Abstract
Background
The aim of this study was to obtain a more accurate assessment of the potential association between nine genetic polymorphisms and the risk of coronary heart disease (CHD).
Methods
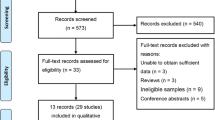
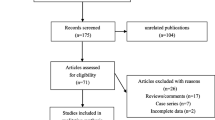
A literature search was performed in PubMed, Embase, OVID, Web of Science, Wanfang, and Chinese National Knowledge Infrastructure (CNKI) databases to identify eligible studies. We analyzed the odds ratios (OR) and 95% confidence intervals (CI) to assess the strength of the associations.
Results
A significant association was found between the PON1 −108C/T polymorphism and CHD risk (TT vs. CC: OR = 1.67, 95% CI = 1.14–2.47, p = 0.009; CT vs. CC: OR = 1.47, 95% CI = 1.17–1.85, p = 0.001; [TT + CT] vs. CC: OR = 1.56, 95% CI = 1.18–2.06, p = 0.002; T allele vs. C allele: OR = 1.28, 95%CI = 1.06–1.54, p = 0.011). There was a significant association between the hOGG1 +1245C/G polymorphism and CHD (GG vs. CC: OR = 2.33, 95% CI: 1.19–4.56, p = 0.014; CG vs. CC: OR = 1.36, 95% CI: 1.01–1.83, p = 0.046; [GG + CG] vs. CC: OR = 1.46, 95% CI: 1.10–1.94, p = 0.010; GG vs. [CC+CG]: OR = 2.11, 95% CI: 1.08–4.10, p = 0.028; G allele vs. C allele: OR = 1.45, 95% CI: 1.14–1.84, p = 0.002). The results also showed a statistically significant association of the SCARB1 +1050C/T polymorphism with CHD (TT vs. CC: OR = 1.30, 95% CI = 1.04–1.62, p = 0.022). Meta-analyses of the other six polymorphisms suggested a lack of any association with CHD risk.
Conclusion
Our results show that the susceptibility to CHD was associated with three polymorphisms: PON1 −108C/T, hOGG1 +1245C/G, and SCARB1 +1050C/T.
Zusammenfassung
Ziel
Ziel der vorliegenden Studie war es, eine genauere Beurteilung des möglichen Zusammenhangs zwischen 9 genetischen Polymorphismen und dem Risiko für eine koronare Herzkrankheit (KHK) zu erhalten.
Methoden
Um geeignete Studien zu finden, wurde eine Literatursuche in den Datenbanken PubMed, Embase, OVID, Web of Science, Wanfang und Chinese National Knowledge Infrastructure (CNKI) durchgeführt. Die Autoren berechneten die Odds Ratio (OR) und das 95%-Konfidenzintervall (95%-KI), um die Stärke des Zusammenhangs zu ermitteln.
Ergebnisse
Es wurde ein signifikanter Zusammenhang zwischen dem PON1−108C/T-Polymorphismus und dem KHK-Risiko festgestellt (TT vs. CC: OR = 1,67; 95%-KI = 1,14–2,47; p = 0,009; CT vs. CC: OR = 1,47; 95%-KI = 1,17–1,85; p = 0,001; [TT + CT] vs. CC: OR = 1,56; 95%-KI = 1,18–2,06; p = 0,002; T‑Allel vs. C‑Allel: OR = 1,28; 95%-KI = 1,06–1,54; p = 0,011). Auch bestand ein signifikanter Zusammenhang zwischen dem hOGG1+1245C/G-Polymorphismus und KHK (GG vs. CC: OR = 2,33; 95%-KI: 1,19–4,56; p = 0,014; CG vs. CC: OR = 1,36; 95%-KI: 1,01–1,83; p = 0,046; [GG + CG] vs. CC: OR = 1,46; 95%-KI: 1,10–1,94; p = 0,010; GG vs. [CC+CG]: OR = 2,11; 95%-KI: 1,08–4,10; p = 0,028; G‑Allel vs. C‑Allel: OR = 1,45; 95%-KI: 1,14–1,84; p = 0,002). Die Ergebnisse zeigten auch einen statistisch signifikanten Zusammenhang des SCARB1+1050C/T-Polymorphismus mit KHK (TT vs. CC: OR = 1,30; 95%-KI = 1,04–1,62; p = 0,022). Metaanalysen der übrigen 6 Polymorphismen ergaben keine Hinweise auf einen Zusammenhang mit dem KHK-Risiko.
Schlussfolgerung
Die vorliegenden Ergebnisse zeigen, dass die Anfälligkeit für eine KHK mit 3 Polymorphismen in Zusammenhang stand: PON1 −108C/T, hOGG1 +1245C/G und SCARB1 +1050C/T.




Similar content being viewed by others
References
Kingsbury KJ, Bondy G (2003) Understanding the essentials of blood lipid metabolism. Prog Cardiovasc Nurs 18(1):13–18
Saedi M, Vaisi-Raygani A, Khaghani S, Shariftabrizi A, Rezaie M, Pasalar P, Rahimi Z, Pourmotabbed T (2012) Matrix metalloproteinase-9 functional promoter polymorphism 1562C>T increased risk of early-onset coronary artery disease. Mol Biol Rep 39(1):555–562. https://doi.org/10.1007/s11033-011-0770-x
Zhou X, Chen J, Xu W (2012) Association between C1431T polymorphism in peroxisome proliferator-activated receptor-γ gene and coronary artery disease in Chinese Han population. Mol Biol Rep 39(2):1863–1868. https://doi.org/10.1007/s11033-011-0931-y
Vogel LK, Saebo M, Hoyer H et al (2014) Intestinal PTGS2 mRNA levels, PTGS2 gene polymorphisms, and colorectal carcinogenesis. PLoS ONE 9(8):e105254. https://doi.org/10.1371/journal.pone.0105254
Mackness B, Turkie W, Mackness M (2013) Paraoxonase-1 (PON1) promoter region polymorphisms, serum PON1 status and coronary heart disease. Arch Med Sci 9(1):8–13. https://doi.org/10.5114/aoms.2013.33189
Aviram MRM, Bisgaier CL, Newton RS, Primo-Parmo SL, La Du BN (1998) Paraoxonase inhibits high-density lipoprotein oxidation and preserves its functions. A possible peroxidative role for paraoxonase. J Clin Invest 101(8):1581–1590
Hirata K, Dichek HL, Cioffi JA, Choi SY, Leeper NJ, Quintana L, Kronmal GS, Cooper AD, Quertermous T (1999) Cloning of a unique lipase from endothelial cells extends the lipase gene family. J Biol Chem 274(20):14170–14175
Jaye M, Lynch KJ, Krawiec J, Marchadier D, Maugeais C, Doan K, South V, Amin D, Perrone M, Rader DJ (1999) A novel endothelial-derived lipase that modulates hdl metabolism. Nat Genet 21(4):424–428
Maugeais C, Tietge UJF, Broedl UC, Marchadier D, Cain W, McCoy MG, Lund-Katz S, Glick JM, Rader DJ (2003) Dose-dependent acceleration of high-density lipoprotein catabolism by endothelial lipase. Circulation 108(17):2121–2126
Edmondson AC, Brown RJ, Kathiresan S, Cupples LA, Demissie S, Manning AK, Jensen MK, Rimm EB, Wang J, Rodrigues A, Bamba V, Khetarpal SA, Wolfe ML, Derohannessian S, Li M, Reilly MP, Aberle J, Evans D, Hegele RA, Rader DJ (2009) Loss-of-function variants in endothelial lipase are a cause of elevated HDL cholesterol in humans. J Clin Invest 119(4):1042–1050
Brown RJ, Lagor WR, Sankaranaravanan S, Yasuda T, Quertermous T, Rothblat GH, Rader DJ (2010) Impact of combined deficiency of hepatic lipase and endothelial lipase on the metabolism of both high-density lipoproteins and apolipoprotein b‑containing lipoproteins. Circ Res 107(3):357–364
Goode EL, Ulrich CM, Potter JD (2002) Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev 11(12):1513–1530
Hayes JD, McLellan LI (1999) Glutathione and glutathione-dependent enzymes represent a co-ordinately regulated defence against oxidative stress. Free Radic Res 31(4):273–300
Ketterer B (1988) Protective role of glutathione and glutathione transferases in muta-genesis and carcinogenesis. Mutat Res 202(2):343–361
Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M (1996) Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science 271(5248):518–520
Rigotti A, Trigatti BL, Penman M, Rayburn H, Herz J, Krieger M (1997) A targeted mutation in the murine gene encoding the high density lipoprotein (HDL) receptor scavenger receptor class B type I reveals its key role in HDL metabolism. Proc Natl Acad Sci USA 94(23):12610–12615
Varban ML, Rinninger F, Wang N, Fairchild-Huntress V, Dunmore JH, Fang Q, Gosselin ML, Dixon KL, Deeds JD, Acton SL, Tall AR, Huszar D (1998) Targeted mutation reveals a central role for SR-BI in hepatic selective uptake of high density lipoprotein cholesterol. Proc Natl Acad Sci USA 95(8):4619–4624
West M, Greason E, Kolmakova A, Jahangiri A, Asztalos B, Pollin TI, Rodriguez A (2009) Scavenger receptor class B type I protein as an independent predictor of high-density lipoprotein cholesterol levels in subjects with hyperalphalipoproteinemia. J Clin Endocrinol Metab 94(4):1451–1457
Knutti D, Kralli A (2001) PGC-1, a versatile coactivator. Trends Endocrinol Metab 12(8):360–365
Huss JM, Kelly DP (2004) Nuclear receptor signaling and cardiac energetics. Circ Res 95(6):568–578
Arany Z, Novikov M, Chin S, Ma Y, Rosenzweig A, Spiegelman BM (2006) Transverse aortic constriction leads to accelerated heart failure in mice lacking PPAR-γ coactivator 1α. Proc Natl Acad Sci USA 103(26):10086–10091
Stang A (2010) Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 25(9):603–605
Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21(11):1539–1558. https://doi.org/10.1002/sim.1186
Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22(4):719–748
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7(3):177–188
Begg CB, Mazumdar M (1994) Operating characteristics of a rank correlation test for publication bias. Biometrics 50(4):1088–1101
Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315(7109):629–634
James RW, Leviev I, Ruiz J, Passa P, Froguel P, Garin MC (2000) Promoter polymorphism T(-107)C of the paraoxonase PON1 gene is a risk factor for coronary heart disease in type 2 diabetic patients. Diabetes 49(8):1390–1393
Huo Z, Zhou X, Zheng F, Ye GM, He X, Liu F, Li X (2002) Relationship between a polymorphism in the promoter region of Chinese Hans paraoxonase gene and coronary heart diseases. Basic Med Sci Clin 22(6):534–537
Wang X, Fan Z, Huang J et al (2003) Extensive association analysis between polymorphisms of PON gene cluster with coronary heart disease in Chinese Han population. Arterioscler Thromb Vasc Biol 23(2:328–334
Blatter Garin M‑C, Moren X, James RW (2006) Paraoxonase-1 and serum concentrations of HDL-cholesterol and apoA-I. J Lipid Res 47(3):515–520. https://doi.org/10.1194/jlr.M500281-JLR200
Shao HQ, Zhang Y (2006) The analysis of CAD with PON1−108C/T−162A/G polymorphism. Chin J Gerontol 26(1):13–16
Shano HQ, Qin B, Zhang Y (2007) Study on relationship between paraoxonase-1 and type 2 diabetes mellitus complicated by coronary heart disease. China J Mod Med 17(15):1841–1846
Saeed M, Perwaiz Iqbal M, Yousuf FA et al (2007) Interactions and associations of paraoxonase gene cluster polymorphisms with myocardial infarction in a Pakistani population. Clin Genet 71(3):238–244. https://doi.org/10.1111/j.1399-0004.2007.00753.x
Ahmad I, Narang R, Venkatraman A, Das N (2011) Frequency distribution of the single-nucleotide −108C/T polymorphism at the promoter region of the PON1 gene in Asian Indians and its relationship with coronary artery disease. J Community Genet 2(1):27–32. https://doi.org/10.1007/s12687-011-0037-1
Gupta N, Singh S, Maturu VN, Sharma YP, Gill KD (2011) Paraoxonase 1 (PON1) polymorphisms, haplotypes and activity in predicting cad risk in North-West Indian Punjabis. PLoS ONE 6(5):e17805. https://doi.org/10.1371/journal.pone.0017805
Gupta N, Binu KB, Singh S et al (2012) Low serum PON1 activity: An independent risk factor for coronary artery disease in North-West Indian type 2 diabetics. Gene 498(1):13–19. https://doi.org/10.1016/j.gene.2012.01.091
Cai GJ, He GP, Huang ZY, Qi CP (2014) Lack of association between a common polymorphism of the endothelial lipase gene and early-onset coronary artery disease in a Chinese Han population. Genet Mol Res 13(1):1059–1069. https://doi.org/10.4238/2014.February.20.7
Xie L, Sun Y, Tong Y, Liu Y, Deng Y (2015) Association of endothelial lipase gene-384A/C with coronary artery disease in Han Chinese people. BMJ Open 5(6):e7621. https://doi.org/10.1136/bmjopen-2015-007621
Cai GJ, He GP, Qi CP (2012) Association of endothelial lipase-384A/C gene polymorphism with acute coronary syndrome and blood lipids in Chinese elderly. Chin J Mult Organ Dis Elderly 11(11):821–824
Cai GJ, He GP, Qi CP (2014) Association of endothelial lipase-384A/C gene polymorphism with acute coronary syndrome and blood lipids. Chin J Emerg Med 23(2):216–219
Gokkusu C, Cakmakoglu B, Dasdemir S et al (2013) Association between genetic variants of DNA repair genes and coronary artery disease. Genet Test Mol Biomarkers 17(4):307–313. https://doi.org/10.1089/gtmb.2012.0383
Kadıoğlu E, Taçoy G, Özçağlı E et al (2016) The role of oxidative DNA damage and GSTM1, GSTT1, and hOGG1 gene polymorphisms in coronary artery disease risk. Anatol J Cardiol 16(12):931–938. https://doi.org/10.14744/AnatolJCardiol.2016.6697
Ramprasath T, Murugan PS, Kalaiarasan E, Gomathi P, Rathinavel A, Selvam GS (2012) Genetic association of glutathione peroxidase-1 (GPx-1) and NAD(P)H:quinone oxidoreductase 1(NQO1) variants and their association of CAD in patients with type-2 diabetes. Mol Cell Biochem 361(1/2):143–150. https://doi.org/10.1007/s11010-011-1098-5
Yu X, Liu J, Zhu H et al (2014) Synergistic association of DNA repair relevant gene polymorphisms with the risk of coronary artery disease in northeastern Han Chinese. Thromb Res 133(2):229–234. https://doi.org/10.1016/j.thromres.2013.11.017
Ramprasath T, Senthil Murugan P, Prabakaran AD, Gomathi P, Rathinavel A, Selvam GS (2011) Potential risk modifications of GSTT1, GSTM1 and GSTP1 (glutathione-S-transferases) variants and their association to CAD in patients with type-2 diabetes. Biochem Biophys Res Commun 407(1):49–53. https://doi.org/10.1016/j.bbrc.2011.02.097
Singh N, Sinha N, Kumar S, Pandey CM, Agrawal S (2011) Glutathione S‑transferase gene polymorphism as a susceptibility factor for acute myocardial infarction and smoking in the North Indian population. Cardiology 118(1):16–21. https://doi.org/10.1159/000324066
Phulukdaree A, Khan S, Moodley D, Chuturgoon AA (2012) GST polymorphisms and early-onset coronary artery disease in young South African Indians. S Afr Med J 102(7):627–630
Yeh HL, Kuo LT, Sung FC, Chiang CW, Yeh CC (2013) GSTM1, GSTT1, GSTP1, and GSTA1 genetic variants are not associated with coronary artery disease in Taiwan. Gene 523(1):64–69. https://doi.org/10.1016/j.gene.2013.02.052
Wu DF, Yin RX, Cao XL et al (2013) Scavenger receptor class B type 1 gene rs5888 single nucleotide polymorphism and the risk of coronary artery disease and ischemic stroke: A case-control study. Int J Med Sci 10(12):1771–1777. https://doi.org/10.7150/ijms.7044
Goodarzynejad H, Boroumand M, Behmanesh M, Ziaee S, Jalali A (2016) The rs5888 single nucleotide polymorphism in scavenger receptor class B type 1 (SCARB1) gene and the risk of premature coronary artery disease: A case-control study. Lipids Health Dis 15:7. https://doi.org/10.1186/s12944-016-0176-9
Ayhan H, Gormus U, Isbir S, Yilmaz SG, Isbir T (2017) SCARB1 gene polymorphisms and HDL subfractions in coronary artery disease. In Vivo 31(5):873–876
Zeng TT, Tang DJ, Ye YX, Su J, Jiang H (2017) Influence of SCARB1 gene SNPs on serum lipid levels and susceptibility to coronary heart disease and cerebral infarction in a Chinese population. Gene 626:319–325. https://doi.org/10.1016/j.gene.2017.05.020
Zhang Y, Xu W, Li X et al (2008) Association between PPARGC1A gene polymorphisms and coronary artery disease in a Chinese population. Clin Exp Pharmacol Physiol 35(10):1172–1177. https://doi.org/10.1111/j.1440-1681.2008.04988.x
Nikitin AG, Chistiakov DA, Minushkina LO, Zateyshchikov DA, Nosikov VV (2010) Association of the CYBA, PPARGC1A, PPARG3, and PPARD gene variants with coronary artery disease and metabolic risk factors of coronary atherosclerosis in a Russian population. Heart Vessels 25(3):229–236. https://doi.org/10.1007/s00380-009-1159-9
Yongsakulchai P, Settasatian C, Settasatian N et al (2016) Association of combined genetic variations in PPARgamma, PGC-1alpha, and LXRalpha with coronary artery disease and severity in Thai population. Atherosclerosis 248:140–148. https://doi.org/10.1016/j.atherosclerosis.2016.03.005
Munafò MR, Clark TG, Flint J (2004) Assessing publication bias in genetic association studies: Evidence from a recent meta-analysis. Psychiatry Res 129(1):39–44
Leviev I, James RW (2000) Promoter polymorphisms of human paraoxonase PON1 gene and serum paraoxonase activities and concentrations. Arterioscler Thromb Vasc Biol 20(2):516–521
Suehiro T, Nakamura T, Inoue M, Shiinoki T, Ikeda Y, Kumon Y, Shindo M, Tanaka H, Hashimoto K (2000) A polymorphism upstream from the human paraoxonase gene and its association with PON1 expression. Atherosclerosis 150(2):295–298
Brophy VH, Jampsa RL, Clendenning JB, McKinstry LA, Jarvik GP, Furlong CE (2001) Effects of 5′ regulatory-region polymorphisms on paraoxonase-gene(PON1) expression. Am J Hum Genet 68(6):1428–1436
deLemos AS, Wolfe ML, Long CJ, Sivapackianathan R, Rader DJ (2002) Identification of genetic variants in endothelial lipase in persons with elevated high-density lipoprotein cholesterol. Circulation 106(11):1321–1326
Kuehl BL, Paterson JWE, Peacock JW, Paterson MC, Rauth AM (1995) Presence of a heterozygous substitution and its relationship to DTdiaphorase activity. Br J Cancer 72(3):555–561
Moran JL, Siegel D, Ross D (1999) A potential mechanism underlying the increased susceptibility of individuals with a polymorphism in NAD(P)H quinone oxidoreductase 1 (NQO1) to benzene toxicity. Proc Natl Acad Sci USA 96(14):8150–8155
Constantineau J, Greason E, West M, Filbin M, Kieft JS, Carletti MZ, Christenson LK, Rodriguez A (2010) A synonymous variant in scavenger receptor, class B, type I gene is associated with lower SR-BI protein expression and function. Atherosclerosis 210(1):177–182
Muller YL, Bogardus C, Pedersen O, Baier L (2003) A Gly482Ser missense mutation in the peroxisome proliferator-activated receptor gamma coactivator-1 is associated with altered lipid oxidation and early insulin secretion in Pima Indians. Diabetes 52(3):895–898
Xie X, Shi X, Xun X, Rao L (2017) Association between microRNA polymorphisms and coronary heart disease: A meta-analysis. Herz 42(6):593–603. https://doi.org/10.1007/s00059-016-4495-4
Author information
Authors and Affiliations
Contributions
Q. Du and Q. Li conceived and designed the experiments. Q. Li performed the experiments. Q. Li and Q. Du analyzed the data.
Corresponding author
Ethics declarations
Conflict of interest
Q. Li and Q. Du declare that they have no competing interests.
For this article no studies with human participants or animals were performed by any of the authors. All studies performed were in accordance with the ethical standards indicated in each case.
Caption Electronic Supplementary Material
59_2019_4806_MOESM1_ESM.tif
Figure S1. A. Sensitivity analysis of PON1 -108C/T polymorphism and CHD risk in the overall analysis under homozygote comparison. B. Sensitivity analysis of PON1 -108C/T polymorphism and CHD risk in the overall analysis under heterozygote comparison
59_2019_4806_MOESM2_ESM.tif
Figure S2. A. Sensitivity analysis of PON1 -162G/A polymorphism and CHD risk in the overall analysis under homozygote comparison. B. Sensitivity analysis of PON1 -162G/A polymorphism and CHD risk in the overall analysis under heterozygote comparison
59_2019_4806_MOESM3_ESM.doc
Supplementary Table 1. Results of Newcastle–Ottawa scale (NOS) assessment for the included studies and full electronic search strategy for electronic databases
Rights and permissions
About this article
Cite this article
Li, Q., Du, Q. Associations between nine candidate genetic polymorphisms with coronary heart disease. Herz 45 (Suppl 1), 15–28 (2020). https://doi.org/10.1007/s00059-019-4806-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00059-019-4806-7