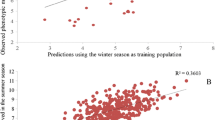
Abstract
Key message
Weighted outperformed unweighted genomic prediction using an unbalanced dataset representative of a commercial breeding program. Moreover, the use of the two cycles preceding predictions as training set achieved optimal prediction ability.
Abstract
Predicting the performance of untested single-cross hybrids through genomic prediction (GP) is highly desirable to increase genetic gain. Here, we evaluate the predictive ability (PA) of novel genomic strategies to predict single-cross maize hybrids using an unbalanced historical dataset of a tropical breeding program. Field data comprised 949 single-cross hybrids evaluated from 2006 to 2013, representing eight breeding cycles. Hybrid genotypes were inferred based on their parents’ genotypes (inbred lines) using single-nucleotide polymorphism markers obtained via genotyping-by-sequencing. GP analyses were fitted using genomic best linear unbiased prediction via a stage-wise approach, considering two distinct cross-validation schemes. Results highlight the importance of taking into account the uncertainty regarding the adjusted means at each step of a stage-wise analysis, due to the highly unbalanced data structure and the expected heterogeneity of variances across years and locations of a commercial breeding program. Further, an increase in the size of the training set was not always advantageous even in the same breeding program. The use of the two cycles preceding predictions achieved optimal PA of untested single-cross hybrids in a forward prediction scenario, which could be used to replace the first step of field screening. Finally, in addition to the practical and theoretical results applied to maize hybrid breeding programs, the stage-wise analysis performed in this study may be applied to any crop historical unbalanced data.




Similar content being viewed by others
References
Albrecht T, Auinger HJ, Wimmer V, Ogutu JO, Knaak C, Ouzunova M, Piepho HP, Schön CC (2014) Genome-based prediction of maize hybrid performance across genetic groups, testers, locations, and years. Theor Appl Genet 127(6):1375–1386
Auinger HJ, Schönleben M, Lehermeier C, Schmidt M, Korzun V, Geiger HH, Piepho HP, Gordillo A, Wilde P, Bauer E et al (2016) Model training across multiple breeding cycles significantly improves genomic prediction accuracy in rye (secale cereale l.). Theor Appl Genet 129(11):2043–2053
Bernal-Vasquez AM, Utz HF, Piepho HP (2016) Outlier detection methods for generalized lattices: a case study on the transition from anova to reml. Theor Appl Genet 129(4):787–804
Bernal-Vasquez AM, Gordillo A, Schmidt M, Piepho HP (2017) Genomic prediction in early selection stages using multi-year data in a hybrid rye breeding program. BMC Genet 18:1–17
Bernardo R (1991) Correlation between testcross performance of lines at early and late selfing generations. Theor Appl Genet 82(1):17–21
Bernardo R (1994) Prediction of maize single-cross performance using rflps and information from related hybrids. Crop Sci 34:20–25
Browning BL, Browning SR (2016) Genotype imputation with millions of reference samples. Am J Hum Genet 98(1):116–126
Burgueño J, de los Campos G, Weigel K, Crossa J (2012) Genomic prediction of breeding values when modeling genotype × environment interaction using pedigree and dense molecular markers. Crop Sci 52(2):707–719
Butler DG, Cullis BR, Gilmour AR, Gogel BJ (2009) ASReml-R reference manual
de los Campos G, Hickey JM, Pong-Wong R, Daetwyler HD, Calus MP (2013) Whole-genome regression and prediction methods applied to plant and animal breeding. Genetics 193(2):327–345
Charcosset A, Lefort-Buson M, Gallais A (1991) Relationship between heterosis and heterozygosity at marker loci: a theoretical computation. Theor Appl Genet 81(5):571–575
Cros D, Tchounke B, Nkague-Nkamba L (2018) Training genomic selection models across several breeding cycles increases genetic gain in oil palm in silico study. Mol Breed 38(7):1–12
Crossa J, Beyene Y, Kassa S, Pérez P, Hickey JM, Chen C, De Los Campos G, Burgueño J, Windhausen VS, Buckler E, et al (2013) Genomic prediction in maize breeding populations with genotyping-by-sequencing. G3: Genes, Genomes, Genetics pp 1903–1926
Crossa J, Pérez-Rodríguez P, Cuevas J, Montesinos-López O, Jarquín D, de los Campos G, Burgueño J, González-Camacho JM, Pérez-Elizalde S, Beyene Y et al (2017) Genomic selection in plant breeding: methods, models, and perspectives. Trends Plant Sci 22(11):961–975
Cullis BR, Smith A, Hunt C, Gilmour A (2000) An examination of the efficiency of australian crop variety evaluation programmes. J Agric Sci 135(3):213–222
Damesa TM, Möhring J, Worku M, Piepho HP (2017) One step at a time: stage-wise analysis of a series of experiments. Agron J 109(3):845–857
Desrousseaux D, Sandron F, Siberchicot A, Cierco-Ayrolles C, Mangin B (2017) LDcorSV: linkage disequilibrium corrected by the structure and the relatedness. R package version 1.3.2. https://CRAN.R-project.org/package=LDcorSV
Dias KODG, Gezan SA, Guimares CT, Nazarian A, Silva LC, Parentoni SN, Guimares PEdO, Anoni CdO, Noda RW, Ribeiro CAG, Magalhes JV, Garcia AAF, Souza JC, Guimares LJM, Pastina MM (2018) Improving accuracies of genomic predictions for drought tolerance in maize by joint modeling of additive and dominance effects in multi-environment trials. Heredity 121:24–37
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (gbs) approach for high diversity species. Plos One 6(5):e19379
Fristche-Neto R, Akdemir D, Jannink JL (2018) Accuracy of genomic selection to predict maize single-crosses obtained through different mating designs. Theor Appl Genet 131(5):1153–1162
Fritsche-Neto R, Gonçalves MC, Vencovsky R, de Souza Junior CL (2010) Prediction of genotypic values of maize hybrids in unbalanced experiments. Crop Breed Appl Biotechnol 10(1):32–39
Garcia AA, Benchimol LL, Barbosa AM, Geraldi IO, Souza CL Jr, Souza APd (2004) Comparison of rapd, rflp, aflp and ssr markers for diversity studies in tropical maize inbred lines. Genet Mol Biol 27(4):579–588
Garrick DJ, Taylor JF, Fernando RL (2009) Deregressing estimated breeding values and weighting information for genomic regression analyses. Genet Sel Evol 41:1–8
Gezan SA, de Carvalho MP, Sherrill J (2017) Statistical methods to explore genotype-by-environment interaction for loblolly pine clonal trials. Tree Genet Genomes 13(1):1–11
Gianola D, de los Campos G, Hill WG, Manfredi E, Fernando R (2009) Additive genetic variability and the bayesian alphabet. Genetics 183(1):347–363
Gilmour AR, Thompson R, Cullis BR (1995) Average information reml: an efficient algorithm for variance parameter estimation in linear mixed models. Biometrics 51:1440–1450
Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, Buckler ES (2014) Tassel-gbs: a high capacity genotyping by sequencing analysis pipeline. Plos One 9(2):e90346
Gorjanc G, Gaynor RC, Hickey JM (2018) Optimal cross selection for long-term genetic gain in two-part programs with rapid recurrent genomic selection. Theor Appl Genet 131(9):1953–1966
Granato IS, Galli G, de Oliveira Couto EG, e Souza MB, Mendonça LF, Fritsche-Neto R (2018) snpready: a tool to assist breeders in genomic analysis. Mol Breed 38(8):1–7
Guo T, Yu X, Li X, Zhang H, Zhu C, Flint-Garcia S, McMullen MD, Holland JB, Szalma SJ, Wisser RJ et al (2019) Optimal designs for genomic selection in hybrid crops. Mol plant 12(3):390–401
Habier D, Fernando R, Dekkers J (2007) The impact of genetic relationship information on genome-assisted breeding values. Genetics 177(4):2389–2397
Habier D, Fernando RL, Garrick DJ (2013) Genomic blup decoded: a look into the black box of genomic prediction. Genetics 194(3):597–607
Kleinknecht K, Möhring J, Singh K, Zaidi P, Atlin G, Piepho H (2013) Comparison of the performance of best linear unbiased estimation and best linear unbiased prediction of genotype effects from zoned indian maize data. Crop Sci 53(4):1384–1391
Li H, Durbin R (2009) Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25(14):1754–1760
Marulanda JJ, Mi X, Melchinger AE, Xu JL, Würschum T, Longin CFH (2016) Optimum breeding strategies using genomic selection for hybrid breeding in wheat, maize, rye, barley, rice and triticale. Theor Appl Genet 129(10):1901–1913
Mendiburu F (2017) Agricolae: statistical procedures for agricultural research. R package version 1.2-8. https://CRAN.R-project.org/package=agricolae
Meuwissen THE, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157(4):1819–1829
Möhring J, Williams ER, Piepho HP (2015) Inter-block information: to recover or not to recover it? Theor Appl Genet 128(8):1541–1554
Piepho H, Büchse A, Emrich K (2003) A hitchhiker’s guide to mixed models for randomized experiments. J Agron Crop Sci 189(5):310–322
Piepho H, Büchse A, Truberg B (2006) On the use of multiple lattice designs and \(\alpha\)-designs in plant breeding trials. Plant Breed 125(5):523–528
Piepho HP (2009) Ridge regression and extensions for genomewide selection in maize. Crop Sci 49(4):1165–1176
Piepho HP, Möhring J (2006) Selection in cultivar trialsis it ignorable? Crop Sci 46(1):192–201
Piepho HP, Möhring J (2007) Computing heritability and selection response from unbalanced plant breeding trials. Genetics 177(3):1881–1888
Piepho HP, Möhring J, Schulz-Streeck T, Ogutu JO (2012) A stage-wise approach for the analysis of multi-environment trials. Biom J 54(6):844–860
R Core Team (2018) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria, http://www.R-project.org/, ISBN 3-900051-07-0
Rincent R, Laloë D, Nicolas S, Altmann T, Brunel D, Revilla P, Rodriguez VM, Moreno-Gonzales J, Melchinger AE, Bauer E, et al (2012) Maximizing the reliability of genomic selection by optimizing the calibration set of reference individuals: comparison of methods in two diverse groups of maize inbreds (zea mays l.). Genetics pp 715–728
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard R (1984) Ribosomal dna spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci 81(24):8014–8018
dos Santos JPR, de Castro Vasconcellos RC, Pires LPM, Balestre M, Von Pinho RG (2016) Inclusion of dominance effects in the multivariate gblup model. Plos One 11(4):e0152045
Schmidt P, Hartung J, Rath J, Piepho HP (2019) Estimating broad-sense heritability with unbalanced data from agricultural cultivar trials. Crop Sci 59(2):525–536
Schopp P, Müller D, Technow F, Melchinger AE (2017) Accuracy of genomic prediction in synthetic populations depending on the number of parents, relatedness, and ancestral linkage disequilibrium. Genetics 205(1):441–454
Schrag TA, Möhring J, Melchinger AE, Kusterer B, Dhillon BS, Piepho HP, Frisch M (2010) Prediction of hybrid performance in maize using molecular markers and joint analyses of hybrids and parental inbreds. Theor Appl Genet 120(2):451–461
Schrag TA, Schipprack W, Melchinger AE (2018) Across-years prediction of hybrid performance in maize using genomics. Theor Appl Genet 131:1–14
Schulz-Streeck T, Ogutu JO, Piepho HP (2013) Comparisons of single-stage and two-stage approaches to genomic selection. Theor Appl Genet 126(1):69–82
Shull GH (1908) The composition of a field of maize. J Hered 4:296–301
Smith A, Cullis B, Gilmour A (2001) Applications: the analysis of crop variety evaluation data in australia. Aust New Zealand J Stat 43(2):129–145
Smith O (1986) Covariance between line per se and testcross performance. Crop Sci 26(3):540–543
Technow F, Schrag TA, Schipprack W, Bauer E, Simianer H, Melchinger AE (2014) Genome properties and prospects of genomic prediction of hybrid performance in a breeding program of maize. Genetics 197(4):1343–1355
VanRaden PM (2008) Efficient methods to compute genomic predictions. J Dairy Sci 91(11):4414–4423
Viana JMS, Pereira HD, Mundim GB, Piepho HP, e Silva FF (2018) Efficiency of genomic prediction of non-assessed single crosses. Heredity 120(4):283
Vieira I, Dos Santos J, Pires L, Lima B, Gonçalves F, Balestre M (2017) Assessing non-additive effects in gblup model. Genet Mol Res 16(2):1–21
Vitezica ZG, Varona L, Legarra A (2013) On the additive and dominant variance and covariance of individuals within the genomic selection scope. Genetics 195(4):1223–1230
Welham SJ, Gogel BJ, Smith AB, Thompson R, Cullis BR (2010) A comparison of analysis methods for late-stage variety evaluation trials. Aust New Zealand J Stat 52(2):125–149
Westhues M, Schrag TA, Heuer C, Thaller G, Utz HF, Schipprack W, Thiemann A, Seifert F, Ehret A, Schlereth A et al (2017) Omics-based hybrid prediction in maize. Theor Appl Genet 130(9):1927–1939
Windhausen VS, Wagener S, Magorokosho C, Makumbi D, Vivek B, Piepho HP, Melchinger AE, Atlin GN (2012) Strategies to subdivide a target population of environments: Results from the cimmyt-led maize hybrid testing programs in africa. Crop Sci 52(5):2143–2152
Acknowledgements
This research was supported by FAPEMIG (Fundação de Amparo á Pesquisa de Minas Gerais), CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, program PREMIO 2045/2014, Grant 23038.007195/2012-39), and Embrapa (Brazilian Agricultural Research Corporation). K.O.G. Dias received a Grant from FAPESP (Fundação de Amparo á Pesquisa do Estado de São Paulo, Grant 2016/12977-7 and 2018/00634-3). AAFG has a productivity scholarship from CNPq. The authors thank Jhonathan Santos, Paul Schmidt and Jens Hartung for careful reading and suggestions on the early draft of the manuscript.
Author information
Authors and Affiliations
Contributions
KOGD: Conceptualization, data curation, methodology, formal analysis, writing the original draft. HPP: conceptualization, methodology, formal analysis, revision and editing. LJMG: conceptualization, funding acquisition, resources, data curation, revision and editing. PEOG: funding acquisition, resources, data curation. SNP: funding acquisition, resources, data curation. MOP: funding acquisition, resources, data curation. RWN: resources, data curation. JVM: funding acquisition, resources, data curation, revision and editing. CTG: funding acquisition, resources, data curation, revision and editing. AAFG: supervision, conceptualization, resources, revision and editing. MMP: supervision, conceptualization, funding acquisition, resources, data curation, revision and editing.
Corresponding authors
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Communicated by Matthias Frisch.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dias, K.O.G., Piepho, H.P., Guimarães, L.J.M. et al. Novel strategies for genomic prediction of untested single-cross maize hybrids using unbalanced historical data. Theor Appl Genet 133, 443–455 (2020). https://doi.org/10.1007/s00122-019-03475-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-019-03475-1