Abstract
Key message
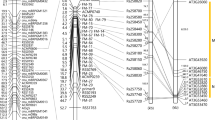
Two major QTL associated with resistance to Fusarium wilt (FW) were identified using whole-genome resequencing. Sequence variations and gene expression level differences suggest that TIR-NBS and LRR-RLK are candidate genes associated with FW-resistance.
Abstract
Fusarium wilt (FW) caused by Fusarium oxysporum f. sp. raphani is an important disease in radish, leading to severe decrease in yield and quality. YR4 as a novel genetic source to resistant to FW was confirmed through screening with five pathogen isolates. We have generated F2 and F2:3 populations segregated with FW resistance using YR4 and YR18 inbred lines. The disease symptom was evaluated in F2:3 population (n = 180) in three independent studies over two years. We identified 4 QTL including the two major QTL (FoRsR7.159A and FoRsR9.359A). FoRsR7.159A and FoRsR9.359A were detected in three replicated experiments. FoRsR7.159A was delimited to the 2.18-Mb physical interval on chromosome R07, with a high LOD value (5.17–12.84) and explained phenotypic variation (9.34%–27.97%). The FoRsR9.359A represented relatively low LOD value (3.38–4.52) and explained phenotypic variation (6.24%–8.82%). On the basis of the re-sequencing data for the parental lines, we identified five putative resistance-related genes and 13 unknown genes with sequence variations at the gene and protein levels. A semi-quantitative RT-PCR analysis revealed that Rs382940 (TIR-NBS) and Rs382200 (RLK) were expressed only in ‘YR4’ from 0 to 6 days after the inoculation. Moreover, Rs382950 (TIR-NBS-LRR) was more highly expressed in ‘YR4’ from 3 to 6 days after the inoculation. These three genes might be important for FW-resistance in radish. We identified several markers based on these potential candidate genes. The marker set should be useful for breeding system to introduce the FW resistance loci from ‘YR4’ to improve tolerance to FW.







Similar content being viewed by others
References
Branham SE, Wechter WP, Ling KS, Chanda B, Massey L, Zhao G, Guner N, Bello M, Kabelka E, Fei Z, Levi A (2020) QTL mapping of resistance to Fusarium oxysporum f. sp. niveum race 2 and Papaya ringspot virus in Citrullus amarus. Theor Appl Genet 133:677–687
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10:421
Catanzariti AM, Do HT, Bru P, de Sain M, Thatcher LF, Rep M, Jones DA (2017) The tomato I gene for Fusarium wilt resistance encodes an atypical leucine-rich repeat receptor-like protein whose function is nevertheless dependent on SOBIR 1 and SERK 3/BAK 1. The Plant J 89:1195–1209
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Cole SJ, Diener AC (2013) Diversity in receptor-like kinase genes is a major determinant of quantitative resistance to Fusarium oxysporum f. sp. matthioli. New Phytol 200:172–184
Collard BC, Mackill DJ (2007) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos Trans Roy Soc B Biol Sci 363:557–572
Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML (2011) Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet 12:499
Dean R, van Kan JAL, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, Foster GD (2012) The top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 13:414–430
Diener AC (2013) Routine mapping of Fusarium wilt resistance in BC1 populations of Arabidopsis thaliana. BMC Plant Biol 13:171
Diener AC, Ausubel FM (2005) RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 171:305–321
Fink M, Kofoet A (2005) A two-dimensional stochastic model of downy mildew of radish. Ecol Model 181:139–148
Fuentes-Pardo AP, Ruzzante DE (2017) Whole-genome sequencing approaches for conservation biology: advantages, limitations and practical recommendations. Mol Ecol 26:5369–5406
Garibaldi A, Gilardi G, Gullino ML (2006) Evidence for an expanded host range of Fusarium oxysporum f. sp. raphani. Phytoparasitica 34:115–121
Hamilton JP, Robin Buell C (2012) Advances in plant genome sequencing. Plant J 70:177–190
Husaini AM, Sakina A, Cambay SR (2018) Host–pathogen interaction in Fusarium oxysporum infections: where do we stand? Mol Plant Microbe in 31:889–898
Jaganathan D, Bohra A, Thudi M, Varshney RK (2020) Fine mapping and gene cloning in the post-NGS era: Advances and prospects. Theor Appl Genet 133:1791–1810
Jeong Y-M, Kim N, Ahn BO, Oh M, Chung W-H, Chung H, Jeong S, Lim K-B, Hwang Y-J, Kim G-B (2016) Elucidating the triplicated ancestral genome structure of radish based on chromosome-level comparison with the Brassica genomes. Theor Appl Genet 129:1357–1372
Jing Y, Shen N, Zheng X, Fu A, Zhao F, Lan W, Luan S (2020) Danger-associated peptide regulates root immune responses and root growth by affecting ros formation in Arabidopsis. Int J Mol Sci 21:4590
Kato T, Hatakeyama K, Fukino N, Matsumoto S (2013) Fine mapping of the clubroot resistance gene CRb and development of a useful selectable marker in Brassica rapa. Breed Sci 63:116–124
Kelley DR, Schatz MC, Salzberg SL (2010) Quake: quality-aware detection and correction of sequencing errors. Genome Biol 11:R116
Kim H, Hwang SM, Lee JH, Oh M, Han JW, Choi GJ (2017) Specific PCR detection of Fusarium oxysporum f. sp. raphani: a causal agent of Fusarium wilt on radish plants. Lett Appl Microbiol 65(2):133–140
Kitashiba H, Li F, Hirakawa H, Kawanabe T, Zou Z, Hasegawa Y, Tonosaki K, Shirasawa S, Fukushima A, Yokoi S (2014) Draft sequences of the radish (Raphanus sativus L.) genome. DNA Res 21:481–490
Kulwal P, Ishikawa G, Benscher D, Feng Z, Yu L-X, Jadhav A, Mehetre S, Sorrells ME (2012) Association mapping for pre-harvest sprouting resistance in white winter wheat. Theor Appl Genet 125:793–805
Landa BB, Navas-Cortés JA, Hervás A, Jiménez-Díaz RM (2001) Influence of temperature and inoculum density of Fusarium oxysporum f. sp. ciceris on suppression of Fusarium wilt of chickpea by rhizosphere bacteria. Phytopathology 91:807–816
Lande R, Thompson R (1990) Efficiency of marker-assisted selection in the improvement of quantitative traits. Genetics 124:743–756
Le T-N, Schumann U, Smith NA, Tiwari S, Au PCK, Zhu Q-H, Taylor JM, Kazan K, Llewellyn DJ, Zhang R (2014) DNA demethylases target promoter transposable elements to positively regulate stress responsive genes in Arabidopsis. Genome Biol 15:1–18
Lee SM, Lee JH, Jang KS, Choi YH, Kim H, Choi GJ (2020) Resistance of commercial radish cultivars to isolates of Fusarium oxysporum f. sp. raphani. Hortic Sci Technol 38(1):97–106
Leeman M, Van Pelt J, Den Ouden F, Heinsbroek M, Bakker P, Schippers B (1995) Induction of systemic resistance against Fusarium wilt of radish by lipopolysaccharides of Pseudomonas fluorescens. Phytopathology 85:1021–1027
Li X, Ramchiary N, Choi SR, Van Nguyen D, Hossain MJ, Yang HK, Lim YP (2010) Development of a high density integrated reference genetic linkage map for the multinational Brassica rapa Genome Sequencing Project. Genome 53:939–947
Li F, Hasegawa Y, Saito M, Shirasawa S, Fukushima A, Ito T, Fujii H, Kishitani S, Kitashiba H, Nishio T (2011) Extensive chromosome homoeology among Brassiceae species were revealed by comparative genetic mapping with high-density EST-based SNP markers in radish (Raphanus sativus L.). DNA Res 18:401–411
Lingling W (2018) Identification and control of radish alternaria leaf spot, black rot, soft rot, virus disease and hollowness. Plant Dis Pests 9 (3–4):8–11
Liu L, Li Y, Li S, Hu N, He Y, Pong R, Lin D, Lu L, Law M (2012) Comparison of next-generation sequencing systems. J Biomed Biotechnol 251364. https://doi.org/10.1155/2012/251364
Liu Z, Xie J, Wang H, Zhong X, Li H, Yu J, Kang J (2019) Identification and expression profiling analysis of NBS–LRR genes involved in Fusarium oxysporum f sp conglutinans resistance in cabbage. 3 Biotech 9:202
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408
Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:2047-2217X-2041-2018. https://doi.org/10.1186/2047-217X-1-18
Lv H-h, Yang L-m, Kang J-g, Wang Q-b, Wang X-w, Fang Z-y, Liu Y-m, Zhuang M, Zhang Y-y, Lin Y (2013) Development of InDel markers linked to Fusarium wilt resistance in cabbage. Mol Breeding 32:961–967
Lv H, Fang Z, Yang L, Zhang Y, Wang Q, Liu Y, Zhuang M, Yang Y, Xie B, Liu B (2014) Mapping and analysis of a novel candidate Fusarium wilt resistance gene FOC1 in Brassica oleracea. BMC Genomics 15:1094
Ma Y, Chhapekar SS, Lu L, Oh S, Singh S, Kim CS, Kim S, Choi GJ, Lim YP, Choi SR (2021) Genome-wide identification and characterization of NBS-encoding genes in Raphanus sativus L. and their roles related to Fusarium oxysporum resistance. BMC Plant Biol 21:1–17
Marçais G, Kingsford C (2011) A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27:764–770
Matić S, Gilardi G, Gullino ML, Garibaldi A (2018) Evidence for an expanded host range of Fusarium oxysporum f. sp. chrysanthemi. Journal of Plant Pathology 100:97–104
Meienberg J, Bruggmann R, Oexle K, Matyas G (2016) Clinical sequencing: is WGS the better WES? Hum Genet 135:359–362
Mendy B, Wang’ombe MW, Radakovic ZS, Holbein J, Ilyas M, Chopra D, Holton N, Zipfel C, Grundler FM, Siddique S (2017) Arabidopsis leucine-rich repeat receptor–like kinase NILR1 is required for induction of innate immunity to parasitic nematodes. PLoS Pathog 13:e1006284
Meng L, Li H, Zhang L, Wang J (2015) QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J 3:269–283
Mitsui Y, Shimomura M, Komatsu K, Namiki N, Shibata-Hatta M, Imai M, Katayose Y, Mukai Y, Kanamori H, Kurita K (2015) The radish genome and comprehensive gene expression profile of tuberous root formation and development. Sci Rep 5:10835
Mun J-H, Chung H, Chung W-H, Oh M, Jeong Y-M, Kim N, Ahn BO, Park B-S, Park S, Lim K-B (2015) Construction of a reference genetic map of Raphanus sativus based on genotyping by whole-genome resequencing. Theor Appl Genet 128:259–272
Murray M, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4326
Nandety RS, Caplan JL, Cavanaugh K, Perroud B, Wroblewski T, Michelmore RW, Meyers BC (2013) The role of TIR-NBS and TIR-X proteins in plant basal defense responses. Plant Physiol 162:1459–1472
Niu Y, Hu B, Li X, Chen H, Takáč T, Šamaj J, Xu C (2018) Comparative digital gene expression analysis of tissue-cultured plantlets of highly resistant and susceptible banana cultivars in response to Fusarium oxysporum. Int J Mol Sci 19:350
Pu Z-j, Shimizu M, Zhang Y-j, Nagaoka T, Hayashi T, Hori H, Matsumoto S, Fujimoto R, Okazaki K (2012) Genetic mapping of a fusarium wilt resistance gene in Brassica oleracea. Mol Breeding 30:809–818
Pu Z, Ino Y, Kimura Y, Tago A, Shimizu M, Natsume S, Sano Y, Fujimoto R, Kaneko K, Shea DJ (2016) Changes in the proteome of xylem sap in Brassica oleracea in response to Fusarium oxysporum stress. Front Plant Sci 7:31
Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80:847–857
Ramchiary N, Nguyen VD, Li X, Hong CP, Dhandapani V, Choi SR, Yu G, Piao ZY, Lim YP (2011) Genic microsatellite markers in Brassica rapa: development, characterization, mapping, and their utility in other cultivated and wild Brassica relatives. DNA Res 18:305–320
Ryan CA, Huffaker A, Yamaguchi Y (2007a) New Insights into Innate Immunity in Arabidopsis. Cellular Microbiol 9(8):1902–1908
Ryan CA, Huffaker A, Yamaguchi Y (2007b) New insights into innate immunity in Arabidopsis. Cell Microbiol 9:1902–1908
Saito M, Kubo N, Matsumoto S, Suwabe K, Tsukada M, Hirai M (2006) Fine mapping of the clubroot resistance gene, Crr3, in Brassica rapa. Theor Appl Genet 114:81
Schumann U, Lee J, Kazan K, Ayliffe M, Wang M-B (2017) DNA-demethylase regulated genes show methylation-independent spatiotemporal expression patterns. Front Plant Sci 8:1449
Shen Y, Diener AC (2013) Arabidopsis thaliana RESISTANCE TO FUSARIUM OXYSPORUM 2 implicates tyrosine-sulfated peptide signaling in susceptibility and resistance to root infection. PLoS Genet 9:e1003525
Shimizu M, Fujimoto R, Ying H, Pu Z-j, Ebe Y, Kawanabe T, Saeki N, Taylor JM, Kaji M, Dennis ES (2014) Identification of candidate genes for fusarium yellows resistance in Chinese cabbage by differential expression analysis. Plant Mol Biol 85:247–257
Shimizu M, Pu Z-j, Kawanabe T, Kitashiba H, Matsumoto S, Ebe Y, Sano M, Funaki T, Fukai E, Fujimoto R (2015) Map-based cloning of a candidate gene conferring Fusarium yellows resistance in Brassica oleracea. Theor Appl Genet 128:119–130
Shirasawa K, Oyama M, Hirakawa H, Sato S, Tabata S, Fujioka T, Kimizuka-Takagi C, Sasamoto S, Watanabe A, Kato M (2011) An EST-SSR linkage map of Raphanus sativus and comparative genomics of the Brassicaceae. DNA Res 18:221–232
Simpson GG, Dean C (2002) Arabidopsis, the Rosetta stone of flowering time? Science 296:285–289
Snyder W, Bardin K (1940) Occurrence of Fusarium wilt in a market. garden crop of Radishes. Plant Dis Rep 33
Stuber CW, Polacco M, Senior ML (1999) Synergy of empirical breeding, marker-assisted selection, and genomics to increase crop yield potential. Crop Sci 39:1571–1583
Untergrasser A, Cutcutache I, Koressaar T, Ye J, Faircloth B, Remm M, Rozen S (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40:e115
Van Ooijen JW (1999) LOD significance thresholds for QTL analysis in experimental populations of diploid species. Heredity 83(5):613–624
Van Bueren ETL, Struik PC, van Eekeren N, Nuijten E (2018) Towards resilience through systems-based plant breeding. A review. Agron Sustain Dev 38:42
Van Dijk EL, Auger H, Jaszczyszyn Y, Thermes C (2014) Ten years of next-generation sequencing technology. Trends Genet 30:418–426
Varshney RK (2016) Exciting journey of 10 years from genomes to fields and markets: some success stories of genomics-assisted breeding in chickpea, pigeonpea and groundnut. Plant Sci 242:98–107
Voorrips R, Jongerius M, Kanne H (1997) Mapping of two genes for resistance to clubroot (Plasmodiophora brassicae) in a population of doubled haploid lines of Brassica oleracea by means of RFLP and AFLP markers. Theor Appl Genet 94:75–82
Wang S, Basten C, Zeng Z (2012) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC (http://statgen.ncsu.edu/qtlcart/WQTLCart.htm)
Yamada K, Yamashita-Yamada M, Hirase T, Fujiwara T, Tsuda K, Hiruma K, Saijo Y (2016) Danger peptide receptor signaling in plants ensures basal immunity upon pathogen-induced depletion of BAK 1. EMBO J 35:46–61
Yu X, Choi SR, Ramchiary N, Miao X, Lee SH, Sun HJ, Kim S, Ahn CH, Lim YP (2013) Comparative mapping of Raphanus sativus genome using Brassica markers and quantitative trait loci analysis for the Fusarium wilt resistance trait. Theor Appl Genet 126:2553–2562
Yu X, Choi SR, Lim YP (2017) Molecular mapping of disease resistance genes. The radish genome. Springer, pp 165–175
Yu X, Kang DH, Choi SR, Ma Y, Lu L, Oh SH, Chhapekar SS, Lim YP (2018) Isolation and characterization of fusarium wilt resistance gene analogs in radish. 3 Biotech 8:255
Yu X, Lu L, Ma Y, Chhapekar SS, Yi SY, Lim YP, Choi SR (2020) Fine-mapping of a major QTL (Fwr1) for fusarium wilt resistance in radish. Theor Appl Genet 133:329–340
Acknowledgements
This research was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries through Golden Seed Project, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (213006-05-5-SBO20, 213006-05-5-SB110).
Author information
Authors and Affiliations
Contributions
SRC and YPL and designed the study. YM carried out experiments, generated data. SRC, YM analyzed all data, and drafted manuscript. SSC participated in candidate gene identification, writing and editing of the manuscript. LL did marker survey and genotyping. LL, SK, GJC, SML, THG were participated in phenotype evaluations. YPL provided plant materials, conceived the study, and finalized the manuscript. SRC conceived and designed the study, participated as a director, and modified the manuscript. All authors read and approved the final manuscript. All the authors declare that they have no conflicts of interest.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Standards
The experiments comply with the laws of the USA, the country in which the study was performed, and the ethical standards of the respective university and employers of the authors.
Additional information
Communicated by Amnon Levi.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
122_2021_3937_MOESM7_ESM.pdf
Symptoms of plants infected with Fusarium oxysporum f. sp. raphani pathotypes 59A, 57A, 147A, HN, JHW at 25 °C and 60% relative humidity, with a 12-h light condition cycle in a culture room at the Korea Research Institute of Chemical Technology (KRICT) (PDF 1194 kb)
122_2021_3937_MOESM9_ESM.pdf
QTL for Fusarium wilt resistant traits to Fusarium oxysporum f. sp. raphani pathotype 59A in F2:3 population of R. sativus by two different software, WinQTL 2.5 and IciMapping 4.1. (a), (b) represented the results of 9 chromosome scane overall radish genome. (c), (d) represented the QTL regions identified on R7 in two different analysis methods by WinQTL 2.5 based on Composite Interval Mappin (CIM) and Inclusive Composite Interval Mapping (ICIM). Tree different replicates were performed during 2 years in culture room and glasshouse (PDF 635 kb)
122_2021_3937_MOESM12_ESM.pdf
Box plots of disease index (DI) variation at different haplotypes of alleles. The central line of box means median, and box limits are the upper and lower quartiles. The significance of difference was analyzed with T-Test and one way ANOVA and in all haplotypes significant difference were observed (P<0.001) (PDF 771 kb)
Rights and permissions
About this article
Cite this article
Ma, Y., Chhapekar, S.S., Lu, L. et al. QTL mapping for Fusarium wilt resistance based on the whole-genome resequencing and their association with functional genes in Raphanus sativus. Theor Appl Genet 134, 3925–3940 (2021). https://doi.org/10.1007/s00122-021-03937-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-021-03937-5