 8a,b). It is worth noting that the prediction accuracy for ZSV was ~ 20% (~ 81%) higher than that of HFN (~ 59%): this is consistent with the theory, where prediction accuracies of the traits that are less complex tend to be higher as compared to those with highly complex genetic architecture. RKHSR—mainly used to observe if there exists epistatic interaction among loci—yielded results on par with other models suggesting that epistatic interactions may not be pervasive for investigated traits.
Accuracy of the genome-wide prediction (GP) for (a) Zeleny sedimentation values and (b) Hagberg-Perten falling number based on five genomic selection models viz. genomic best linear unbiased prediction (GBLUP), BayesA–C, and reproducing kernel Hilbert space regression (RKHSR) evaluated by 50 random runs of fivefold cross-validation cycles. Symbols \(\mu\) and \(\sigma\) denote the corresponding model's mean prediction accuracy and standard deviation. n and \(\widehat{p}\) (markers including the unmapped) denote the number of varieties and the quality markers used in the GP analyses
Discussion
Wheat grain’s baking quality class determines the monetary benefits for growers and, thus, in addition to improved grain yield and disease resistance, fine-tuning and improving the quality trait profiles remains a major target for breeding programs. Most grain quality parameters are influenced by crop management (esp. fertilizer application) practices and the environment. However, there exists variation among wheat varieties that could be ascribed to the underlying genetic factors. Because of the German Fertilizer Ordinance’s obligatory regulations for reduced fertilizer applications (Justiz 2017), exploiting the genetic approaches become vital for sustainable grain quality and climate-resilient agriculture at large. To this end, in this study, we evaluated the phenotypic variation, genetic control, genetic trends, and promises of genomics-assisted breeding for two important grain quality parameters, viz. Zeleny sedimentation values (ZSV) and Hagberg–Perten falling number (HFN) in a set of 372 wheat varieties that were released in previous decades and evaluated over several environments.
Genotypic variance, genetic trends, and association among grain quality and yield reveal stable breeding for baking quality
For both ZSV and HFN, we observed a large genotypic variance and broad-sense heritability estimates—this is consistent with the previous reports (Kristensen et al. 2018; Reif et al. 2011; Würschum et al. 2016). Albeit the genotypic variance for both traits was significant, a high environmental and residual variance was observed, especially for HFN—a trait that is influenced by pre- and post-grain physiological maturity environmental conditions. Most recently, Fradgley et al. (2022) reported a large genotype-by-environment interaction for HFN in the UK winter wheat.
The effect of year-of-registration on traits under consideration provides an overall genetic trend or selection progress. Our analyses revealed, over the years, a significant increase in GY (+ 0.36%) while GPC decreased (– 0.17%) significantly. Cormier et al. (2013) observed 0.33 dt ha−1 increase in GY during a similar period, predominantly in French varieties. Ahlemeyer and Friedt (2011) reported a 0.34 dt ha−1 yearly GY increase for German varieties. More recently, Laidig et al. (2017) reported an increase in grain yield while a decrease in GPC in German variety trials. Interestingly, no statistically significant trend was observed for ZSV, HFN, and GH pointing to the likelihood that although selection pressure on GY took a toll on GPC, the traits concerning protein/gluten quality (ZSV), α-amylase activity (HFN), and grain texture (GH)—which control baking and milling quality—have sustainably been bred. While assessing long-term breeding progress in German variety trials, Laidig et al. (2017) reported positive trends for all ZSV, HFN, and GH. Our findings are also in line with Würschum et al. (2016), who reported that the sedimentation values seem to counterbalance the significant negative genetic trend for GPC while staying at consistent levels and, thus, offsetting the potential monetary losses for growers. The genetic trend for ZSV/GPC ratios further strengthens this inference (Fig. 2e).
Wheat quality traits’ negative association with grain yield is historically well known (Malloch and Newton 1934; Neatby and McCalla 1938; Oury et al. 2003; Shewry 2009). We also observed that quality traits, viz., ZSV, HFN, GPC, and GH bore a significant negative correlation with GY. ZSV and GPC showed a more pronounced negative association with GY than HFN and GH. Albeit relatively weaker, HFN showed significant correlations with the other four traits. ZSV, on the other hand, showed stronger correlations with GPC, GH, and GY. This led to an inference of a possible dissimilar genetic control of ZSV and HFN. Path analysis, nonetheless, showed no direct genetic effects of ZSV and HFN on GY. Here, GPC was the only trait having direct relationship with all other traits, including the GY. This result is usable in practical breeding programs where GPC can be used as a selection criterion for other complementary (ZSV, HFN, and GH) and antagonistic (GY) traits. In relatively early breeding generations, GPC as a trait can be scored noninvasively with high throughput via near-infrared reflectance (NIR) techniques. In addition to GPC, NIR analyses usually provide GH and grain moisture content. Thus, GPC and GH values after calibration at ~ 14% grain moisture could be suitable proxy traits to select ZSV and HFN. Also, since early generation lines are usually evaluated in micro or observation (e.g., two-row) plots and yield estimation is not possible, GPC (lower values) could be employed as one of the negative selection traits for GY when breeding for low-quality (German C class) wheat is undesirable. However, this comment on negative selection for C-class should be taken carefully since this could cause selection bias in practical breeding. For example, the stands are homogeneous if the breeding nursery is treated with fungicides, herbicides, plant growth regulators, and fertilizers. Also, in case of a few plants per plot or border effect, plants realize more nitrogen uptake which results in potentially more GPC and, consequently, deselection of potential high-yielding lines. Conversely, in untreated conditions—which is usually the case in early generations in practical breeding—higher GPC is correlated with lower disease resistance. Consequently, removing lower GPC material could also result in removing the best disease resistant material. Recently, Gogna et al. (2022) showed negative correlations of protein content with major wheat diseases such as Septoria tritici blotch, Drechslera tritici-repentis, and Fusarium head blight in the same GABI wheat panel.
Genetic control of sedimentation values is simpler than falling number
The genome-wide association scan resulted in the detection of four ZSV and two HFN main-effect QTL. Comparison of QTL locations with previous mapping studies is often not possible because of the different use of: (1) marker systems (e.g., microsatellites, SNP arrays, etc.), (2) chromosome maps (e.g., genetic (cM) or physical (bp) positions), and (3) nature of mapping populations (e.g., bi-parental, synthetic, diverse released varieties, and breeding lines, etc.). Moreover, the studies on the association mapping for ZSV and HFN are limited and do not provide marker sequences, especially in winter wheats. Hence, we compare our QTL to the recent and relevant studies in the following.
Of the four QTL, three, viz. QZsv.ipk-1A, -1B, and -1D explained > 25% whereas the MTA representing the fourth ZSV-QTL (QZsv.ipk-5D) corresponding to Pinb-D1 gene explained 11.39% of the genotypic variance. Würschum et al. (2016) also detected four medium- to large-effect QTL that explained ~ 60% of the genotypic variance. This suggests that the large-effect marker-based genetic gain on ZSV QTL could be beneficial in terms of time- and cost-efficiency. The ZSV-QTL QZsv.ipk-1A harbored the gene for peroxidases (POD). To our knowledge, this is the first GWAS that identified an SNP linked with POD for ZSV—most possibly because PODs can improve the physical characteristics of gluten (Geng et al. 2019): this, however, needs further genetic and functional validation in independent populations. Another ZSV-QTL QZsv.ipk-1D harbored markers significantly associated with Glu-D1—a known HMW glutenin locus that influences the gluten and eventually viscoelastic properties of wheat flour doughs (Gale 2005). In addition to Glu-D1, QZsv.ipk-1D’s MTA BS00022768_51 explaining 26.05% of the genotypic variance corresponded to globulin—a non-gluten protein that, albeit present in minor quantities in the wheat endosperm, influences the flour processing (Gupta et al. 1992; Osborne 1907; Pence et al. 1954). Since the extent of linkage disequilibrium (the non-random association between different loci) plays a vital role in GWAS, the MTA BS00022768_51—because of its physical location on the long arm (412.2 Mbp)—is likely to be linked with Glu-D1, and, thus, may constitute the same QTL. Yu et al. (2021) recently colocalized several mixograph parameters with Glu-D1 locus on the same physical position (412–14 Mbp). Also, Fradgley et al. (2022) identified QTL harboring Glu-D1 to be associated with dough rheology traits in the UK winter wheat. Therefore, this locus seems to control several bread-making quality traits in wheat. Pin-1 genes affect wheat’s milling quality by primarily controlling the grain texture (Morris 2002). QZsv.ipk-5D harbored an MTA BS00000020_51 corresponding to Puroindoline-b (Pinb-D1). Pinb-D1’s association with ZSV suggests its possible role in controlling the ZSV—several previous studies have shown this gene’s association with other quality traits, including ZSV (Bhave and Morris 2008; Kristensen et al. 2018; Mohler et al. 2012). By dividing the panel based on Pin-1 genes' profiles, we also observed highly significant differences between the soft and hard wheats for all investigated quality traits, including ZSV. As described elsewhere, breeders in Europe have—compared to Pina-D1—mainly tapped the genetic variation of Pinb-D1 mutant alleles to fine-tune or maintain hardness or the grain texture. The association of this locus with ZSV further confirms the inference mentioned earlier that although GPC have reduced over the years, sustained ZSV and GH values helped sustain/improve bread-making properties. To achieve registration, in terms of GY, a line must outperform the existing check varieties (depending upon the quality class) by a certain margin and thus, the new high-yielding varieties inevitably show sustained/improved baking parameters. At this juncture, it can be safely assumed that wheat breeders seem to have exploited the ZSV/GPC ratio for which a stable and slightly positive genetic trend was observed in our study.
For HFN, seven MTA present on individual wheat chromosomes were identified. Of these seven, only two explained ~ 10% of the variance, and, thus, were designated as HFN-QTL. The QHfn.ipk-1A’s significant marker (AX-94430348; 360.5 Mbp) explained 12.74% of the genotypic variance and corresponded to the albumin protein (Fig. S8). Albumin, like globulin, is a non-gluten minor protein in wheat endosperm that has a significant bearing on wheat’s quality (Belderok et al. 2000). For example, Gupta et al. (1992) reported that dough quality is significantly related to the gluten and other monomeric proteins (e.g., globulin, and albumin). Žilić et al. (2011) identified albumins and globulins as major enzymes involved in metabolic processing. Most recently, Dallinger et al. (2023), via using a genotyping-by-sequencing platform in European winter wheat varieties, identified a QTL on chromosome 1A but at 313.5 Mbp and explaining 4% of the variation. The other HFN-QTL QHfn.ipk-5B (MTA: wsnp_Ku_rep_c71565_71299640; 618.1 Mbp) identified in this study explained 9.38% of the genotypic variance. To our knowledge, no previous reports, especially in winter wheats, identified HFN-QTL on 5B at or near 618.1 Mbp, thus, making it a novel QTL.
Taken together, our GWAS showcased ZSV’s genetic control to be less complex than HFN as its QTL showed more genotypic variance and were more significant than HFN—most possibly because of the action of known quality-related genes, e.g., Glu-D1 and Pinb-D1. Nevertheless, although helpful in breeding programs for speedy selection gain, the QTL and corresponding candidate genes reported in this study need further genetic and functional validation.
Haplotype analysis point toward suitable allelic combinations of major loci for breeding better grain quality and yield
Gluten (glutenin and gliadin) form the majority of the wheat storage proteins and the glutenin (Glu) loci bear a strong effect on the dough and baking quality (Gale 2005). Similarly, Pin-1 genes are the major loci to control grain texture (Morris 2002). We investigated the effect of three HMW Glu-1 loci (Glu-A1, -B1, and -D1) using candidate markers for the corresponding genes. In addition, the Rht-D1 and Pin-1 (Pina-D1 and Pinb-D1) loci were evaluated to observe their allelic influence on the investigated traits. Of the investigated loci, Glu-A1 alleles (Ax-null and Ax2*) did not significantly differentiate the ZSV; however, the presence of Ax2* resulted in a significant decrease in both HFN and GY. Glu-D1 locus (Liu et al. 2008) that differentiated Dx5 + Dy10 (responsible for strong doughs) and Dx2 + Dy12 (weak doughs) subunits was significantly associated with ZSV. Würschum et al. (2016) also found a significant association of Glu-1 loci with sedimentation values evaluated by the SDS-PAGE method. Contrary to Mohler et al. (2014) and Gooding et al. (2012), the Rht-D1’s effect on HFN in our study was statistically insignificant—possibly due to population size and the genetic background as the authors used bi-parental populations. In addition to Glu-D1, the Pinb-D1 gene was highly significantly associated with ZSV.
In the context of using major quality loci, it is vital to observe how breeders have exploited combination of alleles (haplotypes). As stated elsewhere, new varieties—depending on the quality class—must be superior to the checks (current top-yielding varieties) in terms of GY, disease resistance, and grain quality. In this regard, our haplotype analyses shed light on some of the breeders’ “favorite” haplotypes to sustain the bread-making quality while improving grain yield in wheat (Fig. 6). For example, 36 varieties harboring Hap-1 showed significantly increased values for ZSV, HFN, and GH. At the same time, the difference of GY compared to the overall population mean was not significant. Similarly, Hap-6 showed no significant differences in GY while showing improved values for all four quality traits. The most noticeable difference for GY values was observed in Hap-8 where varieties showed 3.51 dt ha−1 more GY than the population mean. In contrast, all other quality traits showed, predictably, lower values—probably because both wild-type forms for Pin-1 loci, Glu-D1 Dx2 + Dy12, and Glu-A1 Ax-null subunits were represented in Hap-8. In contrast, Hap-2 showed significantly decreased GY, harbored better quality subunits/alleles of major loci, and consequently significantly higher quality trait values. It should be noticed that Glu-A1 and -B1 subunits viz. Ax2* and Bx7OE were present only in 10.56 and 6.18% and thus could be a reason for not appearing in any major haplotype: all ten haplotypes harbored Ax-null and Bx7NE subunits. Similarly, from Pin-1 loci, the Pina-D1b null allele was present in only 7.57% of the varieties (Fig. 5) indicating that quality profiles have mainly been shaped by Pinb-D1 mutant alleles (Pinb-D1b–d). The presence of Rht-D1a resulted in either decreased or commensurable values for GY. However, varieties harboring Rht-D1b alleles showed either increased or on par GY values. Taken together, our analyses show that Bx7orBx17, Dx5 + Dy10, and Pinb-D1b–d subunits/alleles have been necessary for improved and sustainable breeding for GY and quality traits. Hence, these can be used in two to three combinations depending upon the target GY and quality class. Also, Hap-2 and Hap-8 may be used to breed for targeted E (high-quality elite) and C (cookies or stock feed) class GY groups, respectively.
The prospects of predictive breeding for sedimentation values and falling number in applied wheat breeding programs
We employed high-density SNP arrays, SSRs, and diagnostic markers with multi-environment robust phenotypic data for GWAS. However, the genotypic variance imparted by the identified MTA amounted to roughly 40% for ZSV and HFN. Hence, the remaining ~ 60% unexplained genotypic variance point to many small-effect maker loci, each explaining less than 10% genotypic variance. The prospects of genome-wide prediction—to predict the total genetic value of a trait based on all maker loci irrespective of their effect size—could be utilized (Meuwissen et al. 2001). In this study, based on five different models, predictive accuracies for ZSV and HFN suggested that these traits could be used for efficient genomic selection. Reports on genomic selection for both ZSV and HFN are scarce. Our results align with recent reports of Würschum et al. (2016) and Kristensen et al. (2018), who, while studying European wheat varieties/lines, reported high predictive abilities for ZSV and HFN. Here, it is worth noting that RKHSR—a model used to assess epistatic interactions among loci—did not outperform other models that primarily exploit the additive effects. This is in line with Würschum et al. (2016) where the authors did not find any significant epistatic QTL that could be exploited in marker-assisted breeding. Most recently, Schwarzwälder et al. (2022) reported that (1) selection on sedimentation values could help improve the baking volume, and (2) lines possessing high per se quality traits should help develop good hybrids: this is because most of the quality traits are additive, and there exists a high correlation between the mid-parent and hybrid performance. In this context, the mean prediction accuracies suggest that genomic selection could help select the lines with better genetic merit for ZSV and HFN. This could help achieve better genetic gains in both line and hybrid breeding programs.
Data availability
The authors declare that all data are contained within the paper and supplementary information. The phenotypic and marker datasets were published in Gogna et al (2022).
References
Ahlemeyer J, Friedt W (2011) Progress in winter wheat yield in Germany-what's the share of the genetic gain? Tagungsband der 61 Jahrestagung der Vereinigung der Pflanzenzüchter und Saatgutkaufleute Österreichs, 23–25 November 2010, Raumberg-Gumpenstein, Österreich, pp 19–24
Allen AM, Winfield MO, Burridge AJ, Downie RC, Benbow HR, Barker GL, Wilkinson PA, Coghill J, Waterfall C, Davassi A, Scopes G, Pirani A, Webster T, Brew F, Bloor C, Griffiths S, Bentley AR, Alda M, Jack P, Phillips AL, Edwards KJ (2017) Characterization of a Wheat Breeders’ Array suitable for high-throughput SNP genotyping of global accessions of hexaploid bread wheat (Triticum aestivum). Plant Biotechnol J 15:390–401
Bates D, Mächler M, Bolker B, Walker S (2015) Fitting linear mixed-effects models using lme4. J Stat Softw 67:1–48
Belderok B, Mesdag J, Mesdag H, Donner DA (2000) Bread-making quality of wheat: a century of breeding in Europe. Springer Science & Business Media
Bhave M, Morris CF (2008) Molecular genetics of puroindolines and related genes: allelic diversity in wheat and other grasses. Plant Mol Biol 66:205–219
Boden S, McIntosh R, Uauy C, Krattinger SG, Dubcovsky J, Rogers W, Xia X, Badaeva E, Bentley A, Brown-Guedira G, Caccamo M, Cattivelli L, Chhuneja P, Cockram J, Contreras-Moreira B, Dreisigacker S, Edwards D, González F, Guzmán C, Ikeda T, Karsai I, Nasuda S, Pozniak C, Prins R, Sen T, Silva P, Simkova H, Zhang Y, Initiative TW (2023) Updated guidelines for gene nomenclature in wheat. Theor Appl Genet 136:72
Brown J, Flavell R (1981) Fractionation of wheat gliadin and glutenin subunits by two-dimensional electrophoresis and the role of group 6 and group 2 chromosomes in gliadin synthesis. Theor Appl Genet 59:349–359
Butow B, Ma W, Gale K, Cornish G, Rampling L, Larroque O, Morell M, Békés F (2003) Molecular discrimination of Bx7 alleles demonstrates that a highly expressed high-molecular-weight glutenin allele has a major impact on wheat flour dough strength. Theor Appl Genet 107:1524–1532
Cormier F, Faure S, Dubreuil P, Heumez E, Beauchêne K, Lafarge S, Praud S, Le Gouis J (2013) A multi-environmental study of recent breeding progress on nitrogen use efficiency in wheat (Triticum aestivum L.). Theor Appl Genet 126:3035–3048
Dallinger HG, Löschenberger F, Azrak N, Ametz C, Michel S, Bürstmayr H (2023) Genome‐wide association mapping for pre‐harvest sprouting in European winter wheat detects novel resistance QTL, pleiotropic effects, and structural variation in multiple genomes. Plant Genome e20301
Endelman JB (2011) Ridge regression and other kernels for genomic selection with R package rrBLUP. The Plant Genome 4
Flintham J, Börner A, Worland A, Gale M (1997) Optimizing wheat grain yield: effects of Rht (gibberellin-insensitive) dwarfing genes. J Agric Sci 128:11–25
Fradgley NS, Gardner K, Kerton M, Swarbreck SM, Bentley AR (2022) Trade-offs in the genetic control of functional and nutritional quality traits in UK winter wheat. Heredity 128:420–433
Gale K (2005) Diagnostic DNA markers for quality traits in wheat. J Cereal Sci 41:181–192
Geng H, Shi J, Fuerst EP, Wei J, Morris CF (2019) Physical mapping of peroxidase genes and development of functional markers for TaPod-D1 on bread wheat chromosome 7D. Front Plant Sci 10:523
Gianola D, Van Kaam JB (2008) Reproducing kernel Hilbert spaces regression methods for genomic assisted prediction of quantitative traits. Genetics 178:2289–2303
Gogna A, Schulthess AW, Röder MS, Ganal MW, Reif JC (2022) Gabi wheat a panel of European elite lines as central stock for wheat genetic research. Scientific Data 9:538
Gooding MJ, Uppal RK, Addisu M, Harris KD, Uauy C, Simmonds JR, Murdoch AJ (2012) Reduced height alleles (Rht) and Hagberg falling number of wheat. J Cereal Sci 55:305–311
Gupta R, Batey I, MacRitchie F (1992) Relationships between protein composition and functional properties of wheat flours. Cereal Chem 69:125–131
Habier D, Fernando RL, Dekkers JC (2007) The impact of genetic relationship information on genome-assisted breeding values. Genetics 177:2389–2397
Hedden P (2003) The genes of the green revolution. Trends Genet 19:5–9
Huang X-Q, Röder MS (2005) Development of SNP assays for genotyping the puroindoline b gene for grain hardness in wheat using pyrosequencing. J Agric Food Chem 53:2070–2075
Justiz Bd (2017) Verordnung über die Anwendung von Düngemitteln, Bodenhilfsstoffen, Kultursubstraten und Pflanzenhilfsmitteln nach den Grundsätzen der guten fachlichen Praxis beim Düngen (Düngeverordnung-DüV). Ausfertigungsdatum: 26-05-2017
Kollers S, Rodemann B, Ling J, Korzun V, Ebmeyer E, Argillier O, Hinze M, Plieske J, Kulosa D, Ganal MW, Röder MS (2013) Whole genome association mapping of Fusarium head blight resistance in European winter wheat (Triticum aestivum L.). PLoS ONE 8:57500
Kristensen PS, Jahoor A, Andersen JR, Cericola F, Orabi J, Janss LL, Jensen J (2018) Genome-wide association studies and comparison of models and cross-validation strategies for genomic prediction of quality traits in advanced winter wheat breeding lines. Front Plant Sci 9:69
Laidig F, Piepho H-P, Rentel D, Drobek T, Meyer U, Huesken A (2017) Breeding progress, environmental variation and correlation of winter wheat yield and quality traits in German official variety trials and on-farm during 1983–2014. Theor Appl Genet 130:223–245
Liu S, Chao S, Anderson JA (2008) New DNA markers for high molecular weight glutenin subunits in wheat. Theor Appl Genet 118:177–183
Liu G, Zhao Y, Gowda M, Longin CFH, Reif JC, Mette MF (2016) Predicting hybrid performances for quality traits through genomic-assisted approaches in Central European wheat. PLoS ONE 11:e0158635
Lukow OM, Payne PI, Tkachuk R (1989) The HMW glutenin subunit composition of Canadian wheat cultivars and their association with bread-making quality. J Sci Food Agric 46:451–460
Malloch J, Newton R (1934) The relation between yield and protein content of wheat. Can J Res 10:774–779
Meuwissen TH, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Mohler V, Schmolke M, Paladey E, Seling S, Hartl L (2012) Association analysis of Puroindoline-D1 and Puroindoline b-2 loci with 13 quality traits in European winter wheat (Triticum aestivum L.). J Cereal Sci 56:623–628
Mohler V, Albrecht T, Mrva K, Schweizer G, Hartl L (2014) Genetic analysis of falling number in three bi-parental common winter wheat populations. Plant Breeding 133:448–453
Morris CF (2002) Puroindolines: the molecular genetic basis of wheat grain hardness. Plant Mol Biol 48:633–647
Muqaddasi QH, Pillen K, Plieske J, Ganal MW, Röder MS (2017) Genetic and physical mapping of anther extrusion in elite European winter wheat. PLoS ONE 12:e0187744
Muqaddasi QH, Zhao Y, Rodemann B, Plieske J, Ganal MW, Röder MS (2019) Genome-wide association mapping and prediction of adult stage Septoria tritici blotch infection in European winter wheat via high-density marker arrays. The Plant Genome 12:180029
Muqaddasi QH, Brassac J, Ebmeyer E, Kollers S, Korzun V, Argillier O, Stiewe G, Plieske J, Ganal MW, Röder MS (2020) Prospects of GWAS and predictive breeding for european winter wheat’s grain protein content, grain starch content, and grain hardness. Sci Rep 10:1–17
Neatby K, McCalla A (1938) Correlation between yield and protein content of wheat and barley in relation to breeding. Can J Res 16:1–15
Osborne TB (1907) The proteins of the wheat Kernel. Carnegie Institution of Washington, 119
Oury F-X, Berard P, Brancourt-Hulmel M, Heumez E, Pluchard P, Rousset M, Doussinault G, Rolland B, Trottet M, Giraud A (2003) Yield and grain protein concentration in bread wheat: a review and a study of multi-annual data from a French breeding program [Triticum aestivum L.]. J Genet Breed 57:59–68
Payne PI (1987) Genetics of wheat storage proteins and the effect of allelic variation on bread-making quality. Annu Rev Plant Physiol 38:141–153
Payne PI, Lawrence GJ (1983) Catalogue of alleles for the complex gene loci, Glu-A1, Glu-B1, and Glu-D1 which code for high-molecular-weight subunits of glutenin in hexaploid wheat. Cereal Res Commun, pp 29–35
Pence JW, Weinstein N, Mecham D (1954) The albumin and globulin contents of wheat flour and their relationship to protein quality. Cereal Chem 31:303–311
Pérez P, de Los Campos G (2014) Genome-wide regression and prediction with the BGLR statistical package. Genetics 198:483–495
Perten H (1964) Application of the falling number method for evaluating alpha-amylase activity. Cereal Chem 41:127–140
Reif JC, Gowda M, Maurer HP, Longin C, Korzun V, Ebmeyer E, Bothe R, Pietsch C, Würschum T (2011) Association mapping for quality traits in soft winter wheat. Theor Appl Genet 122:961–970
Rosseel Y (2012) Lavaan: an R package for structural equation modeling and more. Version 0.5–12 (BETA). J Stat Softw 48:1–36
Sandhu KS, Aoun M, Morris CF, Carter AH (2021) Genomic selection for end-use quality and processing traits in soft white winter wheat breeding program with machine and deep learning models. Biology 10:689
Schwarz G, Felsenstein F, Wenzel G (2004) Development and validation of a PCR-based marker assay for negative selection of the HMW glutenin allele Glu-B1-1d (Bx-6) in wheat. Theor Appl Genet 109:1064–1069
Schwarzwälder L, Thorwarth P, Zhao Y, Reif JC, Longin CFH (2022) Hybrid wheat: quantitative genetic parameters and heterosis for quality and rheological traits as well as baking volume. Theor Appl Genet 135:1131–1141
Shewry PR (2009) Wheat. J Exp Bot 60:1537–1553
Singh N, Shepherd K (1988) Linkage mapping of genes controlling endosperm storage proteins in wheat: 1. Genes on the short arms of group 1 chromosomes. Theor Appl Genet 75:628–641
Sorrells ME, Gustafson JP, Somers D, Chao S, Benscher D, Guedira-Brown G, Huttner E, Kilian A, McGuire PE, Ross K, Tanaka J, Wenzl P, Williams K, Qualset CO (2011) Reconstruction of the Synthetic W7984 × Opata M85 wheat reference population. Genome 54:875–882
Sortenliste B (2021) Bundessortenamt. Landbuch-Verlag, Hannover
Streiner DL (2005) Finding our way: an introduction to path analysis. Can J Psychiatry 50:115–122
Suhr D (2008) Step your way through path analysis. Western users of SAS software conference proceedings, p 2017
Sun C, Dong Z, Zhao L, Ren Y, Zhang N, Chen F (2020) The Wheat 660K SNP array demonstrates great potential for marker-assisted selection in polyploid wheat. Plant Biotechnol J 18:1354–1360
Team RC (2013) R: a language and environment for statistical computing
Utz HF, Melchinger AE, Schön CC (2000) Bias and sampling error of the estimated proportion of genotypic variance explained by quantitative trait loci determined from experimental data in maize using cross validation and validation with independent samples. Genetics 154:1839–1849
van Oort M, Hennink H, Schenkels P, Laane C (2000) Peroxidases in breadmaking. Wheat Struct Biochem Function 212:350
VanRaden PM (2008) Efficient methods to compute genomic predictions. J Dairy Sci 91:4414–4423
Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, Maccaferri M, Salvi S, Milner SG, Cattivelli L, Mastrangelo AM, Whan A, Stephen S, Barker G, Wieseke R, Plieske JJJ, Consortium IWGS, Lillemo M, Mather D, Appels R, Dolferus R, Brown-Guedira G, Korol A, Akhunova AR, Feuillet C, Salse J, Morgante M, Pozniak C, Luo M-C, Dvorak J, Morell M, Dubcovsky J, Ganal M, Tuberosa R, Lawley C, Mikoulitch I, Cavanagh C, Edwards KJ, Hayden M, Akhunov E (2014) Characterization of polyploid wheat genomic diversity using a high-density 90000 single nucleotide polymorphism array. Plant Biotechnol J 12:787–796
Würschum T, Leiser WL, Kazman E, Longin CFH (2016) Genetic control of protein content and sedimentation volume in European winter wheat cultivars. Theor Appl Genet 129:1685–1696
Yu J, Pressoir G, Briggs WH, Bi IV, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, Kresovich S, Buckler ES (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38:203–208
Yu S, Assanga SO, Awika JM, Ibrahim AM, Rudd JC, Xue Q, Guttieri MJ, Zhang G, Baker JA, Jessup KE, Liu S (2021) Genetic mapping of quantitative trait loci for end-use quality and grain minerals in hard red winter wheat. Agronomy 11:2519
Zanke C, Ling J, Plieske J, Kollers S, Ebmeyer E, Korzun V, Argillier O, Stiewe G, Hinze M, Beier S, Ganal MW, Röder MS (2014) Genetic architecture of main effect QTL for heading date in European winter wheat. Front Plant Sci 5:217
Zeleny L (1962) Wheat sedimentation test. 40th Annual Agricultural Outlook Conference, Washington, D.C.
Zhang L-L, Guan E-Q, Yang Y-L, Liu Y-X, Zhang T-J, Bian K (2021) Impact of wheat globulin addition on dough rheological properties and quality of cooked noodles. Food Chem 362:130170
Zhou Z, Guan H, Liu C, Zhang Z, Geng S, Qin M, Li W, Shi X, Dai Z, Lei Z (2021) Identification of genomic regions affecting grain peroxidase activity in bread wheat using genome-wide association study. BMC Plant Biol 21:1–13
Žilić S, Barać M, Pešić M, Dodig D, Ignjatović-Micić D (2011) Characterization of proteins from grain of different bread and durum wheat genotypes. Int J Mol Sci 12:5878–5894
Acknowledgements
The genotyping data were produced in the projects GABI-WHEAT funded by the German Federal Ministry of Education and Research (BMBF; project number 0315067). The first author thanks Damien Dupeu for proofreading the manuscript. We thank the editor and two reviewers whose comments helped improve the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. Costs for open access publishing were partially funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation, grant 491250510).
Author information
Authors and Affiliations
Contributions
Q.H.M. and M.S.R. conceived the idea of writing this manuscript; Q.H.M. analyzed the data, interpreted the results, and wrote the manuscript; R.K.M. contributed to data analyses; E.E. and O.A. contributed to field designs and phenotypic data collection; M.W.G. contributed to genotypic data production; Q.H.M., R.K.M., V.K., E.E., V.M., and J.C.R. revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest. Q.H.M., E.E., V.K., O.A., V.M., and M.W.G. are members of various companies. However, this does not limit the availability or sharing of data and materials.
Ethical standards
On behalf of all co-authors, the corresponding author states that the work described is original, previously unpublished research. All the authors listed have approved the manuscript.
Additional information
Communicated by Susanne Dreisigacker.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Muqaddasi, Q.H., Muqaddasi, R.K., Ebmeyer, E. et al. Genetic control and prospects of predictive breeding for European winter wheat’s Zeleny sedimentation values and Hagberg-Perten falling number. Theor Appl Genet 136, 229 (2023). https://doi.org/10.1007/s00122-023-04450-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00122-023-04450-7