Abstract
Cultivated tomato (Solanum lycopersicum, syn. Lycopersicon esculentum) is susceptible to the necrotrophic ascomycete and causal agent of gray mold, Botrytis cinerea. Resistance to this fungal pathogen is elevated in wild relatives of tomato, including Solanum lycopersicoides. An introgression line population (IL) containing chromosomal segments of S. lycopersicoides within the background of tomato cv. VF36 was used to screen the genome for foliar resistance and susceptibility to B. cinerea. Based on this screen, putative quantitative trait loci (QTL) were identified, five for resistance and two for susceptibility. Four resistance QTL decreased infection frequency while the fifth reduced lesion diameter. One susceptibility QTL increased infection frequency whereas the other increased lesion diameter. Overlapping chromosomal segments provided strong evidence for partial resistance on chromosomes 1 and 9 and for elevated susceptibility on chromosome 11. Segregation analysis confirmed the major resistance QTL on the long arm of chromosome 1 and susceptibility on chromosome 11. Linkage of partial resistance to chromosome 9 could not be confirmed. The usefulness of these data for resistance breeding and for map-based cloning of foliar resistance to B. cinerea is discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Botrytis cinerea (teleomorph: Botryotina fuckeldiana (de Bary) Whetzel) is a necrotrophic ascomycete that attacks more than 200 host species (Elad et al. 2004). This fungus infects various fruit, flower, and vegetable crops in the field and/or after harvest. The cultivated tomato, Solanum lycopersicum (formerly Lycopersicon esculentum), is an agronomically important host, but variation for resistance to B. cinerea among cultivars is small. Sources for resistance to B. cinerea have therefore been sought among wild relatives of tomato. Solanum lycopersicoides exhibits high resistance to B. cinerea (Rick 1987; Rick and Chetelat 1995, Chetelat et al. 1997). Importantly, accession LA2951, which was used to develop a population of introgression lines (ILs) (Canady et al. 2005), proved to express strong foliar resistance to B. cinerea (Guimaraes et al. 2004).
Other resistance traits are important for tomato production. Specifically for greenhouse production, stem resistance is a critical trait because pruning provides sites for fungal penetration. Hence, Solanum habrochaites (formerly L. hirsutum) and Solanum neorickii (formerly L. parviflorum) have been used to identify quantitative trait loci (QTL) for stem resistance to B. cinerea (Finkers et al. 2007, 2008). QTL mapping has also been done in Arabidopsis thaliana (Denby et al. 2004; Rowe and Kliebenstein 2008) and, although the genes that are responsible for quantitative resistance to B. cinerea remain at large, camalexin has been shown to contribute to defense against this fungus (Kliebenstein et al. 2005; Rowe and Kliebenstein 2008). Approximately half of the resistance QTL recently identified were associated with camalexin accumulation (Rowe and Kliebenstein 2008). Arabidopsis studies provide valuable background information that may aid in the identification of candidate genes that control resistance QTL in crop species such as tomato. The relationship to induced phytoalexin biosynthesis is particular intriguing in light of rishitin’s role in UV protection of fruits against B. cinerea (Charles et al. 2008).
The objectives of the present study were to locate resistance factors from S. lycopersicoides using a nearly complete set of ILs. Based on a genome-wide screen for foliar resistance to B. cinerea, three chromosomal regions were selected for further characterization in segregating progeny. The results presented herein establish the ILs as a potentially useful resource for B. cinerea resistance breeding in tomato.
Materials and methods
Biological material
Introgression lines (ILs) of an original backcross Solanum lycopersicum cv. ‘VF36’ × Solanum lycopersicoides ‘LA2951’ were used for linkage mapping of resistance to B. cinerea. The development and description of this population of ILs have been reported (Canady et al. 2005). Seeds of the IL’s and parental lines as well as in vitro-propagated clones of the F1 hybrid were provided by the Tomato Genetics Resource Center (TGRC), University of California, Davis. B. cinerea strain B05.10 was a gift of Jan van Kan (Wageningen University, The Netherlands).
Growth conditions
Seeds were treated with bleach (Guimaraes et al. 2004) and sown in soil (Sunshine Mix SB40, Sungrow). Plants were grown in a greenhouse with supplemental lighting. Day-time and night-time temperatures were set at 21 and 16°C, respectively. Slow release fertilizer (Osmocote 15-9-12) was added to the soil after 1 month of growth. Seedlings were grown for a minimum of 6–8 weeks prior to leaf removal for inoculation trials. A completely randomized design was used to grow seedlings in 5 × 5 flats consisting of 0.5 l pots. Multiple flats were used per experiment. Consecutive numbers were given to the seedlings that were planted. It was, therefore possible to conduct “blind” assays because identification of genotypes was avoided.
B. cinerea was grown on potato dextrose agar from conidia stored at −80°C. The fungus was grown in darkness for 3–4 days and then exposed to fluorescent light (34 μmol m−2 s−1) to promote sporulation. Conidia were harvested according to published protocols (Benito et al. 1998; Guimaraes et al. 2004).
Foliar inoculation
For the genome wide screen, a minimum of five and ten individuals per homozygous and heterozygous IL were screened for resistance to B. cinerea, respectively. The youngest, fully expanded leaves (typically the fourth leaf from the apex) were detached from the plant and inserted into moist florist foam to maintain turgidity. Prior to inoculation, conidia (106 ml−1) were incubated in Gamborg’s medium containing 10 mM sucrose and 10 mM potassium phosphate, pH 6 for 2–3 h at room temperature. A total of ten droplets (103 conidia inoculum−1) were applied per leaf. Inoculated leaves were maintained in clear plastic boxes at room temperature under saturating humidity and a 12 h light/12 h dark regime at 34 μmol m−2 s−1. Each box contained both S. lycopersicum ‘VF36’ and S. lycopersicoides ‘LA2951’ as controls. Lesions were evaluated 2–4 days after inoculation based on two parameters. First, the frequency of lesion expansion was based on the ability of the fungus to spread beyond the initial necrotic region that is confined to a diameter of 2 mm. This assessment provides an indicator of healthy versus diseased leaf tissues. Second, lesion diameters were measured using a caliper.
For the marker cosegregation analysis a minimum of 50 segregating progeny of the IL’s were inoculated using the same inoculation procedure, but with two modifications. A total of ten droplets (5 × 102 conidia inoculum−1) were applied to each leaf, and the frequency and diameter of lesions expanding more than 3 mm were recorded.
Statistical analysis
The SAS (Cary, NC, USA) program package was used to analyze all of the data. Data analysis of the frequency of expanding lesions was based on a binomial distribution using a generalized linear model (GENMOD). Average numbers of infections per leaf over 2–3 days period (3–4 days for the cosegregation test) were used as response variables per ten inoculations. IL was used as a covariate and the model was based on a logistic regression. The DSCALE option was used to limit overdispersion of the model.
The fixed effect model of an analysis of variance (ANOVA) was used to determine differences in lesion diameter among ILs. Levene’s Test was used to assess the homogeneity of variances and only if the data passed this test, contrasts and least squares means (LSMEANS) were used to identify significant differences among means. Each date of evaluation was tested separately.
DNA Extraction
For Southern blot and CAPS marker analysis, DNA extraction was performed according to the protocol of Miklas et al. (1993) with minor modifications. DNA samples were quantified using a UV fluorimeter (Hoefer).
Marker analysis
Southern blots were prepared by digesting DNA from the ILs, VF36, LA2951, and F1 with EcoRI followed by electrophoresis on 1% agarose gels. PCR primers derived from cloned and sequenced tomato RFLP’s (Table 3) were used to amplify fragments from the VF36 parent. Amplification conditions were optimized using a gradient thermocycler (BioRad) to determine appropriate annealing temperatures (Table 3). The PCR profile consisted of a 3 min denaturing at 94°C followed by 35 cycles of 30 s at 94°C, 45 s annealing, and 2 min at 72°C, a final extension step at 72°C for 7 min, and then held at 4°C. Fragments were gel purified and labeled with the AlkPhos labeling kit (Amersham Biosciences). Hybridization and signal detection were performed using the manufacturer’s instructions.
CAPS markers were generated by PCR amplification of the parents and F1 using the same RFLP primers, followed by restriction digests of aliquots of the amplification products with enzymes, such as BamHI, BsrDI, DraIII, HaeIII, HindIII, NotI, and XhoI. After visualization on agarose gels, the restriction enzymes that allowed identification of the parental and F1 genotypes were used for subsequent genotyping of the IL progeny (Table 3).
Results
Screening of the S. lycopersicoides genome for foliar resistance to B. cinerea
S. lycopersicoides ILs were screened for resistance to B. cinerea over a period of more than 2 years (Table 1). Seven trials were conducted to evaluate all of the ILs, which together represent more than 96% of the map units in the S. lycopersicoides genome (Canady et al. 2005). A total of 58 ILs were tested, 49 of those belonging to a ‘primary set’ of ILs (Canady et al. 2005). This allowed for complete coverage of the S. lycopersicoides genome with the exception of a region encompassing the marker TG65 on chromosome 4 (Fig. 1) and two additional small segments on chromosomes 2 and 3 (Canady et al. 2005). Given that S. lycopersicoides restricts the formation of expanding lesions (Guimaraes et al. 2004), all of the seven trials included data on frequencies of lesion expansion (Table 1). In addition, diameters of expanding lesions were recorded during the last four trials (Table 2).
Screen of the S. lycopersicoides genome for resistance to B. cinerea. Introgression lines of the a primary and b secondary set were screened. Columns indicate chromosomes with markers (not to scale), rows indicate introgression lines. Lines in bold were tested at least twice. Italicized lines in bold were only scored as hyper-resistant once out of two trials (IL-2) or twice out of three trials (IL-9B). Solid and hatched boxes indicate homozygous and heterozygous segments, respectively. Reduction in frequency of infection and rate of lesion expansion are color-coded red and yellow, respectively. Elevation in frequency of infection and rate of lesion expansion are color-coded blue and purple, respectively. The consensus shown beneath all introgression lines shows the location of resistance and susceptibility QTL. In each of these indicated consensus regions differences in phenotypes outnumbered similarities to S. lycopersicum cv. VF36
Six S. lycopersicoides ILs were significantly more resistant to B. cinerea than S. lycopersicum ‘VF36’ when frequency of infection was used as a variable. Three of these lines were verified in two independent trials. These lines are referred to as IL-1B and IL-1C because they contain introgressed segments from chromosome 1, and as IL-9A with an introgressed segment of chromosome 9. Another line, IL-9B, was more resistant to B. cinerea than ‘VF36’ when 16 individuals were tested during the second trial (Table 1), but this line was not significantly different from the tomato parent when eight or five individuals were tested in 2003 and 2005, respectively. IL-9A and IL-9B share a polymorphism for the marker TG9 and, therefore, contain a region of overlap (Canady et al. 2005). Nevertheless, the phenotype of IL-9A was more robust and the magnitude of the phenotypic effect was larger in this line than in IL-9B. This suggests either that a susceptibility locus is located on the chromosomal segment that was introgressed into IL-9B or that more than one resistance locus is present on the region that was introgressed into IL-9A. The former explanation appears more likely because line IL-9C (LS10-6A) contains part of the chromosomal region of IL-9B and has a phenotype that is similar to tomato ‘VF36’ (Fig. 1). Moreover, IL-3, which shares a region containing TG18 with IL-9A, is resistant to B. cinerea. These data suggest that a quantitative trait locus (QTL) is located near the ‘top’ of chromosome 9. IL-2 was evaluated as resistant to B. cinerea when 16 individuals were analyzed during the summer but not when seven individuals were analyzed during the winter of 2003. It is possible that there is a seasonal influence or resistance at this locus. We have observed that resistance of S. lycopersicoides can weaken during winter months under conditions in Oregon, but we have not analyzed this phenomenon further. Collectively, these data suggest that QTL controlling frequency of infection by B. cinerea are located on chromosomes 1, 2, and 9, and possibly on chromosome 3.
Four S. lycopersicoides ILs were significantly more susceptible to B. cinerea than S. lycopersicum ‘VF36’ when frequency of infection was evaluated. IL-11A and IL-11C both contain introgressed segments located on chromosome 11 and share segments with IL-11B, an IL which is more susceptible to B. cinerea than the tomato parent based on lesion expansion (Table 2). A higher frequency of expanding lesions was also observed for lines IL-1A and IL-8A.
Besides IL-11B, three other lines, IL-5, IL-8B, and IL-12, were more susceptible than tomato ‘VF36’ when lesion expansion was evaluated (Table 2). Based on the consensus of all lines evaluated, a susceptibility QTL may be present on chromosome 5 (Fig. 1), but this inference is solely based on the phenotype of IL-5 because no other line contained the S. lycopersicoides polymorphism for CD64 (Canady et al. 2005).
IL-4 was scored as more resistant to B. cinerea than ‘VF36’ based on lesion expansion data. The consensus data of all the lines suggests a resistance QTL is linked to CT50 on chromosome 4. In hindsight, it would have been advisable to include LA4250 (Canady et al. 2005) for comparison with IL-4 to verify this prediction. However, line LA4250 contains additional introgressed segments from chromosomes 5, 8, and 9. LA4250 was not included in the original screen because the introgressed region was covered by IL-4.
Segregation analysis of a foliar B. cinerea resistance QTL on chromosome 1
Based on genome-wide analysis of resistance to B. cinerea, a QTL altering infection frequency was identified on chromosome 1 (Table 1). Fewer expanding lesions were observed when accession IL-1B was challenged with B. cinerea compared to tomato cv. ‘VF36’. In order to determine whether this difference in frequency of infection was linked to marker TG17 on chromosome 1, a segregation analysis was performed in the progeny of heterozygous IL-1B. TG17 is located at the bottom of an approximately 43 cM-long segment that has been introgressed into IL-1B (Canady et al. 2005). Segments introgressed from S. lycopersicoides recombine at only a very low rate with corresponding tomato chromosomes (Canady et al. 2006). Recombination rates are generally only 0–10% of expected frequencies (i.e. 90–100% suppressed). In the absence of significant recombination, single markers are sufficient to genotype each IL in segregating populations.
Segregation of TG17 was not significantly different from the Mendelian 1:2:1 expectation. Individuals carrying the S. lycopersicoides allele were significantly more resistant to B. cinerea both in terms of infection frequency and in terms of lesion diameter (Table 4). Resistance of the heterozygous progeny to B. cinerea appeared intermediate relative to homozygous segregants. Nevertheless, heterozygotes were significantly more resistant than homozygotes carrying the S. lycopersicum allele (Table 4). Conversely, resistance of heterozygotes to B. cinerea was not significantly different from homozygotes carrying the S. lycopersicoides allele. With respect to lesion diameter, no significant differences in resistance were observed between heterozygous and homozygous progenies, indicating that genetic control of this trait may be different from infection frequency.
Accession IL-1B apparently harbors a major QTL linked to marker TG17. Infection frequency of homozygotes for the S. lycopersicoides allele was reduced by 55% relative to the difference between tomato cv. VF36 and S. lycopersicoides LA2951 (Fig. 2a). This approximation compared favorably with estimates that were obtained during the genome-wide screen for B. cinerea resistance (Table 1). In the latter case, infection frequency was reduced by 45 and 23% during 2003 and 2005 seasons. More importantly, segregation analysis demonstrates that the QTL on chromosome 1 explains approximately 21% of the phenotypic variation (Table 4). By comparison, the effect on lesion size was smaller (11% of the phenotypic variation) and not observed during the genome-wide screen, perhaps a result of the smaller sample size or lower expression of the trait during the 2005 season.
Segregation for resistance to B. cinerea on chromosome 1. a Numbers of expanding lesions and b lesion diameters were scored among progeny of heterozygous IL-1B and compared to the parents LA2951 and VF36. Alleles of S. lycopersicum cv. ‘VF36’ and S. lycopersicoides line LA2951 are abbreviated + and S, respectively. Heterozygous +/S and homozygous +/+ and S/S individuals were identified using the CAPS marker TG17/HindIII. Means, standard errors, and sample sizes are indicated. Significant differences to +/+ genotypes are indicated at P < 0.05 (*) and P < 0.001 (***)
Resistance to B. cinerea is not linked to TG18 in IL-9A
Genome wide analysis of resistance to B. cinerea indicated a QTL decreasing infection frequency on chromosome 9. To determine linkage between marker TG18 and this trait, homozygous IL-9A and cv. VF36 were crossed, then self-pollinated to generate a segregating population. TG18 is located 14 cM from the top of chromosome 9. The segment that has been introduced into IL-9A is approximately 18 cM in size (Canady et al. 2005).
Segregation of TG18 in the F2 progeny of IL-9A differed significantly from the 1:2:1 expectation. Marker segregation was skewed in favor of the allele from cultivated tomato. Infection frequency (Fig. 3a) and lesion diameter (Fig. 3b) did not significantly differ among the three genotypes that were defined by TG18 segregation. Nevertheless, all the three IL-9A genotypes were significantly more resistant to B. cinerea than cv. VF36. Both infection frequency and lesion diameter were reduced (Fig. 3). The magnitude of reduction in an infection frequency was similar to the 50 and 53% reduction observed during the genome-wide screen (Table 1). The progeny was not genotyped based on phenotype, but on molecular marker data. In the absence of discernible phenotypic segregation, the most parsimonious explanation is that this partial resistance locus is not linked to the segment on chromosome 9 but located elsewhere in the S. lycopersicoides genome even though markers used by Canady et al. (2005) did not detect the presence of other introgressed segments in IL-9A.
Segregation analysis for resistance to B. cinerea on chromosome 9. a Numbers of expanding lesions and b log N-transformed lesion diameters were scored among progeny of heterozygous IL-9A and compared to the parents LA2951 and VF36. +/+ is homozygous for the VF36 allele, S/S is homozygous for the LA2951 allele, and +/S is heterozygous based on CAPS marker TG18/BamHI. Means, standard errors, and sample sizes are indicated
Analysis of a putative susceptibility locus linked to TG46 on chromosome 11
Genome wide analysis of the interactions between S. lycopersicoides-derived ILs and B. cinerea indicated a susceptibility locus on chromosome 11. Marker TG46 was used to analyze segregation for susceptibility to B. cinerea in progeny of heterozygous line IL-11C. TG46 is located approximately in the middle of a ca. 50 cM segment introgressed into IL-11C. Surprisingly, seedlings homozygous for the S. lycopersicoides allele were not recovered between germinating and surviving individuals. Segregation of TG46 was not significantly different from an expectation of 1:2:0. Although we do not know whether embryo lethality occurs in this genotype because we did not attempt embryo rescue, homozygosity at this locus still causes a selective disadvantage.
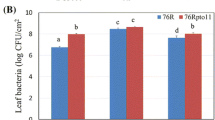
The remaining genotypes were analyzed for susceptibility to B. cinerea (Fig. 4). Frequency of infection was not significantly different between genotypes that were heterozygous or homozygous for the S. lycopersicum allele. Lesion diameters were elevated in the heterozygous genotype compared to individuals homozygous for the S. lycopersicum allele and cv. VF36, but this difference was not statistically significant. It might be argued that a one-tailed test could be used based on the expectation of the genome-wide screen. In this case, the heterozygous genotype would be scored as susceptible. When compared to cv. VF36, susceptibility was clearly elevated (Fig. 4), probably a reflection of the larger number of cultivar seedlings tested. Lesion size was 25% larger in the case of IL-11C when compared in the heterozygous state to cv. VF36. Collectively, these data let us conclude that susceptibility locus exists on chromosome 11.
Segregation for susceptibility to B. cinerea on chromosome 11. Genotypes homozygous for the LA2951 allele were not recovered. a Numbers of expanding lesions and b lesion diameters were scored among the remaining progeny of heterozygous IL-11C and compared to the parents LA2951 and VF36. +/+ is homozygous for the VF36 alelle, and +/S is heterozygous based on the CAPS marker TG46/HaeIII. Means, standard errors, and sample sizes are indicated. Significant difference to VF36 is indicated at P < 0.01 (**)
Discussion
Analysis of foliar susceptibility to B. cinerea among 58 ILs derived from S. lycopersicoides has resulted in identification of four putative resistance QTL for infection frequency and one putative resistance QTL for lesion diameter. In addition, putative susceptibility QTL for infection frequency or lesion diameter was identified. Several of these loci are referred to Fbc for foliar B. cinerea resistance or susceptibility in Fig. 5.
Synopsis of resistance and susceptibility loci mapped to chromosomes 1–5 and to chromosome 11. The genetic map has previously been published (Canady et al. 2005). Solid vertical bars delimit locations of putative foliar B. cinerea (Fbc) resistance QTL on chromosomes 1 through 4 or susceptibility QTL on chromosomes 5 and 11. Approximate locations of previously reported qualitative resistance genes (R genes) or resistance QTL (dashed vertical lines or underlined) are also shown. Resistance loci on chromosomes 3 and 4 have previously been designated (Finkers et al. 2008); QTL3 and 4 were renamed to Snbc3 and 4 in reference to Solanum neorickii. Following references assign remaining resistance loci: Cf9 (Parniske et al. 1997), Rbcq1 and 2 (Finkers et al. 2007), Cm1-1 (Sandbrink et al. 1995), rx-2 (Yu et al. 1995), Tm-1 (Young and Tanksley 1989), Bs4 (Ballvora et al. 2001), Pto and Prf (Salmeron et al. 1996), lb5b and lb11b (Brouwer and St Clair 2004), Sm (Behare et al. 1991), and I2 (Ori et al. 1994)
Colocation of foliar B. cinerea resistance and susceptibility QTL with known resistance loci
Several of the putative foliar B. cinerea resistance QTL identified herein are linked to previously recognized QTL for stem resistance to B. cinerea. The closest association was observed between Fbc3 (Fig. 5) and Snbc3, the latter from S. habrochaites (Finkers et al. 2008). Although we did not find other regions of overlap, Fbc2 and Fbc4 are closely linked to Rbcq2 and Rbcq4b, respectively (Finkers et al. 2007). Homologous QTL for stem resistance to B. cinerea were previously identified (Finkers et al. 2008), suggesting that a limited number of QTL specify resistance to B. cinerea in different Solanum species. Collectively, data on stem resistance (Finkers et al. 2008; Finkers et al. 2007) and on foliar resistance from S. lycopersicoides presented here indicate genomic specialization for B. cinerea resistance on chromosomes 1, 2, 3, 4, and 9 of Solanum. Nevertheless, comparative analysis (Fig. 5) also demonstrates that stem and foliar resistance are controlled by separate QTL. The map locations of resistance genes and QTL controlling interactions with other pathogens are shown in Fig. 5, but due to the large sizes of introgressed S. lycopersicoides fragments, any inferences about potential coincidences would be speculative.
Confirmed QTL for foliar resistance or susceptibility to B. cinerea
Three putative QTL were characterized further because overlapping segments of ILs strongly suggested the presence of loci that have a quantitative influence on interactions with B. cinerea in leaves. Fbc1, a major QTL for foliar resistance to B. cinerea, was identified in this fashion. This QTL was initially identified based on its reduction of infection frequency (Table 1). Fbc1 explains approximately 21% of the phenotypic variation in susceptibility to B. cinerea, which is larger than the magnitudes of previously reported stem resistance QTL (Finkers et al. 2007, 2008). Analysis of a segregating IL-1B population also revealed a significant difference in lesion diameter between homozygous progenies, but this effect was much smaller than the infection frequency (Table 4). The relatively small effect on lesion size probably explains why this trait was not detected in the previous genome-wide screen (Tables 1, 2). Based on the large effect on infection frequency, Fbc1 would make a good candidate for fine-scale mapping and eventual cloning of the resistance gene. Although suppression of recombination in ILs represents a genetic barrier (Canady et al. 2005), S. pennellii has successfully been used as a bridge species to overcome this barrier and increase the rate of recombination between S. lycopersicoides and cultivated tomato (Canady et al. 2006).
A second resistance QTL was examined in detail based on the observation that IL-9A and IL-9B, with overlapping fragments of chromosome 9, were more resistant to B. cinerea. Surprisingly, segregation of resistance was not observed using marker TG18 in progeny of heterozygous IL-9A, although all of the progeny were more resistant than tomato cv. VF36 (Fig. 3). This suggests that the resistance locus either exists somewhere else in the genome or that linkage between TG18 and the QTL has been broken. With respect to the former possibility, it is interesting to note that markers used for mapping ILs were separated by 18.4 cM on average with a range of 2.5–64 cM (Canady et al. 2005). Although Canady et al. (2005) reported introgression of a single segment on chromosome 9 in IL-9A, it can therefore not be excluded that an unlinked small piece was introduced from the S. lycopersicoides genome. This could explain our inability to detect segregation for partial resistance to B. cinerea using TG18.
Genome wide screening for B. cinerea susceptibility indicated that the chromosomal region of S. lycopersicoides in IL-11C increases infection frequency (Table 1). However, segregation analysis showed that Lbc11 significantly increased lesion diameter. Lesion size was not evaluated during the primary screen. An effect on lesion diameter may, therefore, have been missed during this earlier assessment. The population size of the segregating progeny was larger than the number of individuals used for the primary screen. Quantitative differences in lesion diameter are therefore assumed to be reliable. Variability between disease incidence and lesion growth was also observed for QTL9 from S. neorickii (Finkers et al. 2008). Although the authors argued that in their case QTL9 was expected to reduce disease incidence, the cause for different experimental observations in separate screens was not resolved. The size of the phenotypic effect appeared to be relatively small in the case of QTL9 (Finkers et al. 2008). Similarly, Fbc11 appeared to be a relatively minor QTL, at least in comparison to Fbc1. Thus, in the case of minor QTL, large samples appear to be needed to reliable assign resistance or susceptibility traits. However, in the case of Fbc11 no surviving homozygotes for the S. lycopersicoides were recovered, possibly indicating a major defect-related to plant development and defense.
Conclusion
Our studies confirm that S. lycopersicoides is a potent source of foliar resistance to B. cinerea. Several partially resistant ILs were identified in this study. These ILs could be used in combination with marker-assisted selection to develop improved tomato cultivars.
References
Ballvora A, Pierre M, van den Ackerveken G, Schornack S, Rossier O, Ganal M, Lahaye T, Bonas U (2001) Genetic mapping and functional analysis of the tomato Bs4 locus governing recognition of the Xanthomonas campestris pv. vesicatoria AvrBs4 protein. Mol Plant Microbe Interact 14:629–638
Behare J, Laterrot H, Sarfatti M, Zamir D (1991) Restriction-fragment-length-polymorphism mapping of the Stemphylium resistance gene in tomato. Mol Plant Microbe Interact 4:489–492
Benito E, ten Have A, van’t Klooster J, van Kan J (1998) Fungal and plant gene expression during synchronized infection of tomato leaves by Botrytis cinerea. Eur J Plant Pathol 104:207–220
Brouwer DJ, St Clair DA (2004) Fine mapping of three quantitative trait loci for late blight resistance in tomato using near isogenic lines (NILs) and sub-NILs. Theor Appl Genet 108:628–638
Canady MA, Meglic V, Chetelat RT (2005) A library of Solanum lycopersicoides introgression lines in cultivated tomato. Genome 48:685–697
Canady MA, Ji Y, Chetelat RT (2006) Homeologous recombination in Solanum lycopersicoides introgression lines of cultivated tomato. Genetics 174:1775–1788
Charles MT, Mercier J, Makhlouf J, Arul J (2008) Physiological basis of UV-C-induced resistance to Botrytis cinerea in tomato fruit—I. Role of pre- and post-challenge accumulation of the phytoalexin-rishitin. Postharvest Biol Technol 47:10–20
Chetelat RT, Cisneros P, Stamoa L, Rick CM (1997) A male-fertile Lycopersicon esculentum × Solanum lycopersicoides hybrid enables direct backcrossing to tomato at the diploid level. Euphytica 95:99–108
Denby KJ, Kumar P, Kliebenstein DJ (2004) Identification of Botrytis cinerea susceptibility loci in Arabidopsis thaliana. Plant J 38:473–486
Elad Y, Williamson B, Tudzynski P, Delen N (2004) Botrytis: biology, pathology, and control. Kluwer, Dordrecht
Finkers R, van den Berg P, van Berloo R, ten Have A, van Heusden AW, van Kan JAL, Lindhout P (2007) Three QTLs for Botrytis cinerea resistance in tomato. Theor Appl Genet 114:585–593
Finkers R, Bai YL, van den Berg P, van Berloo R, Meijer-Dekens F, ten Have A, van Kan J, Lindhout P, van Heusden AW (2008) Quantitative resistance to Botrytis cinerea from Solanum neorickii. Euphytica 159:83–92
Guimaraes RL, Chetelat RT, Stotz HU (2004) Resistance to Botrytis cinerea in Solanum lycopersicoides is dominant in hybrids with tomato, and involves induced hyphal death. Eur J Plant Pathol 110:13–23
Kliebenstein DJ, Rowe HC, Denby KJ (2005) Secondary metabolites influence Arabidopsis/Botrytis interactions: variation in host production and pathogen sensitivity. Plant J 44:25–36
Miklas PN, Delorme R, Riley R (1993) Identification of QTL conditioning resistance to white mold in snap bean. J Am Soc Hortic Sci 128:564–570
Ori N, Paran I, Aviv D, Eshed Y, Tanksley S, Zamir D, Fluhr R (1994) A genomic search for the gene conferring resistance to Fusarium wilt in tomato. Euphytica 79:201–204
Parniske M, Hammond-Kosack KE, Goldstein C, Thomas CM, Jones DA (1997) Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91:821–832
Rick C (1987) Genetic resources in Lycopersicon. In: Nevins DJ, Jones RA (eds) Plant biology. Alan R. Liss, New York, pp 17–26
Rick C, Chetelat R (1995) Utilization of related wild species for tomato improvement. Acta Hort 412:21–38
Rowe HC, Kliebenstein DJ (2008) Complex genetics control natural variation in Arabidopsis thaliana resistance to Botrytis cinerea. Genetics 180:2237–2250
Salmeron JM, Oldroyd GED, Rommens CMT, Scofield SR, Kim H-S, Lavelle DT, Dahlbeck D, Staskawicz BJ (1996) Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell 86:123–133
Sandbrink JM, van Ooijen JW, Purimahua CC, Vrielink M, Verkerk R, Zabel P, Lindhout P (1995) Localization of genes for bacterial canker resistance in Lycopersicon peruvianum using RFLPs. Theor Appl Genet 90:444–450
Young ND, Tanksley SD (1989) RFLP analysis of the size of chromosomal segments retained around the Tm-2 locus of tomato during backcross breeding. Theor Appl Genet 77:353–359
Yu ZH, Wang JF, Stall RE, Vallejos CE (1995) Genomic localization of tomato genes that control a hypersensitive reaction to Xanthomonas campestris pv. vesicatoria (Doidge) dye. Genetics 141:675–682
Acknowledgments
We thank Rejane Guimarães for advice in resistance screening. The California Tomato Commission provided financial support.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by I. Paran.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Davis, J., Yu, D., Evans, W. et al. Mapping of loci from Solanum lycopersicoides conferring resistance or susceptibility to Botrytis cinerea in tomato. Theor Appl Genet 119, 305–314 (2009). https://doi.org/10.1007/s00122-009-1039-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-009-1039-9