Abstract
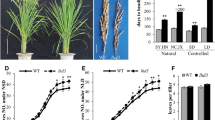
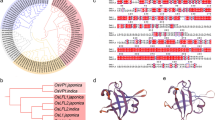
In rice (Oryza sativa), a short-day plant, photoperiod is the most favorable external signal for floral induction because of the constant seasonal change throughout the years. Compared with Arabidopsis, however, a large part of the regulation mechanism of the photoperiodic response in rice still remains unclear due mainly to the lack of induced mutant genes. An induced mutant line X61 flowers 35 days earlier than its original variety Gimbozu under a natural photoperiod in Kyoto (35°01′N). We attempted to identify the mutant gene conferring early heading to X61. Experimental results showed that the early heading of X61 was conferred by a complete loss of photoperiodic response due to a novel single recessive mutant gene se13. This locus interacts with two crucial photoperiod sensitivity loci, Se1 and E1. Wild type alleles at these two loci do not function in coexistence with se13 in a homozygous state, suggesting that Se13 is an upstream locus of the Se1 and E1 loci. Linkage analysis showed that Se13 is located in a 110 kb region between the two markers, INDEL3735_1 and INDEL3735_3 on chromosome 1. A database search suggested that the Se13 gene is identical to AK101395 (=OsHY2), which encodes phytochromobilin synthase, a key enzyme in phytochrome chromophore biosynthesis. Subsequent sequence analysis revealed that X61 harbors a 1 bp insertion in exon 1 of OsHY2, which induces a frame-shift mutation producing a premature stop codon. It is therefore considered that the complete loss of photoperiodic response of X61 is caused by a loss of function of the Se13 (OsHY2) gene involved in phytochrome chromophore biosynthesis.








Similar content being viewed by others
References
Basu D, Dehesh K, Schneider-Poetsch HJ, Harrington SE, McCouch SR, Quail PH (2000) Rice PhyC gene: structure, expression, map-position and evolution. Plant Mol Biol 44:27–42
Clack T, Mathews S, Sharrock RA (1994) The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of phyD and phyE. Plant Mol Biol 25:413–427
Hayama R, Coupland G (2004) The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiol 135:677–684
Ichitani K, Okumoto Y, Tanisaka T (1998) Genetic analyses of low photoperiod sensitivity of rice varieties from the northernmost regions of Japan. Plant Breed 117:543–547
Ichitani K, Inoue H, Nishida H, Okumoto Y, Tanisaka T (2002) Interactive effects of two heading-time loci, Se1 and Ef1. on preflowering developmental phase in rice (Oryza sativa L.). Euphytica 126:227–234
Izawa T, Oikawa T, Tokutomi S, Okuno K, Shimamoto K (2000) Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant). Plant J 22:391–399
Izawa T, Takahashi Y, Yano M (2003) Comparative biology comes into bloom: genomic and genetic comparison of flowering pathways in rice and Arabidopsis. Curr Opin Plant Biol 6:113–120
Kobayashi Y, Weigel D (2007) Move on up, it’s time for change—mobile signals controlling photoperiod-dependent flowering. Genes Dev 21:2371–2384
Kochi T, Mukougawa K, Frankenberg N, Masuda M, Yokota A, Lagarias JC (2001) The Arabidopsis Hy2 gene encodes phytochromobilin synthase, a ferredoxin-dependent biliverdin reductase. Plant Cell 13:425–436
Komari T (1989) Transformation of callus cultures of nine plant species mediated by Agrobacterium. Plant Sci 60:223–229
Koornneef M, Alonso-Blanco C, Peeters AJ, Soppe W (1998) Genetic control of flowering time in Arabidopsis. Annu Rev Plant Physiol Plant Mol Biol 49:345–370
Lin HZ, Liang W, Sasaki T, Yano M (2003) Fine mapping and characterization of quantitative trait loci Hd4 and Hd5 controlling heading date in rice. Breed Sci 53:51–59
Monden Y, Naito K, Okumoto Y, Saito H, Oki N, Tsukiyama T, Ideta O, Nakazaki T, Wessler SR, Tanisaka T (2009) High potential of a transposon mPing as a marker system in japonica x japonica cross in rice. DNA Res 16:131–140
Muramoto T, Kochi T, Yokota A, Hwang I, Goodman HM (1999) The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. Plant Cell 11:335–348
Nishida H, Okumoto Y, Nakagawa H, Ichitani K, Inoue H, Tanisaka T (2001) Analysis of tester lines for rice (Oryza sativa L.) Heading-time genes using reciprocal photoperiodic transfer treatments. Ann Bot 88:527–536
Nishida H, Inoue H, Okumoto Y, Tanisaka T (2002) A novel gene ef1-h conferring an extremely long basic vegetative growth period in rice. Crop Sci 42:348–354
Okumoto Y, Ichitani K, Inoue H, Tanisaka T (1996) Photoperiod insensitivity gene essential to the varieties grown in the northern limit region of paddy rice (Oryza sativa L.) cultivation. Euphytica 92:63–66
Poonyarit M, Mackill DJ, Vergara VS (1989) Genetics of photoperiod sensitivity and critical day length in rice. Crop Sci 29:647–652
Quail PH, Boylan MT, Parks BM, Short TW, Xu Y, Wagner D (1995) Phytochromes: photo sensor perception and signal transduction. Science 268:675–680
Reeves PH, Coupland G (2001) Analysis of flowering time control in Arabidopsis by comparison of double and triple mutants. Plant Physiol 126:1085–1091
Saito H, Yuan Q, Okumoto Y, Doi K, Yoshimura A, Inoue H, Teraishi M, Tsukiyama T, Tanisaka T (2009) Multiple alleles at Early flowering 1 locus making variation in the basic vegetative growth period in rice (Oryza sativa L.). Theor Appl Genet 119:315–323
Sano Y (1992) Genetic comparisons of chromosome 6 between wild and cultivated rice. Jpn J Breed 42:561–572
Sharrock RA, Quail PH (1989) Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution and differential expression of a plant regulatory photoreceptor family. Genes Dev 3:1745–1757
Takano M, Kanegae H, Shinomura T, Miyao A, Hirochika H, Furuya M (2001) Isolation and characterization of rice phytochrome A mutants. Plant Cell 13:521–534
Takano M, Inagaki N, Xie X, Yuzurihara N, Hihara F, Ishizuka T, Yano M, Nishimura M, Miyao A, Hirochika H, Shinomura T (2005) Distinct and cooperative functions of phytochrome A, B and C in the control of de-etiolation and flowering in rice. Plant cell 17:3311–3325
Tanisaka T, Inoue H, Uozu S, Yamagata H (1992) Basic vegetative growth and photoperiod sensitivity of heading-time mutants. Jpn J Breed 42:657–668 (in Japanese with English summary)
Terry MT (1997) Phytochrome chromophore-deficient mutants. Plant Cell Environ 20:740–745
Terry MJ, McDowell MT, Lagarias JC (1995) (3Z)- and (3E)-phytochromobilin are intermediates in the biosynthesis of the phytochrome chromophore. J Biol Chem 270:11111–11118
Tsai KH (1995) Genetic analysis for heading time in wild rice strains. Jpn J Genet 70:555–562
Turck F, Fornara F, Coupland G (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59:573–594
Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303:1003–1006
Wang S, Basten CJ, Zeng ZB (2005) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC
Weller JL, Terry MJ, Reid JB, Kendrick RE (1997) The phytochrome-deficient pcd2 mutant of pea is unable to convert biliverdin IXα to 3Z-phytochromobilin. Plant J 11:1177–1186
Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, Zhang Q (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40:761–767
Yamagata H, Okumoto Y, Tanisaka T (1986) Analysis of genes controlling heading time in Japanese rice. In “Rice genetics”. IRRI, Manila, Philippines, pp 351–359
Yano M, Ebitani T (2002) Development of a series of chromosome segment substitution lines and their utilization in the genetic analysis of quantitative traits in rice. NIAS Annual Report, pp 27–28
Yano M, Harushima Y, Nagamura Y, Kurata N, Minobe Y, Sasaki T (1997) Identification of quantitative trait loci controlling heading date in rice using a high-density linkage map. Theor Appl Genet 95:1025–1032
Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto T, Umehara Y, Nagamura Y, Sasaki T (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12:2473–2484
Yanovsky MJ, Kay SA (2002) Molecular basis of seasonal time measurement in Arabidopsis. Nature 419:308–312
Yokoo M, Okuno K (1993) Genetic analysis of earliness mutations induced in the rice cultivar Norin 8. Jpn J Breed 43:1–11
Yokoo M, Kikuchi F, Nakane A, Fujimaki H (1980) Genetical analysis of heading time by aid of close linkage with blast resistance in rice. Bull Natl Inst Agric Sci D31:95–126 (in Japanese with English summary)
Yuan Q, Saito H, Okumoto Y, Inoue H, Nishida H, Tsukiyama T, Teraishi M, Tanisaka T (2009) Identification of a novel gene ef7 conferring an extremely long basic vegetative growth phase in rice. Theor Appl Genet 119:675–684
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by L. Xiong.
Rights and permissions
About this article
Cite this article
Saito, H., Okumoto, Y., Yoshitake, Y. et al. Complete loss of photoperiodic response in the rice mutant line X61 is caused by deficiency of phytochrome chromophore biosynthesis gene. Theor Appl Genet 122, 109–118 (2011). https://doi.org/10.1007/s00122-010-1426-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-010-1426-2