Abstract
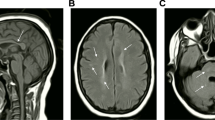
DEAD-box helicase 53 (DDX53) is a member of the DEAD-box protein family of RNA helicases. Unlike other family members that are responsible for RNA metabolism, the biological function of DDX53 and its impact on the human condition are unclear. Herein, we found a full-length DDX53 deletion mutation in a hereditary spastic paraplegia-like (HSP-like) patient with lower extremity spasticity, walking disorder, visual impairment, and lateral ventricular white matter lesions. Bioinformatic analysis revealed that DDX53 was mainly expressed in the cerebellar cortex and may function as a tissue-specific RNA helicase. Transcriptome analysis showed that the expression of multiple brain-associated genes involved in synapse organization, neuron function, and neuromuscular junctions was affected by DDX53 depletion. Moreover, RNA immunoprecipitation sequencing (RIP-seq) analysis showed that DDX53 interacted with 176 genes, and 96 of these genes were associated with the execution of neurofunction, particularly in the regulation of cell projection organization and nervous system development. Collectively, although a more specified cell or animal model is required to fully understand the functional role of DDX53 in the human brain, we report for the first time that the patient with DDX53 defects exhibits HSP-like symptoms and that DDX53 is essential for maintaining neuronal function, with loss-of-function mutation in DDX53 potentially leading to HSP due to impaired RNA metabolism in the nervous system.
Key messages
-
DDX53 deficiency was first reported to be associated with HSP disorder.
-
DDX53 exhibited minimal impact on mitochondrial function.
-
DDX53 impaired RNA metabolism in the nervous system.






Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Shribman S, Reid E, Crosby AH, Houlden H, Warner TT (2019) Hereditary spastic paraplegia: from diagnosis to emerging therapeutic approaches. Lancet Neurol 18(12):1136–1146
Meyyazhagan A, Orlacchio A (2022) Hereditary spastic paraplegia: an update. Int J Mol Sci 23(3):1697
Hedera P (2021) Hereditary spastic paraplegia overview. GeneReviews® [Internet] (Bookshelf ID: NBK1509)
Atorino L, Silvestri L, Koppen M, Cassina L, Ballabio A, Marconi R, Langer T, Casari G (2003) Loss of m-AAA protease in mitochondria causes complex I deficiency and increased sensitivity to oxidative stress in hereditary spastic paraplegia. J Cell Biol 163(4):777–787
Casari G, De Fusco M, Ciarmatori S, Zeviani M, Mora M, Fernandez P, De Michele G, Filla A, Cocozza S, Marconi R et al (1998) Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell 93(6):973–983
Kasher PR, De Vos KJ, Wharton SB, Manser C, Bennett EJ, Bingley M, Wood JD, Milner R, McDermott CJ, Miller CC et al (2009) Direct evidence for axonal transport defects in a novel mouse model of mutant spastin-induced hereditary spastic paraplegia (HSP) and human HSP patients. J Neurochem 110(1):34–44
Xia CH, Roberts EA, Her LS, Liu X, Williams DS, Cleveland DW, Goldstein LS (2003) Abnormal neurofilament transport caused by targeted disruption of neuronal kinesin heavy chain KIF5A. J Cell Biol 161(1):55–66
Spörkel O, Uschkureit T, Büssow H, Stoffel W (2002) Oligodendrocytes expressing exclusively the DM20 isoform of the proteolipid protein gene: myelination and development. Glia 37(1):19–30
Zhao J, Matthies DS, Botzolakis EJ, Macdonald RL, Blakely RD, Hedera P (2008) Hereditary spastic paraplegia-associated mutations in the NIPA1 gene and its Caenorhabditis elegans homolog trigger neural degeneration in vitro and in vivo through a gain-of-function mechanism. J Neurosci 28(51):13938–13951
Ito D, Suzuki N (2009) Seipinopathy: a novel endoplasmic reticulum stress-associated disease. Brain 132(Pt 1):8–15
Hirst J, Irving C, Borner GH (2013) Adaptor protein complexes AP-4 and AP-5: new players in endosomal trafficking and progressive spastic paraplegia. Traffic 14(2):153–164
Linder P, Jankowsky E (2011) From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol 12(8):505–516
Andrisani O, Liu Q, Kehn P, Leitner WW, Moon K, Vazquez-Maldonado N, Fingerman I, Gale M Jr (2022) Biological functions of DEAD/DEAH-box RNA helicases in health and disease. Nat Immunol 23(3):354–357
Salpietro V, Efthymiou S, Manole A, Maurya B, Wiethoff S, Ashokkumar B, Cutrupi MC, Dipasquale V, Manti S, Botia JA et al (2018) A loss-of-function homozygous mutation in DDX59 implicates a conserved DEAD-box RNA helicase in nervous system development and function. Hum Mutat 39(2):187–192
Kellaris G, Khan K, Baig SM, Tsai IC, Zamora FM, Ruggieri P, Natowicz MR, Katsanis N (2018) A hypomorphic inherited pathogenic variant in DDX3X causes male intellectual disability with additional neurodevelopmental and neurodegenerative features. Hum Genomics 12(1):11
Zhan R, Yamamoto M, Ueki T, Yoshioka N, Tanaka K, Morisaki H, Seiwa C, Yamamoto Y, Kawano H, Tsuruo Y et al (2013) A DEAD-box RNA helicase Ddx54 protein in oligodendrocytes is indispensable for myelination in the central nervous system. J Neurosci Res 91(3):335–348
Park S, Lim Y, Lee D, Cho B, Bang YJ, Sung S, Kim HY, Kim DK, Lee YS, Song Y et al (2003) Identification and characterization of a novel cancer/testis antigen gene CAGE-1. Biochim Biophys Acta 1625(2):173–182
Kim H, Kim Y, Jeoung D (2017) DDX53 promotes cancer stem cell-like properties and autophagy. Mol Cells 40(1):54–65
Kim Y, Yeon M, Jeoung D (2017) DDX53 regulates cancer stem cell-like properties by binding to SOX-2. Mol Cells 40(5):322–330
Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS et al (2010) Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466(7304):368–372
Faheem M, Deneault E, Alexandrova R, Rodrigues DC, Pellecchia G, Shum C, Zarrei M, Piekna A, Wei W, Howe JL et al (2023) Disruption of DDX53 coding sequence has limited impact on iPSC-derived human NGN2 neurons. BMC Med Genomics 16(1):5
Lou X, Shi H, Wen S, Li Y, Wei X, Xie J, Ma L, Yang Y, Fang H, Lyu J (2018) A Novel NDUFS3 mutation in a Chinese patient with severe Leigh syndrome. J Hum Genet 63(12):1269–1272
Lou X, Zhou X, Li H, Lu X, Bao X, Yang K, Liao X, Chen H, Fang H, Yang Y et al (2021) Biallelic mutations in ACACA cause a disruption in lipid homeostasis that is associated with global developmental delay, microcephaly, and dysmorphic facial features. Front Cell Dev Biol 9:618492
Wittig I, Braun HP, Schägger H (2006) Blue native PAGE. Nat Protoc 1(1):418–428
Warejko JK, Tan W, Daga A, Schapiro D, Lawson JA, Shril S, Lovric S, Ashraf S, Rao J, Hermle T et al (2018) Whole exome sequencing of patients with steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol 13(1):53–62
Turner TN, Wilfert AB, Bakken TE, Bernier RA, Pepper MR, Zhang Z, Torene RI, Retterer K, Eichler EE (2019) Sex-based analysis of de novo variants in neurodevelopmental disorders. Am J Hum Genet 105(6):1274–1285
Scala M, Bradley CA, Howe JL, Trost B, Salazar NB, Shum C, Reuter MS, MacDonald JR, Ko SY, Frankland PW et al (2023) Genetic variants in DDX53 contribute to autism spectrum disorder associated with the Xp22.11 locus. medRxiv. https://doi.org/10.1101/2023.12.21.23300383
Rocak S, Linder P (2004) DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol 5(3):232–241
Singh RS, Arna AB, Dong H, Yadav M, Aggarwal A, Wu Y (2022) Structure-function analysis of DEAD-box helicase DDX43. Methods 204:286–299
Yadav M, Singh RS, Hogan D, Vidhyasagar V, Yang S, Chung IYW, Kusalik A, Dmitriev OY, Cygler M, Wu Y (2021) The KH domain facilitates the substrate specificity and unwinding processivity of DDX43 helicase. J Biol Chem 296:100085
Verny C, Guegen N, Desquiret V, Chevrollier A, Prundean A, Dubas F, Cassereau J, Ferre M, Amati-Bonneau P, Bonneau D et al (2011) Hereditary spastic paraplegia-like disorder due to a mitochondrial ATP6 gene point mutation. Mitochondrion 11(1):70–75
Pulkrabkova L, Muckova L, Hrabinova M, Sorf A, Kobrlova T, Jost P, Bezdekova D, Korabecny J, Jun D, Soukup O (2023) Differentiated SH-SY5Y neuroblastoma cells as a model for evaluation of nerve agent-associated neurotoxicity. Arch Toxicol 97(8):2209–2217
Hummel T, Krukkert K, Roos J, Davis G, Klämbt C (2000) Drosophila Futsch/22C10 is a MAP1B-like protein required for dendritic and axonal development. Neuron 26(2):357–370
Kaupmann K, Schuler V, Mosbacher J, Bischoff S, Bittiger H, Heid J, Froestl W, Leonhard S, Pfaff T, Karschin A et al (1998) Human gamma-aminobutyric acid type B receptors are differentially expressed and regulate inwardly rectifying K+ channels. Proc Natl Acad Sci USA 95(25):14991–14996
Santana J, Marzolo MP (2017) The functions of Reelin in membrane trafficking and cytoskeletal dynamics: implications for neuronal migration, polarization and differentiation. Biochem J 474(18):3137–3165
Lennox AL, Hoye ML, Jiang R, Johnson-Kerner BL, Suit LA, Venkataramanan S, Sheehan CJ, Alsina FC, Fregeau B, Aldinger KA et al (2020) Pathogenic DDX3X mutations impair RNA metabolism and neurogenesis during fetal cortical development. Neuron 106(3):404–420.e408
Simankova A, Bizen N, Saitoh S, Shibata S, Ohno N, Abe M, Sakimura K, Takebayashi H (2021) Ddx20, DEAD box helicase 20, is essential for the differentiation of oligodendrocyte and maintenance of myelin gene expression. Glia 69(11):2559–2574
Noor A, Whibley A, Marshall CR, Gianakopoulos PJ, Piton A, Carson AR, Orlic-Milacic M, Lionel AC, Sato D, Pinto D et al (2010) Disruption at the PTCHD1 Locus on Xp22.11 in autism spectrum disorder and intellectual disability. Sci Transl Med 2(49):49ra68
Ung DC, Iacono G, Méziane H, Blanchard E, Papon MA, Selten M, van Rhijn JR, Montjean R, Rucci J, Martin S et al (2018) Ptchd1 deficiency induces excitatory synaptic and cognitive dysfunctions in mouse. Mol Psychiatry 23(5):1356–1367
Wells MF, Wimmer RD, Schmitt LI, Feng G, Halassa MM (2016) Thalamic reticular impairment underlies attention deficit in Ptchd1(Y/-) mice. Nature 532(7597):58–63
Ross PJ, Zhang WB, Mok RSF, Zaslavsky K, Deneault E, D’Abate L, Rodrigues DC, Yuen RKC, Faheem M, Mufteev M et al (2020) Synaptic dysfunction in human neurons with autism-associated deletions in PTCHD1-AS. Biol Psychiatry 87(2):139–149
Fink JK (2013) Hereditary spastic paraplegia: clinico-pathologic features and emerging molecular mechanisms. Acta Neuropathol 126(3):307–328
Yamasaki M, Arita N, Hiraga S, Izumoto S, Morimoto K, Nakatani S, Fujitani K, Sato N, Hayakawa T (1995) A clinical and neuroradiological study of X-linked hydrocephalus in Japan. J Neurosurg 83(1):50–55
Schrander-Stumpel C, Höweler C, Jones M, Sommer A, Stevens C, Tinschert S, Israel J, Fryns JP (1995) Spectrum of X-linked hydrocephalus (HSAS), MASA syndrome, and complicated spastic paraplegia (SPG1): clinical review with six additional families. Am J Med Genet 57(1):107–116
Jouet M, Rosenthal A, Armstrong G, MacFarlane J, Stevenson R, Paterson J, Metzenberg A, Ionasescu V, Temple K, Kenwrick S (1994) X-linked spastic paraplegia (SPG1), MASA syndrome and X-linked hydrocephalus result from mutations in the L1 gene. Nat Genet 7(3):402–407
Finckh U, Schröder J, Ressler B, Veske A, Gal A (2000) Spectrum and detection rate of L1CAM mutations in isolated and familial cases with clinically suspected L1-disease. Am J Med Genet 92(1):40–46
Hodes ME, Zimmerman AW, Aydanian A, Naidu S, Miller NR, Garcia Oller JL, Barker B, Aleck KA, Hurley TD, Dlouhy SR (1999) Different mutations in the same codon of the proteolipid protein gene, PLP, may help in correlating genotype with phenotype in Pelizaeus-Merzbacher disease/X-linked spastic paraplegia (PMD/SPG2). Am J Med Genet 82(2):132–139
Dumitrescu AM, Liao XH, Best TB, Brockmann K, Refetoff S (2004) A novel syndrome combining thyroid and neurological abnormalities is associated with mutations in a monocarboxylate transporter gene. Am J Hum Genet 74(1):168–175
Boccone L, Mariotti S, Dessì V, Pruna D, Meloni A, Loudianos G (2010) Allan-Herndon-Dudley syndrome (AHDS) caused by a novel SLC16A2 gene mutation showing severe neurologic features and unexpectedly low TRH-stimulated serum TSH. Eur J Med Genet 53(6):392–395
Mackay-Sim A (2021) Hereditary spastic paraplegia: from genes, cells and networks to novel pathways for drug discovery. Brain Sci 11(3):403
Funding
This work was supported by grants from the National Natural Science Foundation of China-excellent young scientists fund (No. 82222043), the “Pioneer” and “Leading Goose” R&D Program of Zhejiang Province (No. 2024C03152), the Natural Science Foundation of China (Nos. 82172322 and 82302636), the Zhejiang Provincial Natural Science Foundation (No. LQ23H200001), the Scientific Research Fund of Zhejiang Provincial Education Department (No. Y202249698), and the Science and Technology Bureau of Wenzhou (No. Y2023089).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. HF and YY conceived the study and designed the experiments. XY, YW, XL, SZ, DZ, and XL performed the experiments and analyzed data. MW conducted the analysis of transcriptomics and RNA immunoprecipitation sequencing data and provided critical input on the study. The manuscript was written by XY and YW with input from all the authors.
Corresponding authors
Ethics declarations
Ethics approval
Informed consent was obtained from the subjects enrolled in this study, and the study was approved by the Ethics Committee of Peking University First Hospital (No. 2017-217).
Informed consent
Informed consent was obtained from the participant’s legal guardian and family members.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yuan, X., Wang, Y., Li, X. et al. Loss-of-function mutation in DDX53 associated with hereditary spastic paraplegia-like disorder. J Mol Med (2024). https://doi.org/10.1007/s00109-024-02454-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00109-024-02454-4