Abstract
Key message
Calibrating a genomic selection model on a sparse factorial design rather than on tester designs is advantageous for some traits, and equivalent for others.
Abstract
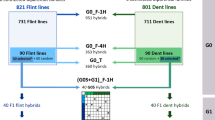
In maize breeding, the selection of the candidate inbred lines is based on topcross evaluations using a limited number of testers. Then, a subset of single-crosses between these selected lines is evaluated to identify the best hybrid combinations. Genomic selection enables the prediction of all possible single-crosses between candidate lines but raises the question of defining the best training set design. Previous simulation results have shown the potential of using a sparse factorial design instead of tester designs as the training set. To validate this result, a 363 hybrid factorial design was obtained by crossing 90 dent and flint inbred lines from six segregating families. Two tester designs were also obtained by crossing the same inbred lines to two testers of the opposite group. These designs were evaluated for silage in eight environments and used to predict independent performances of a 951 hybrid factorial design. At a same number of hybrids and lines, the factorial design was as efficient as the tester designs, and, for some traits, outperformed them. All available designs were used as both training and validation set to evaluate their efficiency. When the objective was to predict single-crosses between untested lines, we showed an advantage of increasing the number of lines involved in the training set, by (1) allocating each of them to a different tester for the tester design, or (2) reducing the number of hybrids per line for the factorial design. Our results confirm the potential of sparse factorial designs for genomic hybrid breeding.






Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Albrecht T, Wimmer V, Auinger H-J et al (2011) Genome-based prediction of testcross values in maize. Theor Appl Genet 123:339–350. https://doi.org/10.1007/s00122-011-1587-7
Andrieu J (1995) Prévision de la digestibilité et de la valeur énergétique du maïs fourrage à l’état frais. INRA Prod Anim 8:273–274. https://doi.org/10.20870/productions-animales.1995.8.4.4136
Argillier O, Barrière Y, Hébert Y (1995) Genetic variation and selection criterion for digestibility traits of forage maize. Euphytica 82:175–184. https://doi.org/10.1007/BF00027064
Baker CW, Givens DI, Deaville ER (1994) Prediction of organic matter digestibility in vivo of grass silage by near infrared reflectance spectroscopy: effect of calibration method, residual moisture and particle size. Anim Feed Sci Technol 50:17–26. https://doi.org/10.1016/0377-8401(94)90006-X
Bernardo R (1994) Prediction of maize single-cross performance using RFLPs and information from related hybrids. Crop Sci 34:20. https://doi.org/10.2135/cropsci1994.0011183X003400010003x
Burdo B, Leon N, Kaeppler SM (2021) Testcross vs randomly paired single-cross progeny tests for genomic prediction of new inbreds and hybrids derived from multiparent maize populations. Crop Sci. https://doi.org/10.1002/csc2.20545
Butler DG, Cullis BR, Gilmour AR, Gogel BG, Thompson R (2017) ASReml-R reference manual version 4. VSN International Ltd, Hemel Hempstead, HP1 1ES, UK
Dias KODG, Gezan SA, Guimarães CT et al (2018) Improving accuracies of genomic predictions for drought tolerance in maize by joint modeling of additive and dominance effects in multi-environment trials. Heredity 121:24–37. https://doi.org/10.1038/s41437-018-0053-6
Fritsche-Neto R, Akdemir D, Jannink J-L (2018) Accuracy of genomic selection to predict maize single-crosses obtained through different mating designs. Theor Appl Genet 131:1153–1162. https://doi.org/10.1007/s00122-018-3068-8
Giraud H, Bauland C, Falque M et al (2017a) Linkage analysis and association mapping QTL detection models for hybrids between multiparental populations from two heterotic groups: application to biomass production in maize (Zea mays L.). G3 Genes Genomes Genet 7:3649–3657. https://doi.org/10.1534/g3.117.300121
Giraud H, Bauland C, Falque M et al (2017b) Reciprocal Genetics: Identifying QTL for general and specific combining abilities in hybrids between multiparental populations from two maize (Zea mays L.) heterotic groups. Genetics 207:1167–1180. https://doi.org/10.1534/genetics.117.300305
Giraud H (2016) Genetic analysis of hybrid value for silage maize in multiparental designs: QTL detection and genomic selection. Thesis, Paris-Saclay
Goering HK, Soest PJV (1970) Forage fiber analyses (apparatus, reagents, procedures, and some applications). U.S. Agricultural Research Service
González-Diéguez D, Legarra A, Charcosset A et al (2021) Genomic prediction of hybrid crops allows disentangling dominance and epistasis. Genetics. https://doi.org/10.1093/genetics/iyab026
Heslot N, Yang H-P, Sorrells ME, Jannink J-L (2012) Genomic selection in plant breeding: a comparison of models. Crop Sci 52:146–160. https://doi.org/10.2135/cropsci2011.06.0297
Jannink J-L, Lorenz AJ, Iwata H (2010) Genomic selection in plant breeding: from theory to practice. Brief Funct Genomics 9:166–177. https://doi.org/10.1093/bfgp/elq001
Kadam DC, Lorenz AJ (2018) Toward redesigning hybrid maize breeding through genomics-assisted breeding. In: Bennetzen J, Flint-Garcia S, Hirsch C, Tuberosa R (eds) The maize genome. Springer International Publishing, Cham, pp 367–388
Kadam DC, Potts SM, Bohn MO et al (2016) Genomic prediction of single crosses in the early stages of a maize hybrid breeding pipeline. G3 Genes Genomes Genet 6:3443–3453. https://doi.org/10.1534/g3.116.031286
Kadam DC, Rodriguez OR, Lorenz AJ (2021) Optimization of training sets for genomic prediction of early-stage single crosses in maize. Theor Appl Genet 134:687–699. https://doi.org/10.1007/s00122-020-03722-w
Lande R, Thompson R (1990) Efficiency of marker-assisted selection in the improvement of quantitative traits. Genetics 124:743
Meuwissen THE, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Molenberghs G, Verbeke G (2007) Likelihood ratio, score, and wald tests in a constrained parameter space. Am Stat 61:22–27. https://doi.org/10.1198/000313007X171322
Peyrat J, Nozière P, Férard A et al (2016) «Prévoir la digestibilité et la valeur énergétique du maïs fourrage : Guide des nouvelles références ». ARVALIS - Institut du végétal - INRA
Powell O, Gaynor RC, Gorjanc G et al (2020) A two-part strategy using genomic selection in hybrid crop breeding programs. bioRxiv 2020.05.24.113258. https://doi.org/10.1101/2020.05.24.113258
Pszczola M, Strabel T, Mulder HA, Calus MPL (2012) Reliability of direct genomic values for animals with different relationships within and to the reference population. J Dairy Sci 95:389–400. https://doi.org/10.3168/jds.2011-4338
R Core Team (2020) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Reif JC, Gumpert F-M, Fischer S, Melchinger AE (2007) Impact of interpopulation divergence on additive and dominance variance in hybrid populations. Genetics 176:1931. https://doi.org/10.1534/genetics.107.074146
Revelle W (2021) psych: procedures for psychological, psychometric, and personality research. Northwestern University, Evanston
Saatchi M, Miraei-Ashtiani SR, Javaremi AN, Moradi-Shahrebabak M (2010) The impact of information quantity and strength of relationship between training set and validation set on accuracy of genomic estimated breeding values. Afr J Biotechnol 9:438–442. https://doi.org/10.5897/AJB09.1024
Schrag TA, Melchinger AE, Sørensen AP, Frisch M (2006) Prediction of single-cross hybrid performance for grain yield and grain dry matter content in maize using AFLP markers associated with QTL. Theor Appl Genet 113:1037–1047. https://doi.org/10.1007/s00122-006-0363-6
Schrag TA, Westhues M, Schipprack W et al (2018) Beyond genomic prediction: combining different types of omics data can improve prediction of hybrid performance in maize. Genetics 208:1373–1385
Self SG, Liang K-Y (1987) Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. J Am Stat Assoc 82:605–610. https://doi.org/10.1080/01621459.1987.10478472
Seye AI, Bauland C, Giraud H et al (2019) Quantitative trait loci mapping in hybrids between Dent and Flint maize multiparental populations reveals group-specific QTL for silage quality traits with variable pleiotropic effects on yield. Theor Appl Genet 132:1523–1542. https://doi.org/10.1007/s00122-019-03296-2
Seye AI, Bauland C, Charcosset A, Moreau L (2020) Revisiting hybrid breeding designs using genomic predictions: simulations highlight the superiority of incomplete factorials between segregating families over topcross designs. Theor Appl Genet 133:1995–2010. https://doi.org/10.1007/s00122-020-03573-5
Seye AI (2019) Prédiction assistée par marqueurs de la performance hybride dans un schéma de sélection réciproque: simulations et évaluation expérimentale pour le maïs ensilage. Thesis, Paris Saclay
Sprague GF, Tatum LA (1942) General vs. specific combining ability in single crosses of corn1. Agron J 34:923–932. https://doi.org/10.2134/agronj1942.00021962003400100008x
Stuber CW, Cockerham CC (1966) Gene effects and variances in hybrid populations. Genetics 54:1279–1286. https://doi.org/10.1093/genetics/54.6.1279
Technow F, Riedelsheimer C, Schrag TA, Melchinger AE (2012) Genomic prediction of hybrid performance in maize with models incorporating dominance and population specific marker effects. Theor Appl Genet 125:1181–1194. https://doi.org/10.1007/s00122-012-1905-8
Technow F, Schrag TA, Schipprack W et al (2014) Genome properties and prospects of genomic prediction of hybrid performance in a breeding program of maize. Genetics 197:1343–1355. https://doi.org/10.1534/genetics.114.165860
VanRaden PM (2008) Efficient methods to compute genomic predictions. J Dairy Sci 91:4414–4423. https://doi.org/10.3168/jds.2007-0980
Varona L, Legarra A, Toro MA, Vitezica ZG (2018) Non-additive effects in genomic selection. Front Genet 9:78. https://doi.org/10.3389/fgene.2018.00078
Vitezica ZG, Varona L, Legarra A (2013) On the additive and dominant variance and covariance of individuals within the genomic selection scope. Genetics 195:1223–1230. https://doi.org/10.1534/genetics.113.155176
Vitezica ZG, Legarra A, Toro MA, Varona L (2017) Orthogonal estimates of variances for additive, dominance, and epistatic effects in populations. Genetics 206:1297–1307. https://doi.org/10.1534/genetics.116.199406
Westhues M, Schrag TA, Heuer C et al (2017) Omics-based hybrid prediction in maize. Theor Appl Genet 130:1927–1939. https://doi.org/10.1007/s00122-017-2934-0
Williams EJ (1959) 136. Query: significance of difference between two non-independent correlation coefficients. Biometrics 15:135. https://doi.org/10.2307/2527608
Williams E, Piepho H-P, Whitaker D (2011) Augmented p-rep designs. Biom J 53:19–27. https://doi.org/10.1002/bimj.201000102
Acknowledgements
We thank Lidea, Limagrain Europe, Maïsadour Semences, Corteva, RAGT 2n, and Syngenta Seeds grouped in the frame of the ProMais SAM-MCR program for the funding, inbred lines development, hybrid production, and phenotyping. We are also grateful to scientists from these companies and to scientists of the INRAE “R2D2” network for helpful discussions on the results. A.L. phD contract was funded by RAGT 2n and ANRT contract n° 2020/0032 and receive support from the "Investissement d’Avenir" project "Amaizing" (Amaizing, ANR-10-BTBR-0001). GQE-Le Moulon benefits from the support of Saclay Plant Sciences-SPS (ANR-17-EUR-0007).
Funding
Lidea, Limagrain Europe, Maïsadour Semences, Corteva, RAGT 2n, and Syngenta Seeds grouped in the frame of the ProMais funded the SAM-MCR project. A.L. phD contract was funded by RAGT 2n and ANRT contract n° 2020/0032 and receive support from the "Investissement d’Avenir" project "Amaizing" (Amaizing, ANR-10-BTBR-0001). GQE-Le Moulon benefits from the support of Saclay Plant Sciences-SPS (ANR-17-EUR-0007).
Author information
Authors and Affiliations
Contributions
CB, AC and LM initiated this project. LM and CB coordinated it with the help of CL and CG. AC, LM and CL supervised this work. SP and CP contributed to the development of the genetic material. AL analyzed the results and prepared the manuscript. All authors discussed the results and contributed to the final manuscript. All authors revised and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Consent to participate
Not relevant for this study on plants.
Consent to publish
Not relevant for this study on plants.
Ethics approval
All usual standards have been respected.
Additional information
Communicated by Matthias Frisch.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lorenzi, A., Bauland, C., Mary-Huard, T. et al. Genomic prediction of hybrid performance: comparison of the efficiency of factorial and tester designs used as training sets in a multiparental connected reciprocal design for maize silage. Theor Appl Genet 135, 3143–3160 (2022). https://doi.org/10.1007/s00122-022-04176-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-022-04176-y