Abstract
Key message
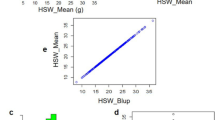
We detected a QTL qHSW-16 undergone strong selection associated with seed weight and identified a novel candidate gene controlling seed weight candidate gene for this major QTL by qRT-PCT.
Abstract
Soybean [Glycine max (L.) Merr.] provides more than half of the world’s oilseed production. To expand its germplasm resources useful for breeding increased yield and oil quality cultivars, it is necessary to resolve the diversity and evolutionary history of this crop. In this work, we resequenced 283 soybean accessions from China and obtained a large number of high-quality SNPs for investigation of the population genetics that underpin variation in seed weight and other agronomic traits. Selective signature analysis detected 78 (~ 25.0 Mb) and 39 (~ 22.60 Mb) novel putative selective signals that were selected during soybean domestication and improvement, respectively. Genome-wide association study (GWAS) identified five loci associated with seed weight. Among these QTLs, qHSW-16, overlapped with the improvement-selective region on chromosome 16, suggesting that this QTL may be underwent strong selection during soybean improvement. Of the 18 candidate genes in qHSW-16, only SoyZH13_16G122400 showed higher expression levels in a large seed variety compared to a small seed variety during seed development. These results identify SoyZH13_16G122400 as a novel candidate gene controlling seed weight and provide foundational insights into the molecular targets for breeding improvement of seed weight and potential seed yield in soybean.






Similar content being viewed by others
References
Caverzan A, Giacomin R, Müller M, Biazus C, Lângaro NC, Chavarria G (2018) How does seed vigor affect soybean yield components? Agron J 110:1318–1327
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13:1194–1202
Chi Z, Shan-Shan D, Jun-Yang X, Wei-Ming H, Tie-Lin Y (2019) PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 10:1786–1788
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R, 1000 Genomes Project Analysis Group (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158
Dashiell K (2005) SOYBEANS: improvement, production, and uses. Third Edition: Boerma, H.R., Specht, J.E. (Eds), American Society of Agronomy, Crop Science Society of America, Soil Science Society of America, Madison, Wisconsin, USA, 2004, 1144 pp. Price: US$155.00 (hardback). ISBN 0-89118-154-7. Agric Syst 83:110–111
Eloy NB, de Freitas Lima M, Van Damme D, Vanhaeren H, Gonzalez N, De Milde L, Hemerly AS, Beemster GT, Inzé D, Ferreira PC (2011) The APC/C subunit 10 plays an essential role in cell proliferation during leaf development. Plant J 68:351–363
Han Y, Li D, Zhu D, Li H, Li X, Teng W, Li W (2012) QTL analysis of soybean seed weight across multi-genetic backgrounds and environments. Theor Appl Genet 125:671–683
Hu Z, Zhang D, Zhang G, Kan G, Hong D, Yu D (2014) Association mapping of yield-related traits and SSR markers in wild soybean (Glycine soja Sieb. and Zucc.). Breeding Sci 63:441–449
Hu D, Zhang H, Du Q, Hu Z, Yang Z, Li X, Wang J, Huang F, Yu D, Wang H, Kan G (2020) Genetic dissection of yield-related traits via genome-wide association analysis across multiple environments in wild soybean (Glycine soja Sieb. and Zucc.). Planta 251:39
Hwang EY, Song Q, Jia G et al (2014) A genome-wide association study of seed protein and oil content in soybean[J]. BMC Genom 15(1):1–12
Kato S, Sayama T, Fujii K, Yumoto S, Kono Y, Hwang T, Kikuchi A, Takada Y, Tanaka Y, Shiraiwa T, Ishimoto M (2014) A major and stable QTL associated with seed weight in soybean across multiple environments and genetic backgrounds. Theor Appl Genet 127:1365–1374
Lam H, Xu X, Liu X, Chen W, Yang G, Wong F, Li M, He W, Qin N, Wang B, Li J, Jian M, Wang J, Shao G, Wang J, Sun SS, Zhang G (2010) Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet 42:1053–1059
Lee G, Crawford GW, Liu L, Sasaki Y, Chen X (2011) Archaeological soybean (Glycine max) in East Asia: does size matter? PLoS ONE 6:e26720
Lee T, Guo H, Wang X, Kim C, Paterson AH (2014) SNPhylo: a pipeline to construct a phylogenetic tree from huge SNP data. BMC Genomics 15:162
Letunic I, Bork P (2019) Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760
Li W, Zheng D (2008) QTL mapping for major agronomic traits across 2 years in soybean. J Crop Sci Biotechnol 11(3):171–190
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome PDPS (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Li YH, Li D, Jiao YQ, Schnable JC, Li YF, Li HH, Chen HZ, Hong HL, Zhang T, Liu B (2020) Identification of loci controlling adaptation in Chinese soya bean landraces via a combination of conventional and bioclimatic GWAS. Plant Biotechnol J 18:389–401
Liang H, Xu L, Yu Y, Yang H, Dong W, Zhang H (2016) Identification of QTLs with main, epistatic and QTL by environment interaction effects for seed shape and hundred-seed weight in soybean across multiple years. J Genet 95:475–477
Lima MDF, Eloy NB, Bottino MC, Hemerly AS, Ferreira PCG (2013) Overexpression of the anaphase-promoting complex (APC) genes in Nicotiana tabacum promotes increasing biomass accumulation. Mol Biol Rep 40:7093–7102
Lin Q, Wang D, Dong H, Gu S, Cheng Z, Gong J, Qin R, Jiang L, Li G, Wang JL, Wu F, Guo X, Zhang X, Lei C, Wang H, Wan J (2012) Rice APC/C-TE controls tillering by mediating the degradation of MONOCULM 1. Nat Commun 3:752
Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z (2012) GAPIT: genome association and prediction integrated tool. Bioinformatics 28:2397–2399
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Lu S, Dong L, Fang C, Liu S, Kong L, Cheng Q, Chen L, Su T, Nan H, Zhang D, Zhang L, Wang Z, Yang Y, Yu D, Liu X, Yang Q, Lin X, Tang Y, Zhao X, Yang X, Tian C, Xie Q, Li X, Yuan X, Tian Z, Liu B, Weller JL, Kong F (2020) Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat Genet 52:428–436
Maughan PJ, Maroof MAS, Buss GR (1996) Molecular-marker analysis of seed-weight: genomic locations, gene action, and evidence for orthologous evolution among three legume species. Theor Appl Genet 93:574–579
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
Mian MAR, Bailey MA, Tamulonis JP, Shipe ER, Carter TE, Parrott WA, Ashley DA, Hussey RS, Boerma HR (1996) Molecular markers associated with seed weight in two soybean populations. Theor Appl Genet 93:1011–1016
Miao L, Yang S, Zhang K, He J, Wu C, Ren Y, Gai J, Li Y (2020) Natural variation and selection in GmSWEET39 affect soybean seed oil content. New Phytol 225:1651–1666
Nielsen R, Williamson S, Kim Y, Hubisz MJ, Clark AG, Bustamante C (2005) Genomic scans for selective sweeps using SNP data. Genome Res 15:1566–1575
Paterson AH, Brubaker CL, Wendel JF (1993) A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol Biol Rep 11:122–127
Penfield S (2017) Seed dormancy and germination. Curr Biol 27:R874–R878
Qi X, Li M, Xie M, Liu X, Ni M, Shao G, Song C, Kay-Yuen Yim A, Tao Y, Wong F, Isobe S, Wong C, Wong K, Xu C, Li C, Wang Y, Guan R, Sun F, Fan G, Xiao Z, Zhou F, Phang T, Liu X, Tong S, Chan T, Yiu S, Tabata S, Wang J, Xu X, Lam H (2014) Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat Commun 5:4340
Rajjou L, Duval M, Gallardo K, Catusse J, Bally J, Job C, Job D (2012) Seed germination and vigor. Annu Rev Plant Biol 63:507–533
Shen Y, Liu J, Geng H, Zhang J, Liu Y, Zhang H, Xing S, Du J, Ma S, Tian Z (2018) De novo assembly of a Chinese soybean genome. Sci China Life Sci 61:871–884
Shin J, Blay S, McNeney B, Graham J (2006) LDheatmap: an R function for graphical display of pairwise linkage disequilibria between single. J Stat Softw 16:1–10
Specht JE, Chase K, Macrander M, Graef GL, Chung J, Markwell JP, Germann M, Orf JH, Lark KG (2001) Soybean response to water: a QTL analysis of drought tolerance. Crop Sci 41:493–509
Tengfei Z, Tingting W, Liwei W, Bingjun J, Caixin Z, Shan Y, Wensheng H, Cunxiang W, Tianfu H, Shi S (2019) A Combined linkage and GWAS analysis identifies QTLs linked to soybean seed protein and oil content. Int J Mol Sci 20:5915
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164
Wang M, Li W, Fang C, Xu F, Liu Y, Wang Z, Yang R, Zhang M, Liu S, Lu S, Lin T, Tang J, Wang Y, Wang H, Lin H, Zhu B, Chen M, Kong F, Liu B, Zeng D, Jackson SA, Chu C, Tian Z (2018) Parallel selection on a dormancy gene during domestication of crops from multiple families. Nat Genet 50:1435
Wu D, Zhan Y, Sun Q, Xu L, Lian M, Zhao X, Han Y, Li W (2018) Identification of quantitative trait loci underlying soybean (Glycine max [L.] Merr.) seed weight including main, epistatic and QTL × environment effects in different regions of Northeast China. Plant Breeding 137:194–202
Xu C, Wang Y, Yu Y, Duan J, Liao Z, Xiong G, Meng X, Liu G, Qian Q, Li J (2012) Degradation of MONOCULM 1 by APC/CTAD1 regulates rice tillering. Nat Commun 3:750
Xu R, Xu J, Wang L, Niu B, Copenhaver GP, Ma H, Zheng B, Wang Y (2019) The Arabidopsis anaphase-promoting complex/cyclosome subunit 8 is required for male meiosis. New Phytol 224:229–241
Yang J, Lee SH, Goddard ME, Visscher PM (2011) GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88:76–82
Zabala G, Vodkin LO (2007) A rearrangement resulting in small tandem repeats in the F3′5′H gene of white flower genotypes is associated with the Soybean W1 Locus. CROP SCI 47:113–124
Zhang W, Liao X, Cui Y, Ma W, Zhang X, Du H, Ma Y, Ning L, Wang H, Huang F, Yang H, Kan G, Yu D (2019) A cation diffusion facilitator, GmCDF1, negatively regulates salt tolerance in soybean. PLoS Genet 15:e1007798
Zheng B, Chen X, McCormick S (2011) Zheng B, Chen X, McCormick S. The anaphase-promoting complex is a dual integrator that regulates both MicroRNA-mediated transcriptional regulation of cyclin B1 and degradation of Cyclin B1 during Arabidopsis male gametophyte development. Plant Cell 23:1033–1046
Zhou Z, Jiang Y, Wang Z, Gou Z, Lyu J, Li W, Yu Y, Shu L, Zhao Y, Ma Y, Fang C, Shen Y, Liu T, Li C, Li Q, Wu M, Wang M, Wu Y, Dong Y, Wan W, Wang X, Ding Z, Gao Y, Xiang H, Zhu B, Lee S, Wang W, Tian Z (2015) Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat Biotechnol 33:125–408
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2018YFE0112200), the Key R&D project of Jiangsu Province (BE2019376).
Author information
Authors and Affiliations
Contributions
ZW, WX, HZ, and SL contributed to field design and phenotypic data collection; ZW and XC performed phenotypic analysis; WX, LS, XL, YZ, and CX assisted in revising the manuscript; ZW analyzed the experimental results; ZW and HT wrote the manuscript. All authors reviewed the manuscript and provided suggestions.
Corresponding authors
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Availability of data and materials
Data provided in supplementary files.
Additional information
Communicated by Istvan Rajcan.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, W., Xu, W., Zhang, H. et al. Comparative selective signature analysis and high-resolution GWAS reveal a new candidate gene controlling seed weight in soybean. Theor Appl Genet 134, 1329–1341 (2021). https://doi.org/10.1007/s00122-021-03774-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-021-03774-6