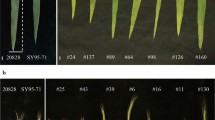
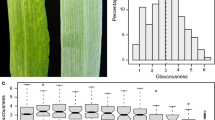
Abstract
Key message
QTLs and candidate gene markers associated with leaf morphological and color traits were identified in two immortalized populations of Brassica rapa, which will provide genetic information for marker-assisted breeding.
Abstract
Brassica rapa is an important leafy vegetable consumed worldwide and morphology is a key character for its breeding. To enhance genetic control, quantitative trait loci (QTLs) for leaf color and plant architecture were identified using two immortalized populations with replications of 2 and 4 years. Overall, 158 and 80 QTLs associated with 23 and 14 traits were detected in the DH and RIL populations, respectively. Among them, 23 common robust-QTLs belonging to 12 traits were detected in common loci over the replications. Through comparative analysis, five crucifer genetic blocks corresponding to morphology trait (R, J&U, F and E) and color trait (F, E) were identified in three major linkage groups (A2, A3 and A7). These might be key conserved genomic regions involved with the respective traits. Through synteny analysis with Arabidopsis, 64 candidate genes involved in chlorophyll biosynthesis, cell proliferation and elongation were co-localized within QTL intervals. Among them, SCO3, ABI3, FLU, HCF153, HEMB1, CAB3 were mapped within QTLs for leaf color; and CYCD3;1, CYCB2;4, AN3, ULT1 and ANT were co-localized in QTL regions for leaf size. These robust QTLs and their candidate genes provide useful information for further research into leaf architecture with crop breeding.





Similar content being viewed by others
References
Albrecht V, Simkova K, Carrie C, Delannoy E, Giraud E, Whelan J, Small ID, Apel K, Badger MR, Pogson BJ (2010) The cytoskeleton and the peroxisomal-targeted snowy cotyledon3 protein are required for chloroplast development in Arabidopsis. Plant Cell 22:3423–3438
Azmach G, Gedil M, Menkir A, Spillane C (2013) Marker-trait association analysis of functional gene markers for provitamin A levels across diverse tropical yellow maize inbred lines. BMC Plant Biol 13:227
Bajaj D, Upadhyaya HD, Khan Y, Das S, Badoni S, Shree T, Kumar V, Tripathi S, Gowda CL, Singh S, Sharma S, Tyagi AK, Chattopdhyay D, Parida SK (2015) A combinatorial approach of comprehensive QTL-based comparative genome mapping and transcript profiling identified a seed weight-regulating candidate gene in chickpea. Sci Rep 5:9264
Barkoulas M, Galinha C, Grigg SP, Tsiantis M (2007) From genes to shape: regulatory interactions in leaf development. Curr Opin Plant Biol 10:660–666
Burroughs SF, Devine DV, Browne G, Kaplan ME (1988) The population of paroxysmal nocturnal hemoglobinuria neutrophils deficient in decay-accelerating factor is also deficient in alkaline phosphatase. Blood 71:1086–1089
Choi SR, Teakle GR, Plaha P, Kim JH, Allender CJ, Beynon E, Piao ZY, Soengas P, Han TH, King GJ, Barker GC, Hand P, Lydiate DJ, Batley J, Edwards D, Koo DH, Bang JW, Park BS, Lim YP (2007) The reference genetic linkage map for the multinational Brassica rapa genome sequencing project. Theor Appl Genet 115:777–792
Chung H, Jeong YM, Mun JH, Lee SS, Chung WH, Yu HJ (2014) Construction of a genetic map based on high-throughput SNP genotyping and genetic mapping of a TuMV resistance locus in Brassica rapa. Mol Genet Genomics 289:149–160
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
De Koeyer D, Douglass K, Murphy A, Whitney S, Nolan L, Song Y, De Jong W (2010) Application of high-resolution DNA melting for genotyping and variant scanning of diploid and autotetraploid potato. Mol Breed 25:67–90
Dekkers BJ, He H, Hanson J, Willems LA, Jamar DC, Cueff G, Rajjou L, Hilhorst HW, Bentsink L (2016) The Arabidopsis DELAY OF GERMINATION 1 gene affects ABSCISIC ACID INSENSITIVE 5 (ABI5) expression and genetically interacts with ABI3 during Arabidopsis seed development. Plant J 85(4):451–465
Dewitte W, Riou-Khamlichi C, Scofield S, Healy JM, Jacqmard A, Kilby NJ, Murray JA (2003) Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15:79–92
Dewitte W, Scofield S, Alcasabas AA, Maughan SC, Menges M, Braun N, Collins C, Nieuwland J, Prinsen E, Sundaresan V, Murray JA (2007) Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc Natl Acad Sci 104:14537–14542
Ge Y, Wang T, Wang N, Wang Z, Liang C, Ramchiary N, Choi SR, Lim YP, Piao ZY (2012) Genetic mapping and localization of quantitative trait loci for chlorophyll content in Chinese cabbage (Brassica rapa ssp. pekinensis). Sci Hortic 147:42–48
Guillemaut P, Maréchal-Drouard L (1992) Isolation of plant DNA: a fast, inexpensive and reliable method. Plant Mol Biol Rep 10:60–65
Guo PG, Baum M, Varshney RK, Graner A, Grando S, Ceccarelli S (2008) QTLs for chlorophyll and chlorophyll fluorescence parameters in barley under post-flowering drought. Euphytica 163:203–214
Horiguchi G, Kim GT, Tsukaya H (2005) The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J 43:68–78
Horiguchi G, Nakayama H, Ishikawa N, Kubo M, Demura T, Fukuda H, Tsukaya H (2011) ANGUSTIFOLIA3 plays roles in adaxial/abaxial patterning and growth in leaf morphogenesis. Plant Cell Physiol 52:112–124
Izhaki A, Bowman JL (2007) KANADI and class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell 19:495–508
Kapoor R, Banga SS, Banga SK (2009) A microsatellite (SSR) based linkage map of Brassica rapa. New Biotechnol 26:239–243
Kebede B, Cheema K, Greenshields DL, Li C, Selvaraj G, Rahman H (2012) Construction of genetic linkage map and mapping of QTL for seed color in Brassica rapa. Genome 55:813–823
Kennard WC, Slocum MK, Figdore SS, Osborn TC (1994) Genetic analysis of morphological variation in Brassica oleracea using molecular markers. Theor Appl Genet 87:721–732
Kim GT, Tsukaya H, Uchimiya H (1998) The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev 12:2381–2391
Kim GT, Tsukaya H, Saito Y, Uchimiya H (1999) Changes in the shapes of leaves and flowers upon overexpression of cytochrome P450 in Arabidopsis. Proc Natl Acad Sci 96:9433–9437
Kim H, Choi SR, Bae J, Hong CP, Lee SY, Hossain MJ, Van Nguyen D, Jin M, Park BS, Bang JW, Bancroft I, Lim YP (2009) Sequenced BAC anchored reference genetic map that reconciles the ten individual chromosomes of Brassica rapa. BMC Genomics 10:432
Kim SM, Suh JP, Lee CK, Lee JH, Kim YG, Jena KK (2014) QTL mapping and development of candidate gene-derived DNA markers associated with seedling cold tolerance in rice (Oryza sativa L.). Mol Gen Genet 289:333–343
Kole C, Kole P, Vogelzang R, Osborn TC (1997) Genetic linkage map of a Brassica rapa recombinant inbred population. J Hered 88:553–557
Kosambi DD (1943) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Li H, Xu L, Wang H, Yuan Z, Cao X, Yang Z, Zhang D, Xu Y, Huang H (2005) The putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and MicroRNA165/166 in Arabidopsis leaf development. Plant Cell 17:2157–2171
Li F, Kitashiba H, Inaba K, Nishio T (2009) A Brassica rapa linkage map of EST-based SNP markers for identification of candidate genes controlling flowering time and leaf morphological traits. DNA Res 16:311–323
Li X, Ramchiary N, Choi SR, Van Nguyen D, Hossain MJ, Yang HK, Lim YP (2010) Development of a high density integrated reference genetic linkage map for the multinational Brassica rapa Genome Sequencing Project. Genome 53:939–947
Li X, Ramchiary N, Dhandapani V, Choi SR, Hur Y, Nou IS, Yoon MK, Lim YP (2013) Quantitative trait loci mapping in Brassica rapa revealed the structural and functional conservation of genetic loci governing morphological and yield component traits in the A, B, and C subgenomes of Brassica species. DNA Res 20:1–16
Lou P, Zhao J, Kim JS, Shen S, Del Carpio DP, Song X, Jin M, Vreugdenhil D, Wang X, Koornneef M, Bonnema G (2007) Quantitative trait loci for flowering time and morphological traits in multiple populations of Brassica rapa. J Exp Bot 58:4005–4016
Lu G, Cao J, Yu X, Xiang X, Chen H (2008) Mapping QTLs for root morphological traits in Brassica rapa L. based on AFLP and RAPD markers. J Appl Genet 49:23–31
Mehrnia M, Balazadeh S, Zanor MI, Mueller-Roeber B (2013) EBE, an AP2/ERF transcription factor highly expressed in proliferating cells, affects shoot architecture in Arabidopsis. Plant Physiol 162:842–857
Mizukami Y, Fischer RL (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci 97:942–947
Mocoeur A, Zhang YM, Liu ZQ, Shen X, Zhang LM, Rasmussen SK, Jing HC (2015) Stability and genetic control of morphological, biomass and biofuel traits under temperate maritime and continental conditions in sweet sorghum (Sorghum bicolour). Theor Appl Genet 128:1685–1701
Panigrahi J, Patnaik A, Kole P, Koleb C (2009) Addition of restriction fragment length polymorphism markers to the genetic linkage map of Brassica rapa L. (syn. campestris). Z. Naturforschung C 64:882–890
Poczai P, Varga I, Laos M, Cseh A, Bell N, Valkonen JP, Hyvonen J (2013) Advances in plant gene-targeted and functional markers: a review. Plant Methods 9:6
Ramchiary N, Nguyen VD, Li X, Hong CP, Dhandapani V, Choi SR, Yu G, Piao ZY, Lim YP (2011) Genic microsatellite markers in Brassica rapa: development, characterization, mapping, and their utility in other cultivated and wild Brassica relatives. DNA Res 18:305–320
Rawson H, Constable G (1980) Carbon production of sunflower cultivars in field and controlled environments. I. Photosynthesis and transpiration of leaves, stems and heads. Funct Plant Biol 7:555–573
Schranz ME, Lysak MA, Mitchell-Olds T (2006) The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci 11:535–542
Shen Q, Wang YT, Tian H, Guo FQ (2013) Nitric oxide mediates cytokinin functions in cell proliferation and meristem maintenance in Arabidopsis. Mol Plant 6:1214–1225
Shi ZY, Wang J, Wan XS, Shen GZ, Wang XQ, Zhang JL (2007) Over-expression of rice OsAGO7 gene induces upward curling of the leaf blade that enhanced erect-leaf habit. Planta 226:99–108
Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews GN, Bowman JL (1999) Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126:4117–4128
Stam P (1993) Construction of integrated genetic linkage maps by means of a new computer package: join map. Plant J 3:739–744
Van Ooijen J (2011) Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Genet Res 93:343–349
Veldboom LR, Lee M, Woodman WL (1994) Molecular marker-facilitated studies in an elite maize population: I. Linkage analysis and determination of QTL for morphological traits. Theor Appl Genet 88:7–16
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wang FH, Wang GX, Li XY, Huang JL, Zheng JK (2008) Heredity, physiology and mapping of a chlorophyll content gene of rice (Oryza sativa L.). J Plant Physiol 165:324–330
Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun JH, Bancroft I, Cheng F, Huang S, Li X, Hua W, Wang J, Wang X, Freeling M, Pires JC, Paterson AH, Chalhoub B, Wang B, Hayward A, Sharpe AG, Park BS, Weisshaar B, Liu B, Li B, Liu B, Tong C, Song C, Duran C, Peng C, Geng C, Koh C, Lin C, Edwards D, Mu D, Shen D, Soumpourou E, Li F, Fraser F, Conant G, Lassalle G, King GJ, Bonnema G, Tang H, Wang H, Belcram H, Zhou H, Hirakawa H, Abe H, Guo H, Wang H, Jin H, Parkin IA, Batley J, Kim JS, Just J, Li J, Xu J, Deng J, Kim JA, Li J, Yu J, Meng J, Wang J, Min J, Poulain J, Wang J, Hatakeyama K, Wu K, Wang L, Fang L, Trick M, Links MG, Zhao M, Jin M, Ramchiary N, Drou N, Berkman PJ, Cai Q, Huang Q, Li R, Tabata S, Cheng S, Zhang S, Zhang S, Huang S, Sato S, Sun S, Kwon SJ, Choi SR, Lee TH, Fan W, Zhao X, Tan X, Xu X, Wang Y, Qiu Y, Yin Y, Li Y, Du Y, Liao Y, Lim Y, Narusaka Y, Wang Y, Wang Z, Li Z, Wang Z, Xiong Z, Zhang Z, Brassica rapa Genome Sequencing Project C (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Gen 43:1035–1039
Wang W, Tang W, Ma T, Niu D, Jin JB, Wang H, Lin R (2016) A pair of light signaling factors FHY3 and FAR1 regulates plant immunity by modulating chlorophyll biosynthesis. J Integr Plant Biol 58:91–103
Yu X, Wang H, Zhong W, Bai J, Liu P, He Y (2013) QTL mapping of leafy heads by genome resequencing in the RIL population of Brassica rapa. PLoS ONE 8:e76059
Yu X, Choi SR, Dhandapani V, Rameneni JJ, Li X, Pang W, Lee JY, Lim YP (2016) Quantitative trait loci for morphological traits and their association with functional genes in Raphanus sativus. Front Plant Sci 7:255
Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468
Zhang J, Lu Y, Yuan Y, Zhang X, Geng J, Chen Y, Cloutier S, McVetty PB, Li G (2009a) Map-based cloning and characterization of a gene controlling hairiness and seed coat color traits in Brassica rapa. Plant Mol Biol 69:553–563
Zhang KP, Fang ZJ, Liang Y, Tian JC (2009b) Genetic dissection of chlorophyll content at different growth stages in common wheat. J Genet 88:183–189
Zhao J, Jamar DC, Lou P, Wang Y, Wu J, Wang X, Bonnema G, Koornneef M, Vreugdenhil D (2008) Quantitative trait loci analysis of phytate and phosphate concentrations in seeds and leaves of Brassica rapa. Plant Cell Environ 31:887–900
Acknowledgments
This research was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) through Golden Seed Project, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (213006-05-1-SB110).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Communicated by Lixi Jiang.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Choi, S.R., Yu, X., Dhandapani, V. et al. Integrated analysis of leaf morphological and color traits in different populations of Chinese cabbage (Brassica rapa ssp. pekinensis). Theor Appl Genet 130, 1617–1634 (2017). https://doi.org/10.1007/s00122-017-2914-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-017-2914-4