Abstract
Key message
A GST for red-spot-petals in Gossypium arboreum was identified as the candidate under the scope of multi-omics approaches.
Abstract
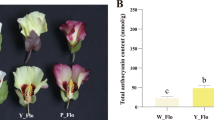
Colored petal spots are correlated with insect pollination efficiency in Gossypium species. However, molecular mechanisms concerning the formation of red spots on Gossypium arboreum flowers remain elusive. In the current study, the Shixiya1-R (SxyR, with red spots) × Shixiya1-W (SxyW, without red spots) segregating population was utilized to determine that the red-spot-petal phenotype was levered by a single dominant locus. This phenotype was expectedly related to the anthocyanin metabolites, wherein the cyanidin and delphinidin derivatives constituted the major partition. Subsequently, this dominant locus was narrowed to a 3.27 Mb range on chromosome 7 by genomic resequencing from the two parents and the two segregated progeny bulks that have spotted petals or not. Furthermore, differential expressed genes generated from the two bulks at either of three sequential flower developmental stages that spanning the spot formation were intersected with the annotated ones that allocated to the 3.27 Mb interval, which returned eight genes. A glutathione S-transferase-coding gene (Gar07G08900) out of the eight was the only one that exhibited simultaneously differential expression among all three developmental stages, and it was therefore considered to be the probable candidate. Finally, functional validation upon this candidate was achieved by the appearance of scattered petal spots with inhibited expression of Gar07G08900. In conclusion, the current report identified a key gene for the red spotted petal in G. arboreum under the scope of multi-omics approaches, such efforts and embedded molecular resources would benefit future applications underlying the flower color trait in cotton.



Similar content being viewed by others
Data availability
Genomic resequencing data were deposited into Sequence read archive (SRA) database under the accession numbers of SAMN28187015 to SAMN28187018, and the transcriptome data were under the accession numbers of SAMN28185984 to SAMN28185989.
References
Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H et al (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 30:174–178. https://doi.org/10.1038/nbt.2095
Abid MA, Wei Y, Meng Z, Wang Y, Ye Y, Wang Y et al (2022) Increasing floral visitation and hybrid seed production mediated by beauty mark in Gossypium hirsutum. Plant Biotechnol J 20:1274–1284. https://doi.org/10.1111/pbi.1380
Ageorges A, Fernandez L, Vialet S, Merdinoglu D, Terrier N, Romieu C (2006) Four specific isogenes of the anthocyanin metabolic pathway are systematically co-expressed with the red colour of grape berries. Plant Sci 170:372–383. https://doi.org/10.1016/j.plantsci.2005.09.007
Alfenito MR, Souer E, Goodman CD, Buell R, Mol J, Koes R et al (1998) Functional complementation of anthocyanin sequestration in the vacuole by widely divergent glutathione S-transferases. Plant Cell 10:1135–1149. https://doi.org/10.1105/tpc.10.7.1135
Cai X, Lin L, Wang X, Xu C, Wang Q (2018) Higher anthocyanin accumulation associated with higher transcription levels of anthocyanin biosynthesis genes in spinach. Genome 61:487–496. https://doi.org/10.1139/gen-2017-0261
Cavallini-Speisser Q, Morel P, Monniaux M (2021) Petal cellular identities. Front Plant Sci 12:745507. https://doi.org/10.3389/fpls.2021.745507
Chen G, He W, Guo X, Pan J (2021) Genome-wide identification, classification and expression analysis of the MYB transcription factor family in petunia. Int J Mol Sci 22:4838. https://doi.org/10.3390/ijms22094838
Chen J, Hu X, Shi T, Yin H, Sun D, Hao Y et al (2020) Metabolite-based genome-wide association study enables dissection of the flavonoid decoration pathway of wheat kernels. Plant Biotechnol J 18:1722–1735. https://doi.org/10.1111/pbi.13335
Chen W, Gong L, Guo Z, Wang W, Zhang H, Liu X et al (2013) A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol Plant 6:1769–1780. https://doi.org/10.1093/mp/sst080
Cui D, Zhao S, Xu H, Allan AC, Zhang X, Fan L et al (2021a) The interaction of MYB, bHLH and WD40 transcription factors in red pear (Pyrus pyrifolia) peel. Plant Mol Biol 106:407–417. https://doi.org/10.1007/s11103-021-01160-w
Cui Y, Fan J, Lu C, Ren J, Qi F, Huang H et al (2021b) ScGST3 and multiple R2R3-MYB transcription factors function in anthocyanin accumulation in Senecio cruentus. Plant Sci 313:111094. https://doi.org/10.1016/j.plantsci.2021.111094
Dossou S, Luo Z, Wang Z, Zhou W, Zhou R, Zhang Y et al (2022) The dark pigment in the sesame (Sesamum indicum L.) seed coat: isolation, characterization, and its potential precursors. Front Nutr 9:858673. https://doi.org/10.3389/fnut.2022.858673
Duan HR, Wang LR, Cui GX, Zhou XH, Duan XR, Yang HS (2020) Identification of the regulatory networks and hub genes controlling alfalfa floral pigmentation variation using RNA-sequencing analysis. BMC Plant Biol 20:110. https://doi.org/10.1186/s12870-020-2322-9
Erpelding JE (2021) Genetic characterization of the red coloured corolla phenotype for Gossypium arboreum accession PI 529731. Plant Breed 140:142–149. https://doi.org/10.1111/pbr.12884
Erpelding JE (2022) Genetic analysis of the Asiatic cotton (Gossypium arboreum) petal spot phenotypes. Plant Breed 141:71–76. https://doi.org/10.1111/pbr.12984
Geng X, Jiang C, Yang J, Wang L, Wu X, Wei W (2016) Rapid identification of candidate genes for seed weight using the SLAF-seq method in Brassica napus. PLoS ONE 211:e0147580. https://doi.org/10.1371/journal.pone.0147580
Gonzalez A, Zhao M, Leavitt J, Lloyd A (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53:814–827. https://doi.org/10.1111/j.1365-313X.2007.03373.x
Han Y, Zhao F, Gao S, Wang X, Wei A, Chen Z et al (2018) Fine mapping of a male sterility gene ms-3 in a novel cucumber (Cucumis sativus L.) mutant. Theor Appl Genet 131:449–460. https://doi.org/10.1007/s00122-017-3013-2
Hou XG, Guo Q, Wei WQ, Guo LL, Guo DL, Zhang L (2018) Screening of genes related to early and late flowering in tree peony based on bulked segregant RNA sequencing and verification by quantitative real-time PCR. Molecules 23:689. https://doi.org/10.3390/molecules23030689
Huang G, Wu Z, Percy RG, Bai M, Li Y, Frelichowski J et al (2020) Genome sequence of Gossypium herbaceum and genome updates of Gossypium arboreum and Gossypium hirsutum provide insights into cotton A-genome evolution. Nat Genet 52:516–524. https://doi.org/10.1038/s41588-020-0607-4
Jaakola L (2013) New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci 18:477–483. https://doi.org/10.1016/j.tplants.2013.06.003
Jiang S, Chen M, He N, Chen X, Wang N, Sun Q et al (2019) MdGSTF6, activated by MdMYB1, plays an essential role in anthocyanin accumulation in apple. Hortic Res 6:40. https://doi.org/10.1038/s41438-019-0118-6
Jiang T, Zhang MD, Wen CX, Xie XL, Tian W, Wen SQ et al (2020) Integrated metabolomic and transcriptomic analysis of the anthocyanin regulatory networks in Salvia miltiorrhiza Bge. flowers. BMC Plant Biol 20:349. https://doi.org/10.1186/s12870-020-02553-7
Jiao F, Zhao L, Wu X, Song Z, Li Y (2020) Metabolome and transcriptome analyses of the molecular mechanisms of flower color mutation in tobacco. BMC Genomics 21:611. https://doi.org/10.1186/s12864-020-07028-5
Jones K (1996) Pollinator behavior and post-pollination reproductive success in alternative floral phenotypes of Clarkia gracilis (Onagraceae). Int J Plant Sci 157:593–599. https://doi.org/10.1086/297396
Kitamura S, Matsuda F, Tohge T, Yonekura-Sakakibara K, Narumi I (2010) Metabolic profiling and cytological analysis of proanthocyanidins in immature seeds of Arabidopsis thaliana flavonoid accumulation mutants. Plant J 62:549–559. https://doi.org/10.1111/j.1365-313X.2010.04174.x
Kitamura S, Shikazono N, Tanaka A (2004) Transparent Testa 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J 37:104–114. https://doi.org/10.1046/j.1365-313x.2003.01943.x
Larsen ES, Alfenito MR, Briggs WR, Walbot V (2003) A carnation anthocyanin mutant is complemented by the glutathione S-transferases encoded by maize Bz2 and petunia An9. Plant Cell Rep 21:900–904. https://doi.org/10.1007/s00299-002-0545-x
Li S, Strid A (2005) Anthocyanin accumulation and changes in CHS and PR-5 gene expression in Arabidopsis thaliana after removal of the inflorescence stem (decapitation). Plant Physiol Bioc 43:521–525
Li AN, Li S, Li HB, Xu DP, Xu XR, Chen F (2014a) Total phenolic contents and antioxidant capacities of 51 edible and wild flowers. J Funct Foods 6:319–330. https://doi.org/10.1016/j.jff.2013.10.022
Li D, Chen G, Ma B, Zhong CG, He NJ (2020) Metabolic profiling and transcriptome analysis of mulberry leaves provide insights into flavonoid biosynthesis. J Agric Food Chem 68:1494–1504. https://doi.org/10.1021/acs.jafc.9b06931
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2089. https://doi.org/10.1093/bioinformatics/btp352
Li JJ, Jiang CR, Brown JB, Huang H, Bickel PJ (2011) Sparse linear modeling of next-generation mRNA sequencing (RNA-Seq) data for isoform discovery and abundance estimation. P Natl Acad Sci USA 108:19867–19872. https://doi.org/10.1073/pnas.1113972108
Li Q, Wang J, Sun HY, Shang X (2014b) Flower color patterning in pansy (Viola×wittrockiana Gams.) is caused by the differential expression of three genes from the anthocyanin pathway in acyanic and cyanic flower areas. Plant Physiol Biochem 84:134–141. https://doi.org/10.1016/j.plaphy.2014.09.012
Liu D, Guo X, Lin Z, Nie Y, Zhang X (2006) Genetic diversity of Asian cotton (Gossypium arboretum L.) in China evaluated by microsatellite analysis. Genet Resour Crop Evol 53:1145–1152. https://doi.org/10.1007/s10722-005-1304-y
Liu G, Zhao T, You X, Jiang J, Xu X (2019) Molecular mapping of the cf-10 gene by combining SNP/InDel-index and linkage analysis in tomato (Solanum lycopersicum). BMC Plant Biol 19:15. https://doi.org/10.1186/s12870-018-1616-7
Luo HF, Dai C, Li YP, Feng J, Liu ZC, Kang CY (2018) Reduced anthocyanins in petioles codes for a GST anthocyanin transporter that is essential for the foliage and fruit coloration in strawberry. J Exp Bot 69:2595–2608. https://doi.org/10.1093/jxb/ery096
Marrs KA, Alfenito MR, Lloyd AM, Walbot V (1995) A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature 375:397–400. https://doi.org/10.1038/375397a0
Martin C, Li J (2017) Medicine is not health care, food is health care: plant metabolic engineering, diet and human health. New Phytol 216:699–719. https://doi.org/10.1111/nph.14730
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. https://doi.org/10.1101/gr.107524.110
Meng J, Wang B, He G, Wang Y, Tang XF, Wang SM et al (2019) Metabolomics integrated with transcriptomics reveals redirection of the phenylpropanoids metabolic flux in Ginkgo biloba. J Agric Food Chem 67:3284–3291. https://doi.org/10.1021/acs.jafc.8b06355
Morimoto H, Narumi-Kawasaki T, Takamura T, Fukai S (2020) Flower color mutation caused by spontaneous cell layer displacement in carnation (Dianthus caryophyllus). Plant Sci 299:110598. https://doi.org/10.1016/j.plantsci.2020.110598
Motamayor JC, Mockaitis K, Schmutz J, Haiminen N, Livingstone D, Cornejo O et al (2013) The genome sequence of the most widely cultivated cacao type and its use to identify candidate genes regulating pod color. Genome Biol 14:r53. https://doi.org/10.1186/gb-2013-14-6-r53
Pu X, Li Z, Tian Y, Gao R, Hao L, Hu Y et al (2020) The honeysuckle genome provides insight into the molecular mechanism of carotenoid metabolism underlying dynamic flower coloration. New Phytol 227:930–943. https://doi.org/10.1111/nph.16552
Reumers J, De RP, Zhao H, Liekens A, Smeets D, Cleary J et al (2012) Optimized filtering reduces the error rate in detecting genomic variants by short-read sequencing. Nat Biotechnol 30:61–68. https://doi.org/10.1038/nbt.2053
Saigo T, Wang T, Watanabe M, Tohge T (2020) Diversity of anthocyanin and proanthocyanin biosynthesis in land plants. Curr Opin Plant Biol 55:93–99. https://doi.org/10.1016/j.pbi.2020.04.001
Saito K, Yonekura SK, Nakabayashi R, Higashi Y, Yamazaki M, Tohge T et al (2013) The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity. Plant Physiol Biol 72:21–34. https://doi.org/10.1016/j.plaphy.2013.02.001
Sasaki N, Nakayama T (2015) Achievements and perspectives in biochemistry concerning anthocyanin modification for blue flower coloration. Plant Cell Physiol 56:28–40. https://doi.org/10.1093/pcp/pcu097
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. https://doi.org/10.1038/nprot.2008.73
Shang Y, Venail J, Mackay S, Bailey PC, Schwinn KE, Jameson PE et al (2011) The molecular basis for venation patterning of pigmentation and its effect on pollinator attraction in flowers of Antirrhinum. New Phytol 189:602–615. https://doi.org/10.1111/j.1469-8137.2010.03498.x
She H, Qian W, Zhang H, Liu Z, Wang X, Wu J et al (2018) Fine mapping and candidate gene screening of the downy mildew resistance gene RPF1 in spinach. Theor Appl Genet 131:2529–2541. https://doi.org/10.1007/s00122-018-3169-4
Sheng Y, Wang Y, Jiao S, Jin Y, Ji P, Luan F (2017) Mapping and preliminary analysis of aborted microspores (AMS) as the candidate gene underlying the male sterility (MS-5) mutant in melon (Cucumis melo L.). Front Plant Sci 8:902. https://doi.org/10.3389/fpls.2017.00902
Sun Y, Li H, Huang JR (2012) Arabidopsis TT19 functions as a carrier to transport anthocyanin from the cytosol to tonoplasts. Mol Plant 5:387–400. https://doi.org/10.1093/mp/ssr110
Tanaka Y, Sasaki N, Ohmiya A (2008) Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J 54:733–749. https://doi.org/10.1111/j.1365-313X.2008.03447.x
Vela-Hinojosa C, Escalona-Buendía HB, Mendoza-Espinoza JA, Villa-Hernández JM, Pérez-Flores LJ (2019) Antioxidant balance and regulation in tomato genotypes of different color. J Am Soc Hortic Sci 144:45–54. https://doi.org/10.21273/JASHS04525-18
Wang G, Chen B, Du H, Zhang F, Zhang H, Wang Y et al (2018) Genetic mapping of anthocyanin accumulation-related genes in pepper fruits using a combination of SLAF-seq and BSA. PLoS ONE 13:e0204690. https://doi.org/10.1371/journal.pone.0204690
Wang Y, Xiao L, Guo S, An F, Du D, Li M (2016) Fine mapping and whole-genome resequencing identify the seed coat color gene in Brassica rapa. PLoS ONE 11:e0166464. https://doi.org/10.1371/journal.pone.0166464
Wang ZR, Cui YY, Vainstein A, Chen SW, Ma HQ (2017) Regulation of Fig (Ficus carica L.) fruit color: metabolomic and transcriptomic analyses of the flavonoid biosynthetic pathway. Front Plant Sci 8:1990. https://doi.org/10.3389/fpls.2017.01990
Weiss MR (1995) Floral color change: a widespread functional convergence. Am J Bot 82:167–185. https://doi.org/10.1002/j.1537-2197.1995.tb11486.x
Yang X, Xu Y, Yang F, Magwanga RO, Cai X, Wang X et al (2019) Genome-wide identification of OSCA gene family and their potential function in the regulation of dehydration and salt stress in Gossypium hirsutum. J Cotton Res 2:87–104. https://doi.org/10.1186/s42397-019-0028-z
Zhang K, Yuan M, Xia H, He L, Ma J, Wang M et al (2022) BSA-seq and genetic mapping reveals AhRt2 as a candidate gene responsible for red testa of peanut. Theor Appl Genet 135:1529–1540. https://doi.org/10.1007/s00122-022-04051-w
Zhang S, Lan Q, Gao X, Yang B, Cai C, Zhang T et al (2016) Mapping of genes for flower-related traits and QTLs for flowering time in an interspecific population of Gossypium hirsutum×G. darwinii. J Genet 95:197–201. https://doi.org/10.1007/s12041-016-0617-2
Zhang X, Li R, Chen L, Niu S, Chen L, Gao J et al (2017) Fine-mapping and candidate gene analysis of the Brassica juncea white-flowered mutant Bjpc2 using the whole-genome resequencing. Mol Genet Genomics 293:359–370. https://doi.org/10.1007/s00438-017-1390-5
Zhang Y, Butelli E, Martin C (2014) Engineering anthocyanin biosynthesis in plants. Curr Opin Plant Biol 19:81–90. https://doi.org/10.1016/j.pbi.2014.05.011
Zhang YL, Fang ZZ, Ye XF, Pan SL (2018) Identification of candidate genes involved in anthocyanin accumulation in the peel of jaboticaba (Myrciaria cauliflora) fruits by transcriptomic analysis. Gene 676:202–213. https://doi.org/10.1016/j.gene.2018.07.039
Zhou ZL (2011) Study on genetic diversity of G. arboreum L. germplasm resources conserved in China. Master Dissertation of Chinese Academy of Agricultural Sciences, 20 (in Chinese). https://doi.org/10.7666/d.Y1932322
Funding
This work was supported by HAAFS Science and Technology Innovation Special Project (2022KJCXZX-MHS-1) and Youth Fund of Hebei Natural Science Foundation (C2021301039).
Author information
Authors and Affiliations
Contributions
JZ, XZ and SZ conceived and designed the experiments. SZ, XC, HW, LT and XL performed the experiments. JC and TJ analyzed the data. JC and SZ wrote the paper. All authors read and approved the final version of the paper.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by David D. Fang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
122_2022_4191_MOESM3_ESM.xlsx
Table S1 Primers used in the current study. Table S2 Chromosomal distribution of the SNPs and InDels. Table S3 Candidate genes involved in the chromosomal intervals mapped by the Euclidean distance and SNP-index association algorithms. Table S4 DEGs at stage II.Table S5 DEGs at stage III. Table S6 DEGs at stage IV. (XLSX 76 KB)
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, S., Chen, J., Jiang, T. et al. Genetic mapping, transcriptomic sequencing and metabolic profiling indicated a glutathione S-transferase is responsible for the red-spot-petals in Gossypium arboreum. Theor Appl Genet 135, 3443–3454 (2022). https://doi.org/10.1007/s00122-022-04191-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-022-04191-z