Abstract
Background
The KIF6 rs20455 polymorphism has been verified as an important genetic factor of coronary heart disease (CHD), but with controversial results. The aim of this study was to explore the association between KIF6 rs20455 polymorphism and susceptibility to CHD.
Methods
All eligible studies were identified by searching Medline (mainly PubMed), EMBASE, the Web of Science, Cochrane Collaboration Database, Chinese National Knowledge Infrastructure, Wanfang Database and China Biological Medicine up to October 5, 2016.Odds ratios (ORs) with 95% confidence interval (CI) were used to explore the association between KIF6 rs20455 polymorphism and CHD risk. Begg’s and Egger’s tests were used to examine the publication bias. Subgroup analysis and sensitivity analysis were performed to test the reliability and stability of the results. All the analyses were carried out by Stata 12.0 software.
Results
A total of 28 publications including 50 individual studies were analyzed in this present work. There are no significant association found between KIF6 rs20455 polymorphism and CHD risk (Homozygote model: OR = 1.007, 95% CI =0.952–1.066, P = 0.801; Heterozygote model: OR = 1.009, 95% CI = 0.968–1.052, P = 0.636; Dominant model: OR = 1.007, 95% CI = 0.966–1.048, P = 0.753; Recessive model: OR = 0.989, 95% CI = 0.943–1.037, P = 0.655; Allele comparison model: OR = 1.00, 95% CI = 0.971–1.030, P = 0.988). Furthermore, subgroup analyses were performed by ethnicity, source of control.
Conclusions
Our result suggests that KIF6 rs20455 polymorphism may not be associated with CHD susceptibility. However, additional very well-designed large-scale studies are warranted to confirm our results.
Similar content being viewed by others
Background
Coronary heart disease (CHD), a multifactorial heart disorder resulting from both environmental and genetic factors [1], is one of the leading causes of disability and death around the world [2]. Epidemiology studies have suggested that hypertension, hyperlipidemia, diabetes mellitus, obesity and smoking are major risk factors for CHD [3]. In recent years, more and more studies reveled that several loci and variants are strongly associated with CHD [4, 5]. It has been estimated that approximately 50% of the variability of the major risk factors for CHD is determined by genetics [6].
The KIF6 protein is one of several molecular components that mediate intracellular transport of organelles, protein complexes, and mRNAs. A common Trp719Arg (rs20455) SNP in exon 19 of the KIF6 gene has been identified as a potential risk factor for CHD [7, 8]. The KIF6 protein belongs to the kinesin superfamily, which is involved in the intracellular transport in a microtubule and ATP-dependent manner [9]. The rs20455 polymorphism replaces the nonpolar ‘Trp’ residue in codon 719 with a basic ‘Arg’ amino acid. This SNP lies near the putative cargo binding taildomain and may alter the cargo activity of KIF6 [10]. Carriers of the 719Arg allele exhibit a 50% increased risk of events compared with non-carriers [8, 11]. Up to now, multiple large prospective and case–control studies have reported the association between KIF6 rs20455 polymorphism and the risk of CHD. However, somestudies have not verified inconsistent results. Published studies have generally been restricted in terms of sample size and ethnic diversity, and individual studies may have in-sufficient power to achieve a comprehensive and reliable conclusion. In view of the discrepancies in the findings of previous published studies, we aimed to perform a meta-analysis of the published studies to clarify the association between KIF6 rs20455 polymorphism and CHD to get a better under-standing of this relationship.
Methods
Literature search
A comprehensive search for all related studies from both electric databases, such as, Medline (mainly PubMed), Embase, Web of science, China National Knowledge Infrastructure (CNKI) et al., and hand search from references of all eligible literatures. Single or combinations of the following keywords were used: “kinesin like protein 6” or “KIF6” or “rs20455” or “719Arg”, “single nucleotide polymorphism, SNP or variation, mutation”, “genetic association” and “coronary heart disease” or “CHD”. No language and sample size were set. When more than one studies of the same population were included in several publications, only the most recent or complete studies were included in this meta-analysis.
Selection criteria
Articles included should meet following criteria: an appropriate description of KIF6 rs20455 polymorphism in CHD cases and healthy controls; results expressed as odds ratio (OR); and studies with a 95% confidence interval (CI) for OR with sufficient data to calculate these numbers. While for the exclusion criteria provided as follows: studies without raw data; case-only studies, family-based studies, case reports, editorials, and review articles (including meta-analyses). In studies with overlapping cases/controls, the study with the higher quality score or the study with more information on the origin of the cases/controls was included in the meta-analysis.
Data extraction
Two researchers extracted important information independently and carefully from all eligible studies according to the criteria listed above. Any disagreement will be resolved by the two authors through discussion or the third author. The following data were extracted from each included study: first author’s surname, year of publication, country, ethnicity, genotyping method, source of control, total number of cases and controls, distributions of KIF6 rs20455 genotypes. Different ethnicity descents were categorized as Caucasian, Asian, and Mixed populations (the original studies didn’t clarify the race of the subjects or mixed races).
Statistical analysis
We adopted poled ORs and corresponding 95% confidence interval (CIs) to detect the association between KIF6 rs20455 polymorphism and CHD risk. Heterogeneity was explored by Q statistic [12], and the P value was <0.05 will be considered statistically significant. Heterogeneity was also assessed using the I2 statistic, which takes values between 0% and 100% with higher values denoting greater degree of heterogeneity (I2 = 0–25%: noheterogeneity; I2 = 25–50%: moderate heterogeneity; I2 = 50–75%: large heterogeneity; I2 = 75–100%: extreme heterogeneity) [13]. Different statistical models will be selected according to the result of heterogeneity. Random (Der Simonian-Laird method) [14] will be used to calculate the precise results when the P value of heterogeneity was <0.05, or the I2 > 50%. Otherwise, fixed effects model (Mantel-Haenszel method) will be adopted [15]. Five genetic comparison model were carried out and calculated as follows: homozygote model (GG vs. AA), heterozygote model (AG vs. AA), recessive model (GG vs. AG + AA), and dominant model (GG + AG vs. AA), and allele comparison model (G-allele vs. A-allele). Hardy–Weinberg equilibrium in the control group was tested by the chi-square test for goodness of fit, and a P value of <0.05 was considered significant. Subgroup analyses were performed by ethnicity, source of control, to confirm if our results were stable and robust [16]. Begg’s funnel plots [17] and Egger’s test [18] were explored to examine if potential publication bias were existed in this study. Sensitivity analysis was carried out by sequentially omitting each study and finding the influence on the overall summary estimate [19]. All the statistical analyses were finished by STATA software (version 12.0; Stata Corporation, College Station, TX). All the P values were two-sided.
Results
Characteristics of all included studies
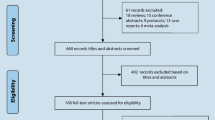
Totally, 209 potential relevant studies were searched through several databases. Based on the including criteria listed above, only 28 articles including 50 separate studies were included finally [8, 20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46]. A flow diagram summarizing the process of study selection was present in Fig. 1. The baseline characteristics ofall included studies were listed in Table 1. Helgadottir et al. contained two individual studies [25], Samani et al. contained two individual studies [26], Assimes et al. contained 20 individual studies [31], and Wu et al. contained two separate studies [41]. Moreover, there were 37 studies from Caucasian decedent, 9 studies from Asian populations and the rest 14 studies from mixed populations. There were 20 population-based (PB) studies, 21 hospital-based (HB) studies and four family based (FB) study, three community based (CB) study, two hospital and community based (H-CB) study. Different ethnicity descents were categorized as Caucasian, Asian and Mix (the original studies didn’t clarify the race of the subjects or mixed races).
Quantitative synthesis
All the eligible data were calculated and significant heterogeneity was detected under homozygote (I2 = 33.9%; Pheterogeneity = 0.012), heterozygote (I2 = 35.5%; Pheterogeneity = 0.008), dominant (I2 = 39.8; Pheterogeneity = 0.002), recessive (I2 = 26.5%; Pheterogeneity = 0.047) and allele comparison model (I2 = 44.2%; Pheterogeneity = 0.001) between this gene variation and the risk of CHD. So, random-effect model was used to calculate the statistical parameters. Overall, there were no significant association existed between KIF6 rs20455 polymorphism and the risk of CHD (Homozygote model: OR = 1.007, 95% CI =0.952–1.066, P = 0.801, Fig. 2; Heterozygote model: OR = 1.009, 95% CI = 0.968–1.052, P = 0.636, Fig. 3; Dominant model: OR = 1.007, 95% CI = 0.966–1.048, P = 0.753, Fig. 4; Recessive model: OR = 0.989, 95% CI = 0.943–1.037, P = 0.655, Fig. 5; Allele comparison model: OR = 1.00, 95% CI = 0.971–1.030, P = 0.988, Fig. 6). Furthermore, we explored the subgroup analyses by ethnicity and source of control. All the results were listed in Table 2.
Sensitivity analysis
The sensitivity analysis was performed to evaluate the influence of each individual study on the pooled OR by omitting every single study. The analysis results reflected that our results were statistically stable and reliable.
Publication bias
There was no significant publication bias found in the meta-analysis, reflected by P values from Begg’s correlation (Heterozygote model: P = 0.089; Dominant model: P = 0.061; Allele comparison model: P = 0.052, Fig. 7) and Egger’s regression (Heterozygote model: P = 0.070; Dominant model: P = 0.058; Allele comparison model: P = 0.066, Fig. 8). However, significant publication bias found in the meta-analysis, reflected by P values from Begg’s correlation (Homozygote model: P = 0.046; Recessive model: P = 0.025) and Egger’s regression (Homozygote model: P = 0.041; Recessive model: P = 0.040). All the results are listed in Table 2.
Discussion
Large sample and unbiased epidemiological studies of predisposition genes polymorphisms could provide insight into the in vivo relationship between candidate genes and complex diseases. Many epidemiological studies have investigated the relationship between the KIF6 rs20455 polymorphism and the risk of CHD, but because of small sample size and the low statistical power of individual studies, results have been contradictory. In this present study, we searched all eligible studies to date and got the precise result if KIF6 rs20455 polymorphism could contribute to the risk of CHD. To the best of our knowledge, our present work was the most comprehensive one through enrolling all eligible studies.
Herein, we included 50 individual studies, including 40,059 cases and 64,032 controls. Overall, there was no association between KIF6 rs20455 polymorphism and CHD risk. Hamidizadeh et al. found that significant association was found between this gene polymorphism and CHD risk among Caucasian populations [43], and the result was verified in another study through enrolling 143,000 subjects [40]. However, no association was found in a meta-analysis, among South Asians, African-Americans, Hispanics, East Asians, and mixed decedent populations [39]. Furthermore, other recent studies were also found no association existed between this gene polymorphism and CHD risk [25, 26, 47,48,49]. When we got the subgroup analyses by ethnicity, there was also no association found among Caucasian and Asian populations. While decreased risk of this gene polymorphism and CHD risk was found among mixed populations. Of note, mixed populations means the original studies didn’t clarify the race of the subjects or mixed races. This result may be not provided some useful information for clinical deeds. So, further studies should be performed with clearly race or ethnicity stated in their work.
Publication bias was found in some genetic models. The explanations might arise from some aspects. First, our meta-analysis took into consideration only fully published data, and the abstract and conference papers were excluded. Second, this meta-analysis only focused on papers published in Chinese and English language, and some eligible studies which were reported in other languages might be missed. Third, positive results tend to be accepted by journals while negative results are often rejected or not even submitted. We should point out that the publication bias might partly account for the results, but which were not affected deeply. When we adjusted the results using the trim and fill method, the adjusted risk estimate was attenuated but remained significant, indicating the stability of our results.
Some limitations of this meta-analysis should be addressed. Firstly, heterogeneity is a potential problem when interpreting all the results of meta-analysis. Although we minimized the likelihood by performing a careful search for published studies, using the explicit criteria for study inclusion, the significant between-study heterogeneity still existed in most of comparison. The presence of heterogeneity can result from differences in the age distribution, selection of controls, prevalence lifestyle factors and so on. Secondly, only published studies were included in this meta-analysis. Therefore, potential publication bias was existed in some genetic models. Despite the limitations, our meta-analysis significantly increased the statistical power based on substantial data from different studies. The sensitivity analyses outcomes reflected that our results were statistically stable and reliable.
In conclusion, this present meta-analysis suggests that carriers of KIF6 rs20455 polymorphism may irrelative to the risk of CHD. We also observed no compelling evidence of an association between the KIF6 rs20455 SNP and CHD in multiple race/ethnic groups. These findings do not support the clinical utility of testing for the KIF6 rs20455 polymorphism in the primary prevention of CHD and indirectly question whether genotype information at this locus is able to identify subjects most likely to benefit from the use of statins.
Abbreviations
- CHD:
-
Coronary heart disease
- CHS:
-
Cardiovascular Health Study
- CI:
-
Confidence interval
- KIF6:
-
Kinesin-like protein 6
- OR:
-
Odds ratio
- PROSPER:
-
Prospective Study of Pravastatin in the Elderly at Risk
- SNPs:
-
Single nucleotide polymorphisms
- WHS:
-
the Women’s Health Study
References
Luo JQ, Wen JG, Zhou HH, Chen XP, Zhang W. Endothelial nitricoxide synthase gene G894T polymorphism and myocardial infarction: a meta-analysis of 34 studies involving 21, 068 subjects. PLoS One. 2014;9:e87196.
Naghavi M, Wang H, Lozano R, Davis A, Liang X, Zhou M, et al. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the global burden of disease study 2013. Lancet. 2015;385:117–71.
Hackam DG, Anand SS. Emerging risk factors for atherosclerotic vascular disease: a critical review of the evidence. JAMA. 2003;290:932–40.
Shiffman D, Sabatine MS, Louie JZ, Kirchgessner TG, Iakoubova OA, Campos H, et al. Effect of pravastatin therapy on coronary events in carriers of the KIF6 719Arg allele from the cholesterol and recurrent events trial. Am J Cardiol. 2010;105(9):1300–5.
Li Y, Iakoubova OA, Shiffman D, Devlin JJ, Forrester JS, Superko HR. KIF6polymorphism as a predictor of risk of coronary events and of clinical eventreduction by statin therapy. Am J Cardiol. 2010;106(7):994–8.
Sekuri C, Cam FS, Ercan E, et al. Renin–angiotensin system gene polymorphisms and premature coronary heart disease. J Renin-Angiotensin-Aldosterone Syst. 2005;6:38–42.
Akao H, Polisecki E, Kajinami K, Trompet S, Robertson M, Ford I, et al. KIF6,LPA, TAS2R50, and VAMP8 genetic variation, low density lipoproteincholesterol lowering response to pravastatin, and heart disease riskreduction in the elderly. Atherosclerosis. 2012;220(2):456–62.
Iakoubova OA, Sabatine MS, Rowland CM, Tong CH, Catanese JJ, Ranade K, et al. Polymorphism in KIF6 gene and benefit from statins after acutecoronary syndromes: results from the PROVE IT-TIMI 22 study. J Am Coll Cardiol. 2008;51(4):449–55.
Miki H, Setou M, Kaneshiro K, Hirokawa N. All kinesin superfamily protein, KIF,genes in mouse and human. Proc Natl Acad Sci U S A. 2001;98:7004–11.
Li Y, Iakoubova OA, Shiffman D, Devlin JJ, Forrester JS, Superko HR. KIF6polymorphism as a predictor of risk of coronary events and of clinical event reductionby statin therapy. Am J Cardiol. 2010;106(7):994–8.
Li Y, Sabatine MS, Tong CH, Ford I, Kirchgessner TG, Packard CJ, et al. Genetic variants in the KIF6 region and coronary event reduction from statin therapy. Hum Genet. 2011;129(1):17–23.
Zintzaras E, Ioannidis JP. HEGESMA: genome search meta-analysisand heterogeneity testing. Bioinformatics. 2005;21:3672–3.
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuringinconsistency in meta-analyses. BMJ. 2003;327:557–60.
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177e188.
Mantel N, Haenszel W. Statistical aspects of the analysis of data fromretrospective studies of disease. J Natl Cancer Inst. 1959;22:719e748.
Zintzaras E, Chatzoulis DZ, Karabatsas CH, Stefanidis I. Therelationship between C677T methylenetetrahydrofolate reductasegene polymorphism and retinopathy in type 2 diabetes: a meta-analysis. J Hum Genet. 2005;50:267–75.
Begg CB, Mazumdar M. Operating characteristics of a rankcorrelation test for publication bias. Biometrics. 1994;50:1088–101.
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34.
Zintzaras E, Chatzoulis DZ, Karabatsas CH, Stefanidis I. The relationship between C677T methylenetetrahydrofolate reductase gene polymorphism and retinopathy in type 2 diabetes: a meta-analysis. J Hum Genet. 2005;50:267–75.
Berglund G, Elmstahl S, Janzon L, Larsson SA. The Malmo diet and cancer study. Design and feasibility. J Intern Med. 1993;233(1):45–51.
Vartiainen E, Jousilahti P, Alfthan G, Sundvall J, Pietinen P, Puska P. Cardiovascular risk factor changes in Finland, 1972–1997. Int J Epidemiol. 2000;29(1):49–56.
Senti M, Tomas M, Marrugat J, Elosua R. Paraoxonase 1–192 polymorphism modulates the nonfatal myocardial infarction risk associated with decreased HDLs. Arterioscler Thromb Vasc Biol. 2001;21(3):415–20.
Yusuf S, Hawken S, Ounpuu S, Dans T, Avezum A, Lanas F, et al. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case–control study. Lancet. 2004;364(9438):937–52.
Low AF, O’Donnell CJ, Kathiresan S, Everett B, Chae CU, Shaw SY, et al. Aging syndrome genes and premature coronary artery disease. BMC Med Genet. 2005;6:38.
Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316(5830):1491–3.
Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357(5):443–53.
Meng W, Hughes A, Patterson CC, Belton C, Kamaruddin MS, Horan PG, et al. Genetic variants of complement factor H gene are not associated with premature coronary heart disease: a family-based study in the Irish population. BMC Med Genet. 2007;8:62.
Meiner V, Friedlander Y, Milo H, Sharon N, Ben-Avi L, Shpitzen S, et al. Cholesteryl ester transfer protein (CETP) genetic variation and early onset of non-fatal myocardial infarction. Ann Hum Genet. 2008;72(Pt 6):732–41.
Serre D, Montpetit A, Pare G, Engert JC, Yusuf S, Keavney B, et al. Correction of population stratification in large multi-ethnic association studies. PLoS One. 2008;3(1):1–11.
Morgan TM, Xiao L, Lyons P, Kassebaum B, Krumholz HM, Spertus JA. Investigation of 89 candidate gene variants for effects on all-cause mortality following acute coronary syndrome. BMC Med Genet. 2008;9(66):1471–2350.
Assimes TL, Knowles JW, Basu A, Iribarren C, Southwick A, Tang H, et al. Susceptibility locus for clinical and subclinical coronary artery disease at chromosome 9p21 in the multi-ethnic ADVANCE study. Hum Mol Genet. 2008;17(15):2320–8.
Vennemann MM, Hummel T, Berger K. The association between smoking and smell and taste impairment in the general population. J Neurol. 2008;255(8):1121–6.
Sutton BS, Crosslin DR, Shah SH, Nelson SC, Bassil A, Hale AB, et al. Comprehensive genetic analysis of the platelet activating factor acetylhydrolase (PLA2G7) gene and cardiovascular disease in case–control and family datasets. Hum Mol Genet. 2008;17(9):1318–28.
Martinelli N, Girelli D, Malerba G, Guarini P, Illig T, Trabetti E, et al. FADS genotypes and desaturase activity estimated by the ratio of arachidonic acid to linoleic acid are associated with inflammation and coronary artery disease. Am J Clin Nutr. 2008;88(4):941–9.
Stewart AF, Dandona S, Chen L, Assogba O, Belanger M, Ewart G, et al. Kinesin family member 6 variant Trp719Arg does not associate with angiographically defined coronary artery disease in the Ottawa heart genomics study. J Am Coll Cardiol. 2009;53(16):1471–2.
Luke MM, Lalouschek W, Rowland CM, Catanese JJ, Bolonick JI, Bui ND, et al. Polymorphisms associated with both noncardioembolic stroke and coronary heart disease: vienna stroke registry. Cerebrovasc Dis. 2009;28(5):499–504.
Bare LA, Ruiz-Narvaez EA, Tong CH, Arellano AR, Rowland CM, Catanese JJ, Sacks FM, Devlin JJ and Campos H. Investigation of KIF6 Trp719Arg in a case-control study of myocardial infarction: a Costa Rican population. Plos one. 2010;5(9):e13081.
Assimes TL, Holm H, Kathiresan S, Reilly MP, Thorleifsson G, Voight BF, Erdmann J, Willenborg C, Vaidya D, Xie C, Patterson CC, Morgan TM, Burnett MS, Li M, Hlatky MA, Knowles JW, et al. Lack of association between the Trp719Arg polymorphism in kinesin-like protein-6 and coronary artery disease in 19 case-control studies. J Am Coll Cardiol. 2010;56(19):1552–63.
Bhanushali AA, Contractor A, Shah VT, Das BR. Investigation of KIF6 Trp719Arg in a case-control study of coronary artery disease in western Indians. Genet Test Mol Biomarkers. 2011;15(12):883–6.
Peng P, Lian J, Huang RS, Xu L, Huang Y, Ba Y, Yang X, Huang X, Dong C, Zhang L, Ye M, Zhou J, Duan S. Meta-analyses of KIF6 Trp719Arg in coronary heart disease and statin therapeutic effect. PLoS One. 2012;7(12):e50126.
Wu G, Li GB, Dai B. Association of KIF6 variant with lipid level and angiographic coronary artery disease events risk in the Han Chinese population. Molecules. 2012;17(9):11269–80.
Wu G, Li GB, Dai B, Zhang DQ. Novel KIF6 polymorphism increases susceptibility to type 2 diabetes mellitus and coronary heart disease in Han Chinese men. J Diabetes Res. 2014;871439(10):31.
Hamidizadeh L, Haji Hosseini Baghdadabadi R, Babaee Baigi MA, Dastsooz H, Khazaei Nejhad A, Fardaei M. Impact of KIF6 polymorphism rs20455 on coronary heart disease risk and effectiveness of Statin therapy in 100 patients from southern Iran. Arch Iran Med. 2015;18(10):683–7.
Vishnuprabu D, Geetha S, Bhaskar LV, Mahapatra NR, Munirajan AK. Genotyping and meta-analysis of KIF6 Trp719Arg polymorphism in south Indian coronary artery disease patients: a case-control study. Meta gene. 2015;5:129–34.
Hubacek JA, Vrablik M, Dlouha D, Stanek V, Gebauerova M, Adamkova V, Ceska R, Dostalova G, Linhart A, Vitek L, Pitha J. Gene variants at FTO, 9p21, and 2q36.3 are age-independently associated with myocardial infarction in Czech men. Clinicachimicaacta Int J Clin Chem. 2016;454:119–23.
Vatte C, Cyrus C, AlShehri AM, Chathoth S, Almansori M, Al-Nafaie A, Al-Ali R, Al-Muhanna F, Asselbergs FW, Al-Ali A. Investigation of KIF6 Trp719Arg gene polymorphism in a case-control study of coronary artery disease and non-fatal myocardial infarction in the Eastern Province of Saudi Arabia. Ann Saudi Med. 2016;36(2):105–11.
Myocardial Infarction Genetics C, Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, Anand S, Engert JC, Samani NJ, Schunkert H, Erdmann J, Reilly MP, Rader DJ, Morgan T, Spertus JA, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41(3):334–41.
Wellcome Trust Case Control C. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–78.
Arsenault BJ, Boekholdt SM, Hovingh GK, Hyde CL, DeMicco DA, Chatterjee A, Barter P, Deedwania P, Waters DD, LaRosa JC, Pedersen TR, Kastelein JJ. The 719Arg variant of KIF6 and cardiovascular outcomes in statin-treated, stable coronary patients of the treating to new targets and incremental decrease in end points through aggressive lipid-lowering prospective studies. Circ Cardiovasc Genet. 2012;5(1):51–7.
Acknowledgements
We thank all our colleagues of this present work.
Funding
Not applicable.
Availability of data and materials
Please contact author for data requests.
Author information
Authors and Affiliations
Contributions
YL, ZC, HS participated in the design of the study. YL, ZC, HS carried out the literature search and data extraction. YL, ZC, HS participated in the analysis of eligible data. YL, ZC, HS wrote the manuscript All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Li, Y., Chen, Z. & Song, H. Association between KIF6 rs20455 polymorphism and the risk of coronary heart disease (CHD): a pooled analysis of 50 individual studies including 40,059 cases and 64,032 controls. Lipids Health Dis 17, 4 (2018). https://doi.org/10.1186/s12944-017-0651-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12944-017-0651-y