Abstract
Transcriptional regulation remains one of the most intriguing and challenging subjects in biomedical research. The catalysis of transcription is a clear example of multiple proteins interacting to orchestrate a biological process, offering a starting point for the study of biological systems. Transcriptional regulation is viewed as one of the principal mechanisms governing the spatial and temporal distribution of gene expression, thus the field of transcriptional regulation provides a natural stage for quantitative studies of multiple gene systems. Building on the body of focused experimental studies and new genomics-driven data, computational biologists are making significant strides in accelerating our understanding of the transcriptional regulatory process in metazoan cells. Recent advances in the computational analysis of the interplay between factors have been fueled by well-defined computational methods for the modeling of the binding of individual transcription factors. We present here an overview of advances in the analysis of regulatory systems and the fundamental methods that underlie the recent developments.




Similar content being viewed by others
References
Anderson JD, Widom J (2000) Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J Mol Biol 296(4):979–987
Arnone MI, Davidson EH (1997) The hardwiring of development: organization and function of genomic regulatory systems. Development 124(10):1851–1864
Bailey TL, Elkan C (1995) Unsupervised learning of multiple motifs in biopolymers using expectation maximization. Machine Learning 21:51–83
Benos PV, Bulyk ML, Stormo GD (2002) Additivity in protein-DNA interactions: how good an approximation is it? Nucleic Acids Res 30(20):4442–4451
Berman BP, Nibu Y, Pfeiffer BD, Tomancak P, Celniker SE, Levine M, Rubin GM, Eisen MB (2002) Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc Natl Acad Sci USA 99(2):757–762
Bird AP (1987) CpG islands as gene markers in the vertebrate nucleus. Trends Genet 3(12):342–347
Blackwood EM, Kadonaga JT (1998) Going the distance: a current view of enhancer action. Science 281:61–63
Blanchette M, Tompa M (2002) Discovery of regulatory elements by a computational method for phylogenetic footprinting. Genome Res 12(5):739–748
Blanchette M, Schwikowski B, Tompa M (2000) An exact algorithm to identify motifs in orthologous sequences from multiple species. Proc Int Conf Intell Syst Mol Biol 8:37–45
Bode J, Benham C, Knopp A, Mielke C (2000) Transcriptional augmentation: modulation of gene expression by scaffold/matrix-attached regions (S/MAR elements). Crit Rev Eukaryot Gene Expr 10(1):73–90
Boutanaev AM, Kalmykova AI, Shevelyov YY, Nurminsky DI (2002) Large clusters of co-expressed genes in the Drosophila genome. Nature 420(6916):666–669
Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA (2001) The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet 10(3):211–219
Brazma A, Jonassen I, Vilo J, Ukkonen E (1998) Predicting gene regulatory elements in silico on a genomic scale. Genome Res 8(11):1202–1215
Bucher P (1990) Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J Mol Biol 212:563–579
Bulyk ML, Johnson PL, Church GM (2002) Nucleotides of transcription factor binding sites exert interdependent effects on the binding affinities of transcription factors. Nucleic Acids Res 30(5):1255–1261
Bussemaker HJ, Li H, Siggia ED (2000) Building a dictionary for genomes: identification of presumptive regulatory sites by statistical analysis. Proc Natl Acad Sci USA 97(18):10096–10100
Caron H, Schaik B van, Mee M van der, Baas F, Riggins G, Sluis P van, Hermus MC, Asperen R van, Boon K, Voute PA, Heisterkamp S, Kampen A van, Versteeg R (2001) The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science 291(5507):1289–1292
Chung I, Bresnick E (1997) Identification of positive and negative regulatory elements of the human cytochrome P4501A2 (CYP1A2) gene. Arch Biochem Biophys 338(2):220–226
Claverie JM (2000) From bioinformatics to computational biology. Genome Res 10:1277–1279
Cohen BA, Mitra RD, Hughes JD, Church GM (2000) A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat Genet 26(2):183–186
Cremer T, Cremer C (2001) Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet 2(4):292–301
Cremer T, Kreth G, Koester H, Fink RH, Heintzmann R, Cremer M, Solovei I, Zink D, Cremer C (2000) Chromosome territories, interchromatin domain compartment, and nuclear matrix: an integrated view of the functional nuclear architecture. Crit Rev Eukaryot Gene Expr 10(2):179–212
Crowley EM, Roeder K, Bina M (1997) A statistical model for locating regulatory regions in genomic DNA. J Mol Biol 268(1):8–14
Davidson EH (2001) Genomic regulatory systems: development and evolution. Academic Press, San Diego
Davuluri RV, Grosse I, Zhang MQ (2001) Computational identification of promoters and first exons in the human genome. Nat Genet 29(4):412–417
Dempsey AA, Pabalan N, Tang HC, Liew CC (2001) Organization of human cardiovascular-expressed genes on chromosomes 21 and 22. J Mol Cell Cardiol 33(3):587–591
Down TA, Hubbard TJ (2002) Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res 12(3):458–461
Dundr M, Misteli T (2001) Functional architecture in the cell nucleus. Biochem J 356(2):297–310
Fickett JW (1996a) Quantitative discrimination of MEF2 sites. Mol Cell Biol 16:437–441
Fickett JW (1996b) Coordinate positioning of MEF2 and myogenin binding sites. Gene 172(1):GC19–32
Fickett JW, Hatzigeorgiou AG (1997) Eukaryotic promoter recognition. Genome Res 7(9):861–878
Fickett JW, Wasserman WW (2000) Discovery and modeling of transcriptional regulatory regions. Curr Opin Biotechnol 11:19–24
Fournier B, Gutzwiller S, Dittmar T, Matthias G, Steenbergh P, Matthias P (2001) Estrogen receptor (ER)-α, but not ER-β, mediates regulation of the insulin-like growth factor I gene by antiestrogens. J Biol Chem 276(38):35444–35449
Frech K, Danescu-Mayer J, Werner T (1997) A novel method to develop highly specific models for regulatory units detects a new LTR in GenBank which contains a functional promoter. J Mol Biol 270(5):674–687
Frisch M, Frech K, Klingenhoff A, Cartharius K, Liebich I, W (2002) In silico prediction of scaffold/matrix attachment regions in large genomic sequences. Genome Res 12(2):349–354
Frith MC, Hansen U, Weng Z (2001) Detection of cis-element clusters in higher eukaryotic DNA. Bioinformatics 17(10):878–889
Frith MC, Spouge JL, Hansen U, Weng Z (2002) Statistical significance of clusters of motifs represented by position-specific scoring matrices in nucleotide sequences. Nucleic Acids Res 30(14):3214–3224
Gardiner-Garden M, Frommer M (1987) CpG islands in vertebrate genomes. J Mol Biol 196(2):261–282
Gregori C, Porteu A, Lopez S, Kahn A, Pichard AL (1998) Characterization of the aldolase B intronic enhancer. J Biol Chem 273(39):25237–25243
GuhaThakurta D, Stormo GD (2001) Identifying target sites for cooperatively binding factors. Bioinformatics 17(7):608–621
Halfon MS, Michelson AM (2002) Exploring genetic regulatory networks in metazoan development methods and models. Physiol Genomics 10(3):131–143
Halfon MS, Grad Y, Church GM, Michelson AM (2002) Computation-based discovery of related transcriptional regulatory modules and motifs using an experimentally validated combinatorial model. Genome Res 12(7):1019–1028
Hannenhalli S, Levy S (2001) Promoter prediction in the human genome. Bioinformatics 17 [suppl 1]:S90–S96
Hannenhalli S, Levy S (2002) Predicting transcription factor synergism. Nucleic Acids Res 30(19):4278–4284
Helden J van, Andre B, Collado-Vides J (1998) Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J Mol Biol 281(5):827–842
Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO (2001) Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409(6819):533–538
Jackson DA (1997) Chromatin domains and nuclear compartments: establishing sites of gene expression within eukaryotic nuclei. Mol Biol Rep 24(3):209–220
Kadonaga JT (1998) Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell 92(3):307–313
Kel AE, Kel-Margoulis OV, Farnham PJ, Bartley SM, Wingender E, Zhang MQ (2001) Computer-assisted identification of cell cycle-related genes: new targets for E2F transcription factors. J Mol Biol 309(1):99–120
Klingenhoff A, Frech K, Quandt K, Werner T (1999) Functional promoter modules can be detected by formal models independent of overall nucleotide sequence similarity. Bioinformatics 15(3):180–186
Kornberg RD, Lorch Y (1999) Twenty-five years of the nucleosome: fundamental particle of the eukaryote chromosome. Cell 98(3):285–294
Kraus RJ, Murray EE, Wiley SR, Zink NM, Loritz K, Celembiuk GW, Mertz JE (1996) Experimentally determined weight matrix definitions of the initiator and TBP binding site elements of promoters. Nucleic Acids Res 24:1531–1539
Krivan W, Wasserman WW (2001) A predictive model for regulatory sequences directing liver-specific transcription. Genome Res 11(9):1559–1666
Kruglyak S, Tang H (2000) Regulation of adjacent yeast genes. Trends Genet 16(3):109–111
Lawrence CE, Reilly A (1990) An EM algorithm for the identification and characterization of common sites in unaligned biopolymers sequence. Proteins 7:41–51
Lawrence CE, Altschul SF, Boguski MS, Liu JS, Neuwald AF, Wootton JC (1993) Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science 262:208–214
Lemon B, Tjian R (2000) Orchestrated response: a symphony of transcription factors for gene control. Genes Dev 14(20):2551–2569
Lercher MJ, Urrutia AO, Hurst LD (2002) Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat Genet 31(2):180–183
Levitsky VG, Podkolodnaya OA, Kolchanov NA, Podkolodny NL (2001) Nucleosome formation potential of eukaryotic DNA: calculation and promoters analysis. Bioinformatics 17(11):998–1010
Levy S, Hannenhalli S (2002) Identification of transcription factor binding sites in the human genome sequence. Mamm Genome 13(9):510–514
Levy S, Hannenhalli S, Workman C (2001) Enrichment of regulatory signals in conserved non-coding genomic sequence. Bioinformatics 17(10):871–877
Liu R, States DJ (2002) Consensus promoter identification in the human genome utilizing expressed gene markers and gene modeling. Genome Res 12(3):462–469
Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, Rubin EM, Frazer KA (2000) Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 288:136–140
Loots GG, Ovcharenko I, Pachter L, Dubchak I, Rubin EM (2002) rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res 12(5): 832–839
Markstein M, Markstein P, Markstein V, Levine MS (2002) Genome-wide analysis of clustered dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc Natl Acad Sci USA 99(2):763–768
McCue L, Thompson W, Carmack C, Ryan MP, Liu JS, Derbyshire V, Lawrence CE (2001) Phylogenetic footprinting of transcription factor binding sites in proteobacterial genomes. Nucleic Acids Res 29(3):774–782
McCue LA, Thompson W, Carmack CS, Lawrence CE (2002) Factors influencing the identification of transcription factor binding sites by cross-species comparison. Genome Res 12(10):1523–1532
McGuire AM, Hughes JD, Church GM (2000) Conservation of DNA regulatory motifs and discovery of new motifs in microbial genomes. Genome Res 10(6):744–757
McNally JG, Muller WG, Walker D, Wolford R, Hager GL (2000) The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287(5456):1262–1265
Michelson AM (2002) Deciphering genetic regulatory codes: a challenge for functional genomics. Proc Natl Acad Sci USA 99:546–548
Ohler U, Niemann H (2001) Identification and analysis of eukaryotic promoters: recent computational approaches. Trends Genet 17(2):56–60
Ohler U, Niemann H, Liao GC, Rubin GM (2001) Joint modeling of DNA sequence and physical properties to improve eukaryotic promoter recognition. Bioinformatics 17 [suppl 1]:S199–S206
Pearce D, Matsui W, Miner JN, Yamamoto KR (1998) Glucocorticoid receptor transcriptional activity determined by spacing of receptor and non-receptor DNA sites. J Biol Chem 273:30081–30085
Pedersen AG, Baldi P, Chauvin Y, Brunak S (1999) The biology of eukaryotic promoter predictiona review. Comput Chem 23(3–4):191–207
Polach KJ, Widom J (1995) Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J Mol Biol 254(2):130–149
Polach KJ, Widom J (1996) A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. J Mol Biol 258(5):800–812
Rajewsky N, Socci ND, Zapotocky M, Siggia ED (2002a) The evolution of DNA regulatory regions for proteo-gamma bacteria by interspecies comparisons. Genome Res 12(2):298–308
Rajewsky N, Vergassola M, Gaul U, Siggia ED (2002b) Computational detection of genomic cis-regulatory modules applied to body patterning in the early Drosophila embryo. BMC Bioinformatics 3(1):30
Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, Volkert TL, Wilson CJ, Bell SP, Young RA (2000) Genome-wide location and function of DNA binding proteins. Science 290(5500):2306–2309
Roth FP, Hughes JD, Estep PW, Church GM (1998) Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat Biotechnol 16(10):939–945
Roulet E, Busso S, Camargo AA, Simpson AJ, Mermod N, Bucher P (2002) High-throughput SELEX SAGE method for quantitative modeling of transcription-factor binding sites. Nat Biotechnol 20(8):831–835
Roy PJ, Stuart JM, Lund J, Kim SK (2002) Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature 418(6901):975–979
Scherf M, Klingenhoff A, Frech K, Quandt K, Schneider R, Grote K, Frisch M, Gailus-Durner V, Seidel A, Brack-Werner R, Werner T (2001) First pass annotation of promoters on human chromosome 22. Genome Res 11(3):333–340
Schneider TD, Stephens RM (1990) Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 18(20):6097–6100
Shannon CE (1948) A mathematical theory of communication. Bell Syst Tech J 27:379–423, 623–56
Shim EY, Woodcock C, Zaret KS (1998) Nucleosome positioning by the winged helix transcription factor HNF3. Genes Dev 12:5–10
Simpson ER, Michael MD, Agarwal VR, Hinshelwood MM, Bulun SE, Zhao Y (1997) Cytochromes P450 11: expression of the CYP19 (aromatase) gene: an unusual case of alternative promoter usage. FASEB J 11(1):29–36
Skalnikova M, Kozubek S, Lukasova E, Bartova E, Jirsova P, Cafourkova A, Koutna I, Kozubek M (2000) Spatial arrangement of genes, centromeres and chromosomes in human blood cell nuclei and its changes during the cell cycle, differentiation and after irradiation. Chromosome Res 8(6):487–499
Smale ST (1997) Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim Biophys Acta 1351(1–2):73–88
Spellman PT, Rubin GM (2002) Evidence for large domains of similarly expressed genes in the Drosophila genome. J Biol 1(1):5
Stormo GD (2000) DNA binding sites: representation and discovery. Bioinformatics 16(1):16–23
Stormo GD, Fields DS (1998) Specificity, free energy and information content in protein-DNA interactions. Trends Biochem Sci 23:109–113
Stormo GD, Tan K (2002) Mining genome databases to identify and understand new gene regulatory systems. Curr Opin Microbiol 5(2):149–153
Tan K, Moreno-Hagelsieb G, Collado-Vides J, Stormo GD (2001) A comparative genomics approach to prediction of new members of regulons. Genome Res 11(4):566–584
Tautz D (2000) Evolution of transcriptional regulation. Curr Opin Genet Dev 10(5):575–579
Thijs G, Moreau Y, De Smet F, Mathys J, Lescot M, Rombauts S, Rouzé P, De Moor B, Marchal K (2002a) INCLUSive: integrated clustering, upstream sequence retrieval and motif sampling. Bioinformatics 18(2):331–332
Thijs G, Marchal K, Lescot M, Rombauts S, De Moor B, Rouze P, Moreau Y (2002b) A Gibbs sampling method to detect overrepresented motifs in the upstream regions of coexpressed genes. J Comput Biol 9(2):447–464
Tronche F, Ringeisen F, Blumenfeld M, Yaniv M, Pontoglio M (1997) Analysis of the distribution of binding sites for a tissue-specific transcription factor in the vertebrate genome. J Mol Biol 266(2):231–245
Udalova IA, Mott R, Field D, Kwiatkowski D (2002) Quantitative prediction of NF-kappa B DNA-protein interactions. Proc Natl Acad Sci USA 99(12):8167–8172
Wagner A (1999) Genes regulated cooperatively by one or more transcription factors and their identification in whole eukaryotic genomes. Bioinformatics 15(10):776–784
Wasserman WW, Fickett JW (1998) Identification of regulatory regions which confer muscle-specific gene expression. J Mol Biol 278(1):167–181
Wasserman WW, Palumbo M, Thompson W, Fickett JW, Lawrence CE (2000) Human–mouse genome comparisons to locate regulatory sites. Nat Genet 26:225–228
Werner T (1999) Models for prediction and recognition of eukaryotic promoters. Mamm Genome 10(2):168–175
Wolberger C (1999) Multiprotein–DNA complexes in transcriptional regulation. Annu Rev Biophys Biomol Struct 28:29–56
Wolffe AP, Guschin D (2000) Review: chromatin structural features and targets that regulate transcription. J Struct Biol 129(2–3):102–122
Workman CT, Stormo GD (2000) ANN-Spec: a method for discovering transcription factor binding sites with improved specificity. Proc Pac Symp Biocomputing 2000:467–478
Wu J, Grunstein M (2000) 25 years after the nucleosome model: chromatin modifications. Trends Biochem Sci 25(12):619–623
Xing EP, Wu W, Karp RM (2003) Capturing characteristic structural features for motif detection using a hierarchical Bayesian Markovian model. Genome Biol
Zhu JL, Kaytor EN, Pao CI, Meng XP, Phillips LS (2000) Involvement of Sp1 in the transcriptional regulation of the rat insulin-like growth factor-1 gene. Mol Cell Endocrinol 164:205–218
Acknowledgements
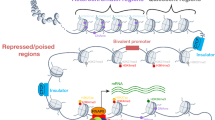
We are grateful for suggestions from David A. Adler, James L. Holloway, Charles E. Lawrence, Richard H. Price, Gary D. Stormo, Marissa Vignali, and the members of the Wasserman research group. We are indebted to Carol Sattler for providing the electron micrograph in the background of Fig. 1, which was created by Edward J. Andrews (SomeLabDesign, Seattle, WA, USA). We also thank David A. Adler for help with Fig. 1.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wasserman, W.W., Krivan, W. In silico identification of metazoan transcriptional regulatory regions. Naturwissenschaften 90, 156–166 (2003). https://doi.org/10.1007/s00114-003-0409-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00114-003-0409-4