Abstract
Oestrogen receptor-negative breast cancer, particularly subtypes such as triple-negative breast cancer (TNBC, around 10–15% of cases), are characterised by poor long-term survival, poor response to therapy and early progression to metastasis. Purine-based compounds represent a privileged scaffold in anticancer drug design, with several clinically approved and experimental agents in clinical development comprising a purine core structure. In this study, a series of new purine-based compounds were synthesised; seven of the new analogues were found to significantly reduce the in vitro viability of TNBC cell lines (MDA-MB-231 and MDA-MB-436) with IC50 values of ≤50 μM. In previous work, we have proposed a new concept for targeting angiogenesis driving TNBC progression, by disrupting the protein–protein interaction between the molecular chaperone αB-crystallin (CRYAB) and VEGF. Since previous clinical studies applying anti-VEGF therapy to TNBC patients have met with limited success, we were interested to test our most promising purine analogues against CRYAB/VEGF, using a custom-designed cell-based CRYAB/VEGF165 interaction assay platform. Analogues 4e and 4f significantly reduced the interaction between CRYAB/VEGF165, and compound 4e (100 μM) was also found to decrease the levels of soluble VEGF expressed by MDA-MB-231 cells by 40%. In conclusion, these promising early activity profiles warrant further investigation to validate this concept.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Triple-negative breast cancer (TNBC) represents an important and unmet clinical challenge, accounting for around 10–15% of all newly diagnosed breast cancer cases. TNBC is classified as a subpopulation of invasive breast cancer characterised by the lack of expression of the oestrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) (Fosu-Mensah et al. 2015; Lehmann et al. 2011; Mayer et al. 2014). As a subset of breast cancer cases, TNBC is also a “disease of the young”, as it frequently affects pre-menopausal women of minority background, including black and Hispanic women. It is an aggressive disease associated with high histological grade, increased risk of recurrence and metastatic spread and poor prognosis. Currently, the standard of care treatment for TNBC patients involves the use of cytotoxic chemotherapy (e.g., taxanes). However, this treatment lacks long-term efficacy.
Angiogenesis, the development of new blood vessels from existing vasculature, is essential for early stage tumourigenesis and metastasis. This process is regulated by several pro-angiogenic factors, including vascular endothelial growth factor (VEGF). TNBC is highly vascularised as a result of upregulation of intra-tumoural VEGF levels, which in turn is associated with poor prognosis. In this regard, anti-angiogenic therapies were evaluated clinically as potential targeted therapies in TNBC albeit not successfully in terms of long-term survival. Bevacizumab is an anti-angiogenic antibody that has been used in combination with chemotherapy and was first investigated as a therapeutic option due to enhanced angiogenesis in TNBC presentations (Steeg 2006; Kim et al. 2004). This therapy showed promising results in early clinical trials, however, meta-analysis of clinical trial data showed no beneficial effect on overall survival rates (O’Shaughnessy et al. 2009; von Minckwitz et al. 2012; Bear et al. 2012). Most patients were non-responsive to the therapy, while patients who showed initial response eventually developed resistance and relapsed disease. Moreover, in 2011 the FDA revoked its approval of bevacizumab for breast cancer therapy due to reported adverse side effects (Goozner 2011; Ranpura et al. 2011; O’Reilly et al. 2015). There are currently no molecularly targeted therapies that are effective against TNBC in the clinical setting, and therefore there is a high unmet medical need for the development of new effective and efficacious therapies for this poor prognosis patient group (Fosu-Mensah et al. 2015; Chen et al. 2014).
Given the high priority afforded to the further study and treatment of this disease, TNBC has been the subject of intense gene array profiling studies. These studies identified new molecular targets including but not limited to basal epithelial proteins, such as cytokeratin 5 and 6, CK14, CK17, P-cadherin, p53 mutations, epidermal growth factor receptor (EGFR) and αB-crystallin (Lehmann et al. 2011). Ultimately, these studies will provide potential therapeutic targets for drug discovery programmes. Since many of the newly identified TNBC therapeutic targets have established therapies in other disease settings, there are presently numerous clinical studies underway to assess their efficacy in TNBC.
An interesting TNBC-associated target under investigation within our laboratory is αB-crystallin (CRYAB, HspB5). CRYAB is a member of the mammalian small heat-shock protein family that function as molecular chaperones to modulate cell proteostasis, and consequently, cell survival by suppressing aggregation of denatured proteins in stress-induced environments (Tsang et al. 2012; Arrigo et al. 2007; Garrido et al. 2012). CRYAB is ubiquitously expressed in many tissues with varying functions, and upregulation of this protein is observed in many disease pathologies, including neuropathy, myopathy, ischaemia/reperfusion, cataract and various solid tumours (gliomas, prostate, ovarian, colon, liver, head and neck cancer) (Ruan et al. 2011). CRYAB represents a promising therapeutic target owing to its diverse functions as a molecular chaperone. Studies in breast cancers have provided compelling clinical data that showed that CRYAB is constitutively expressed in the most aggressive phenotypes of the disease, including TNBC. Accumulating evidence has revealed that overexpression of CRYAB in TNBC contributes to tumour progression and correlates with the poor survival rates observed in this aggressive tumour type. This can be attributed to its critical role as a metastatic and anti-apoptotic regulator. Thus, CRYAB has been evaluated as a potentially important biomarker in TNBC (Tsang et al. 2012; Ruan et al. 2011). Our previous structure-based molecular docking studies identified 3-methylglutamic acid as a small molecule with the potential to inhibit CRYAB/VEGF interaction. 3-Methylglutamic acid was shown to disrupt CRYAB/VEGF165 interaction and elicit both in vitro and in vivo antitumour, anti-proliferative and anti-angiogenic effects through downregulation of VEGF signalling (Chen et al. 2014).
In recent years, our laboratory has focused on the discovery of new drug candidates for potential application in the TNBC setting (Chen et al. 2014; Haynes et al. 2016). As part of these efforts, we were attracted to the possibilities associated with purine-based compounds, given their long and distinguished history in cancer drug development, including clinically approved anti-leukaemia drugs such as fludarabine (Rodriguez 1994) and clofarabine (Ghanem et al. 2010). Purines represent a privileged heterocycle in drug design, given the ability of purine analogues to bind tightly within hydrophobic folds and inhibit a range of key enzyme targets, such as members of the diverse protein kinase, sulfatase and phosphorylase families. For example, roscovitine (Selciclib) is a cyclin-dependent kinase inhibitory purine analogue in clinical development in a range of cancer, infectious disease and anti-inflammatory settings (De Azevedo et al. 1997). The specific sub-class of thiopurine analogues, such as 6-mercaptopurine and thioguanine, are also used as standard cancer chemotherapeutic agents, principally as anti-leukaemic drugs (Sahasranaman et al. 2008). Figure 1 shows the chemical structures of selected purine analogues in clinical use or advanced development.
During our previous work on the identification of 3-methylglutamic acid in disrupting CRYAB/VEGF165 interaction (Chen et al. 2014), we noted that the thioguanine core structure was also able to potentially bind within the protein–protein interface binding pocket. In this paper, we extend our studies on the identification of novel chemical scaffolds active in models of TNBC through the synthesis and in vitro antitumour evaluation of a series of substituted thiopurines, and test their potential as CRYAB/VEGF inhibitors.
Materials and methods
All solvents and reagents were used as obtained from commercial sources unless otherwise indicated. Final purification of target compounds was carried out by silica gel column chromatography using CH2Cl2/MeOH as eluent. Compound purity was established using 1H and 13C NMR, alongside accurate mass spectrometry. 1H NMR and 13C NMR spectra were recorded in the appropriate deuterated solvents with a Bruker Avance DPX500 spectrometer operating at 500 MHz. Chemical shifts (δ) are reported in parts per million (ppm) using the following abbreviations: s, singlet; d, doublet; t, triplet, q, quartet, quin, quintet; sex, sextet; sep, septet; m, multiplet, br, broad, app.t, apparent triplet. All coupling constants are reported in Hz. Mass spectra were recorded with a Bruker microTOF mass spectrometer using electrospray ionisation (ESI) conditions, or provided as a service at the National Mass Spectrometry Facility at Swansea University, UK. Synthetic procedures and analytical/spectroscopic data for intermediate compounds can be found in Supplementary Information.
General methods for the alkylation of purines
Alkylation method 1
Substituted purine compound (1.0 eq) and potassium carbonate (1.1 eq) were dissolved in DMF (5 mL) at r.t. for 5 min. Bromoethane (1.2 eq) was added dropwise, then the reaction stirred for 3–18 h, after which the reaction was quenched with cold water resulting in precipitation. The precipitate was filtered and washed with water. If no precipitate formed, the product was extracted with chloroform or ethyl acetate, dried over sodium sulphate and concentrated under reduced pressure.
Alkylation method 2
Substituted purine compound (1.0 eq) were dissolved in 1 M potassium hydroxide (1.1 eq). Bromoethane (1.2 eq) was added dropwise, then the reaction stirred for 3–5 h. On completion of the reaction, the product was precipitated with acetic acid, filtered, washed with water and dried.
Alkylation method 3
Substituted purine compound (1.0 eq) and 1,8-diazabicyclo[5.4.0]undec-7-ene (1.1 eq) were dissolved in DMF at r.t. for 5 min. The alkyl halide (1.2 eq) was added dropwise, then the reaction stirred for 3–5 h. The reaction was then quenched with cold water resulting in precipitation. The precipitate was filtered, washed with water and dried prior to purification by column chromatography.
2,6-bis(ethylthio)-7H-purin-8(9H)-one (4a)
According to general method 3 with 2,6-dimercapto-7H-purin-8(9H)-one (200 mg, 1.0 mmol), DMF (2 mL), DBU (0.15 ml, 1.0 mmol) and bromoethane (0.14 ml, 2.0 mmol) The product was purified by column chromatography with CH2Cl2/MeOH 90:10 as an eluent to afford a white solid of yield 90 mg, 35%. 1H-NMR (500 MHz, DMSO-d6): δ 11.70 (s, br, 1H, NH), 11.16 (s, br, 1H, NH), 3.24 (q, 2H, J = 7.5 Hz, SCH2), 3.09 (q, 2H, J = 7.5 Hz, SCH2), 1.32 (td, 6H, J1 = 7.5 Hz, J2 = 3.0 Hz, CH3). 13C-NMR (500 MHz, DMSO-d6): δ 161.45 (C2), 153.78 (C6), 149.40 (C8), 144.08 (C4), 116.81 (C5), 25.09 (SCH2), 23.39 (SCH2), 15.64 (CH3), 15.28 (CH3). m/z (FTMS + ESI) = Found [M]+ (C9H12N4OS2) 256.9068 requires 256.3420.
2,6-bis(methylthio)-7H-purin-8(9H)-one (4b)
According to the general method 3 with 2,6-dimercapto-7H-purin-8(9H)-one (200 mg, 1.0 mmol), DMF (2 ml), DBU (0.15 ml, 1.0 mmol) and iodomethane (0.12 ml, 2.0 mmol). The product was purified by column chromatography with CH2Cl2/MeOH 90:10 as an eluent to afford a white solid of yield 110 mg, 48%. 1H-NMR (500 MHz, DMSO-d6): δ 11.75 (s, br, 1H, NH), 11.23 (s, br, 1H, NH), 2.60 (s, 3H, SCH3), 2.51 (s, 3H, SCH3). 13C-NMR (500 MHz, DMSO-d6): δ 161.98 (C2), 153.78 (C6), 149.13 (C8), 144.58 (C4), 116.59 (C5), 14.24 (SCH3), 11.82 (SCH3). m/z (FTMS + ESI) = Found [M+H]+ (C7H8N4OS2) 229.0212 requires 228.2880.
2,6-bis(propylthio)-7H-purin-8(9H)-one (4c)
According to the general method 3 with 2,6-dimercapto-7H-purin-8(9H)-one (100 mg, 0.5 mmol), DMF (2 ml), DBU (0.08 ml, 0.5 mmol) and 1-bromopropane (0.09 ml, 1.0 mmol). The product was purified by column chromatography with CH2Cl2/MeOH 90:10 as an eluent to afford a light yellow solid of yield 100 mg, 70%. 1H-NMR (500 MHz, DMSO-d6): δ 11.69 (s, br, 1H, NH), 11.15 (s, br, 1H, NH), 3.21 (t, 2H, J = 7.0 Hz, SCH2), 3.06 (t, 2H, J = 7.0 Hz, SCH2), 1.65–1.73 (m, 4H, CH2), 0.97–1.00 (m, 6H, CH3). 13C-NMR (500 MHz, DMSO-d6): δ 161.45 (C2), 153.78 (C6), 149.37 (C8), 144.07 (C4), 116.82 (C5), 32.68 (SCH2), 30.64 (SCH2), 23.41 (CH2), 23.02 (CH2), 13.79 (CH3), δ 13.63 (CH3). m/z (FTMS + ESI) = Found [M]+ (C11H16N4OS2) 284.9649 requires 284.3960.
2,6-bis(isobutylthio)-7H-purin-8(9H)-one (4d)
According to the general method 3 with 2,6-dimercapto-7H-purin-8(9H)-one (200 mg, 1.0 mmol), DMF (2 ml), DBU (0.15 ml, 1.0 mmol) and 1-bromo-2-methylpropane (0.22 ml, 2.0 mmol). The product was purified by column chromatography with CH2Cl2/MeOH 90:10 as eluent to afford a light yellow solid of yield 90 mg, 29%. 1H-NMR (500 MHz, DMSO-d6): δ 11.67 (s, br, 1H, NH), 11.17 (s, br, 1H, NH), 3.18 (d, 2H, J = 7.0 Hz, SCH2), 3.02 (d, 2H, J = 7.0 Hz, SCH2), 1.92 (sep, 2 × 1H, J = 7.0 Hz, CH), 1.00 (d, 12H, J = 7.0 Hz, CH3). 13C-NMR (500 MHz, DMSO-d6): δ 161.45 (C2), 153.81 (C6), 149.36 (C8), 144.00 (C4), 116.81 (C5), 39.20 (SCH2), 36.86 (SCH2), 28.80 (CH), 28.38 (CH), 22.21 (CH3), 22.01 (CH3). m/z (FTMS + ESI) = Found [M+H]+ (C13H20N4OS2) 313.0257 requires 312.4500.
2,6-bis(benzylthio)-7H-purin-8(9H)-one (4e)
According to the general method 3 with 2,6-dimercapto-7H-purin-8(9H)-one (100 mg, 0.5 mmol), DMF (2 ml), DBU (0.08 ml, 0.5 mmol) and benzyl bromide (0.11 ml, 1.0 mmol). The product was purified by column chromatography with CH2Cl2/MeOH 90:10 as an eluent to afford an off white solid of yield 140 mg, 74%. 1H-NMR (500 MHz, DMSO-d6): δ 11.81 (s, br, 1H, NH), 11.76 (s, br, 1H, NH), 7.41 (d, 2H, J = 7.2 Hz, CHBz), 7.37 (d, 2H, J = 7.2 Hz, CHBz), 7.29–7.32 (m, 4H, CHBz), 7.23–7.27 (m, 2H, CHBz), 4.51 (s, 2H, SCH2), 4.40 (s, 2H, SCH2). 13C-NMR (500 MHz, DMSO-d6): δ 161.12 (C2), 153.77 (C6), 149.55 (C8), 143.52 (C4), 138.24 (CBz), 138.02 (CBz), 129.34 (CHBz), 129.23 (CHBz), 128.97 (CHBz), 128.89 (CHBz), 127.70 (CHBz), 127.50 (CHBz), 116.76 (C5), 34.95 (CH2), 32.73 (CH2). m/z (FTMS + ESI) = Found [M+H]+ (C19H16N4OS2) 381.0796 requires 380.4840.
2,6-bis(phenethylthio)-7H-purin-8(9H)-one (4f)
According to the general method 3 with 2,6-dimercapto-7H-purin-8(9H)-one (100 mg, 0.5 mmol), DMF (2 ml), DBU (0.08 ml, 0.5 mmol) and 2-bromoethyl benzene (0.14 ml, 1.0 mmol). The product was purified by column chromatography with CH2Cl2/MeOH 90:10 as an eluent to afford a white solid of yield 100 mg, 49%. 1H-NMR (500 MHz, DMSO-d6): δ 11.78 (s, br, 1H, NH), 11.20 (s, br, 1H, NH), 7.19–7.31 (m, 10H, HBz), 3.53 (t, 2H, J = 7.5 Hz, SCH2), 3.36 (t, 2H, J = 7.5 Hz, SCH2), 2.95 (t, 4H, J = 7.5 Hz, CH2). 13C-NMR (500 MHz, DMSO-d6): δ 161.35 (C2), 153.80 (C6), 149.45 (C8), 143.86 (C4), 140.72 (CBz), 140.32 (CBz), 128.98 (CHBz), 128.84 (CHBz), 128.82 (CHBz), 126.82 (CHBz), 126.72 (CHBz), 116.95 (C5), 35.82 (CH2), 35.60 (CH2), 32.04 (SCH2), 30.05 (SCH2). m/z (FTMS + ESI) = Found [M+H]+ (C21H20N4OS2) 409.1150 requires 408.5380.
2,6-bis((3-phenylpropyl)thio)-7H-purin-8(9H)-one (4g)
According to the general method 3 with 2,6-dimercapto-7H-purin-8(9H)-one (100 mg, 0.5 mmol), DMF (2 ml), DBU (0.08 ml, 0.5 mmol) and 1-bromo-3-phenylpropane (0.15 ml, 1.0 mmol). The product was purified by column chromatography with CH2Cl2/MeOH 90:10 as an eluent to afford an off-white solid of yield 110 mg, 50%. 1H-NMR (500 MHz, DMSO-d6): δ 11.71 (s, br, 1H, NH), 11.17 (s, br, 1H, NH), 7.25–7.29 (m, 4H, HPh), 7.16–7.19 (m, 6H, HPh), 3.21 (t, 2H, J = 7.5 Hz, SCH2), 3.06 (t, 2H, J = 7.5 Hz, SCH2), 2.69 (t, 4H, J = 7.0 Hz, CH2), 1.92–1.98 (m, 4H, CH2). 13C-NMR (500 MHz, DMSO-d6): δ 161.33 (C2), 153.78 (C6), 149.44 (C8), 143.88 (C4), 141.67 (CPh), 141.47 (CPh), 128.84 (CHPh), 128.82 (CHPh), 128.76 (CHPh), 128.74 (CHPh), 126.38 (CHPh), 126.33 (CHPh), 116.97 (C5), 34.70 (CH2), 34.57 (CH2), 31.51 (CH2), 31.17 (CH2), 30.33 (SCH2), 28.40 (SCH2). m/z (FTMS + ESI) = Found [M+H]+ (C23H24N4OS2) 437.2875 requires 436.5920.
9-ethyl-2,6-bis(ethylthio)-8,9-dihydro-7H-purine-8-thiol (6a)
According to the general method 3 with 2,6-dimercapto-7,9-dihydro-8H-purine-8-thione (100 mg, 0.46 mmol), DMF (2 ml), DBU (0.07 ml, 0.46 mmol) and bromoethane (0.07 ml, 0.92 mmol). The product was purified by column chromatography with CH2Cl2/MeOH 95:5 as an eluent to afford a light yellow solid of yield 40 mg, 29%. 1H-NMR (500 MHz, DMSO-d6): δ 13.21 (s, br, 1H, NH), 3.27–3.31 (m, 4H, CH2), 3.10–3.16 (m, 2H, CH2), 1.32–1.39 (m, 9H, CH3). 13C-NMR (500 MHz, DMSO-d6): δ 169.99 (C8), 163.58 (C2), 150.73 (C6), 147.44 (C4), 119.40 (C5), 25.82 (NCH2), 25.25 (SCH2), 23.49 (SCH2), 15.43 (CH3), 15.29 (CH3), 15.19 (CH3). m/z (FTMS + ESI) = Found [M+H]+ (C11H16N4S3) 301.0609 requires 300.4570.
9-methyl-2,6-bis(methylthio)-7,9-dihydro-8H-purine-8-thione (6b)
According to the general method 3 with 2,6-dimercapto-7,9-dihydro-8H-purine-8-thione (100 mg, 0.46 mmol) DMF (2 ml), DBU (0.07 ml, 0.46 mmol) and iodomethane (0.06 ml, 0.92 mmol). The product was purified by column chromatography with CH2Cl2/MeOH 95:5 as an eluent to afford the tri-substituted product as an off-white solid of yield 40 mg, 17%. 1H-NMR (500 MHz, DMSO-d6, tri-substituted product): δ 2.54 (s, 3H, CH3), 2.52 (s, 3H, CH3), 2.47 (s, 3H, CH3). 13C-NMR (500 MHz, DMSO-d6): δ 163.84 (C8), 163.19 (C2), 156.40 (C6), 148.74 (C4), 133.58 (C5), 14.31 (CH3), 14.12 (CH3), 11.56 (CH3). m/z (FTMS + ESI) = Found [M+H]+ (C8H10N4S3) 259.0133 requires 258.3760.
The tetra-substituted product (7) was an off-white solid of yield 50 mg, 30%. 1H-NMR (500 MHz, DMSO-d6, tetra-substituted product): δ 3.85 (s, 3H, NCH3), 2.75 (s, 3H, NCH3), 2.64 (s, 3H, SCH3), 2.54 (s, 3H, SCH3). 13C-NMR (500 MHz, DMSO-d6): δ 162.84 (C8), 156.54 (C2), 153.80 (C6), 151.99 (C4), 128.85 (C5), 33.45 (CH3), 29.08 (CH3), 14.09 (CH3), 11.68 (CH3). m/z (FTMS + ESI) = Found [M+H]+ (C9H12N4S3) 273.0334 requires 272.4030.
9-propyl-2,6-bis(propylthio)-8,9-dihydro-7H-purine-8-thiol (6c)
According to general the method 3 with 2,6-dimercapto-7,9-dihydro-8H-purine-8-thione (100 mg, 0.46 mmol), DMF (2 ml), DBU (0.07 ml, 0.46 mmol) and 1-bromopropane (0.08 ml, 0.92 mmol). The product was purified by column chromatography with CH2Cl2/MeOH 95:5 as an eluent to afford a light yellow solid of yield 74 mg, 47%. 1H-NMR (500 MHz, DMSO-d6): δ 3.24–3.29 (m, 4H, SCH2), 3.08–3.13 (m, 2H, NCH2), 1.68–1.76 (m, 6H, CH2), 0.97–1.03 (m, 9H, CH3). 13C-NMR (500 MHz, DMSO-d6): δ 170.03 (C8), 163.56 (C2), 150.72 (C6), 147.41 (C4), 119.47 (C5), 32.88 (SCH2), 32.82 (SCH2), 30.69 (NCH2), 23.20 (CH2), 23.03 (CH2), 22.95 (CH2), 13.78 (CH3), 13.69 (CH3), 13.66 (CH3). m/z (FTMS + ESI) = Found [M+H]+ (C14H22N4S3) 343.1079 requires 342.5380.
9-isobutyl-2,6-bis(isobutylthio)-8,9-dihydro-7H-purine-8-thiol (6d)
According to the general method 3 with 2,6-dimercapto-7,9-dihydro-8H-purine-8-thione (100 mg, 0.46 mmol), DMF (2 ml), DBU (0.07 ml, 0.46 mmol) and 1-bromo-2-methylpropane (0.10 ml, 0.92 mmol). The product was purified by column chromatography with CH2Cl2/MeOH 95:5 as an eluent to afford a light yellow solid of yield 80 mg, 45%. 1H-NMR (500 MHz, DMSO-d6): δ 13.36 (s, br, 1H, NH), 3.22 (dd, 4H, J = 6.8 Hz, SCH2), 3.08 (d, 2H, J = 6.8 Hz, NCH2), 1.90–2.02 (m, 3H, CH), 1.00–1.03 (m, 18H, CH3). 13C-NMR (500 MHz, DMSO-d6): δ 162.43 (C8), 156.04 (C2), 152.58 (C6), 151.88 (C4), 129.41 (C5), 39.51 (S-CH2), 39.43 (SCH2), 36.51 (NCH2), 28.73 (CH), 28.52 (CH), 28.43 (CH), 22.23 (CH3), 22.08 (CH3), 21.95 (CH3). m/z (FTMS + ESI) = Found [M+H]+ (C17H28N4S3) 385.1744 requires 384.6190.
9-benzyl-2,6-bis(benzylthio)-8,9-dihydro-7H-purine-8-thiol (6e)
According to the general method 3 with 2,6-dimercapto-7,9-dihydro-8H-purine-8-thione (100 mg, 0.46 mmol), DMF (2 ml), DBU (0.07 ml, 0.46 mmol) and benzyl bromide (0.11 ml, 0.92 mmol). The precipitate was filtered, washed with water and dried. The product was purified by column chromatography with CH2Cl2/MeOH 95:5 as an eluent to afford a light yellow solid of yield 120 mg, 54%. 1H-NMR (500 MHz, DMSO-d6): δ 13.55 (s, br, 1H, NH), 7.42–7.47 (m, 6H, CHBz), 7.30–7.33 (m, 6H, CHBz), 7.24–7.28 (m, 3H, CHBz), 4.59 (s, 2H, SCH2), 4.56 (s, br, 2H, NCH2), 4.46 (s, 2H, SCH2). 13C-NMR (500 MHz, DMSO-d6): δ 162.29 (C8), 155.76 (C2), 152.73 (C6), 151.56 (C4), 138.21 (CBz), 137.53 (CBz), 129.47 (CHBz), 129.40 (CHBz), 129.28 (CHBz), 128.99 (CHBz), 128.95 (CHBz), 128.91 (CHBz), 127.97 (CHBz), 129.41 (C5), 127.66 (CHBz), 127.53 (CHBz), 35.24 (SCH2), 35.13 (SCH2), 32.38 (NCH2). m/z (FTMS + ESI) = Found [M+H]+ (C26H22N4S3) 487.3201 requires 486.6700.
9-phenethyl-2,6-bis(phenethylthio)-8,9-dihydro-7H-purine-8-thiol (6f)
According to the general method 3 with 2,6-dimercapto-7H-purine-8(9H)-thione (100 mg, 0.46 mmol) DMF (2 ml), DBU (0.07 ml, 0.46 mmol) and 2-bromoethyl benzene (0.13 ml, 0.92 mmol). The precipitate was filtered, washed with water and dried. The product was purified by column chromatography with CH2Cl2/MeOH 95:5 as an eluent to afford a light yellow solid of yield 102 mg, 42%. 1H-NMR (500 MHz, DMSO-d6): δ 13.43 (s, br, 1H, NH), 7.29–7.34 (m, 12H, CHBz), 7.21–7.25 (m, 3H, CHBz), 3.52–3.59 (m, 4H, SCH2), 3.41–3.44 (m, 2H, NCH2), 2.99–3.05 (m, 6H, CH2). 13C-NMR (500 MHz, DMSO-d6): δ 162.37 (C8), 156.00 (C2), 152.71 (C6), 151.60 (C4), 140.76 (CBz), 140.49 (CBz), 140.21 (CBz), 129.55 (C5), 129.09 (CHBz), 129.00 (CHBz), 128.87 (CHBz), 128.82 (CHBz), 126.91 (CHBz), 126.84 (CHBz), 126.73 (CHBz), 35.71 (SCH2), 35.57 (SCH2), 35.33 (NCH2), 32.62 (SCH2), 32.25 (SCH2), 29.79 (NCH2). m/z (FTMS + ESI) = Found [M+H]+ (C29H28N4S3) 529.15 requires 528.75.
9-(3-phenylpropyl)-2,6-bis((3-phenylpropyl)thio)-8,9-dihydro-7H-purine-8-thiol (6g)
According to the general method 3 with 2,6-dimercapto-7,9-dihydro-8H-purine-8-thione (100 mg, 0.46 mmol) DMF (2 ml), DBU (0.07 ml, 0.46 mmol) and 1-bromo-3-phenyl propane (0.14 ml, 0.92 mmol). The reaction was quenched with water resulting in precipitation. The precipitate was filtered, washed with water and dried. The product was purified by column chromatography with CH2Cl2/MeOH 95:5 as an eluent to afford a light yellow solid of yield 116 mg, 44%. 1H-NMR (500 MHz, DMSO-d6): δ 13.40 (s, br, 1H, NH), 7.26–7.31 (m, 6H, CHPh), 7.16–7.24 (m, 9H, CHPh), 3.26–3.29 (m, 4H, SCH2), 3.13 (dd, J1 = 7.5 Hz, J2 = 2.5 Hz, 2H, NCH2), 1.95–2.07 (m, 6H, CH2). 13C-NMR (500 MHz, DMSO-d6): δ 162.33 (C8), 155.96 (C2), 152.66 (C6), 151.66 (C4), 141.68 (CPh), 141.53 (CPh), 141.44 (CPh), 129.54 (C5), 128.84 (CHPh), 128.83 (CHPh), 128.79 (CHPh), 128.77 (CHPh), 126.39 (CHPh), 126.37 (CHPh), 126.34 (CHPh), 34.70 (SCH2), 34.62 (SCH2), 34.39 (NCH2), 31.35 (SCH2), 31.15 (SCH2), 30.97 (NCH2), 30.87 (SCH2), 30.54 (SCH2), 38.00 (NCH2). m/z (FTMS + ESI) = Found [M+H]+ (C34H34N4S3) 571.2016 requires 570.8320.
General method for the hydrolysis of brominated alkyl-guanines: synthesis of 2-amino-6 hydroxy-9-alkyl-7,9-dihydro-8H-purin-8-ones (13a-g)
A mixture of brominated alkyl-guanine (12a-g, 1 eq), sodium acetate (5.3 eq), acetic acid (3 ml) and acetic anhydride (0.5 ml) were heated under reflux and N2 for 15 h. The reaction was cooled and evaporated to dryness. The resulting residue was suspended in water (5 ml) and the pH adjusted to 13 with 10 M NaOH. The mixture was refluxed for 20 min, cooled and the solid formed collected, triturated with potassium phosphate buffer pH 7.5, filtered and dried.
2-amino-6-hydroxy-9-methyl-7,9-dihydro-8H-purin-8-one (13a)
According to the general method with 2-amino-8-bromo-9-methyl-9H-purin-6-ol (100 mg, 0.41 mmol) and sodium acetate (178 mg, 2.17 mg) to afford a tan solid of yield 62 mg, 63%. 1H-NMR (500 MHz, DMSO-d6): δ 10.41 (s, 1H, OH), 6.90 (s, br, 2H, NH2), 3.08 (s, 3H, CH3). 13C-NMR (500 MHz, DMSO-d6): δ 154.69 (C2), 153.02 (C6), 152.08 (C4), 148.78 (C8), 98.55 (C5), 25.90 (CH3). m/z (FTMS + ESI) = Found [M+H]+ (C6H7N5O2) 182.0672 requires 181.1550.
2-amino-9-ethyl-6-hydroxy-7,9-dihydro-8H-purin-8-one (13b)
According to the general method with 2-amino-8-bromo-9-ethyl-9H-purin-6-ol (100 mg, 0.39 mmol) and sodium acetate (170 mg, 2.05 mg) to afford a tan solid of yield 40 mg, 40%. 1H-NMR (500 MHz, DMSO-d6): δ 10.65 (s, br, 1H, NH), 10.49 (s, 1H, OH), 6.48 (s, br, 2H, NH2), 3.62 (q, 2H, J = 7.0 Hz, NCH2), 1.16 (t, 3H, J = 7.0 Hz, CH3). 13C-NMR (500 MHz, DMSO-d6): δ 153.97 (C2), 152.79 (C6), 151.45 (C4), 148.51 (C8), 98.55 (C5), 41.38 (NCH2), 11.48 (CH3). m/z (FTMS + ESI) = Found [M+H]+ (C6H7N5O2) 196.0828 requires 195.1820.
2-amino-6-hydroxy-9-propyl-7,9-dihydro-8H-purin-8-one (13c)
According to the general method with 2-amino-8-bromo-9-propyl-9H-purin-6-ol (100 mg, 0.37 mmol) and sodium acetate (160 mg, 1.95 mg to afford a tan solid of yield 20 mg, 26%. 1H-NMR (500 MHz, DMSO-d6): δ 10.64 (s, br, 1H, NH), 10.53 (s, 1H, OH), 6.49 (s, br, 2H, NH2), 3.54 (t, 2H, J = 7.5 Hz, NCH2), 1.60 (sext, 2H, J = 7.5 Hz, CH2), 0.83 (t, 3H, J = 7.5 Hz, CH3). 13C-NMR (500 MHz, DMSO-d6): δ 153.97 (C2), 152.79 (C6), 151.45 (C4), 148.51 (C8), 98.55 (C5), 41.38 (NCH2), 21.99 (CH2), 11.48 (CH3). m/z (FTMS + ESI) = Found [M+H]+ (C8H11N5O2) 210.0986 requires 209.2090.
2-amino-6-hydroxy-9-isobutyl-7,9-dihydro-8H-purin-8-one (13d)
According to the general method with 2-amino-8-bromo-9-isobutyl-9H-purin-6-ol (100 mg, 0.35 mmol) and sodium acetate (152 mg, 1.85 mg to afford a tan solid of yield 45 mg, 58%. 1H-NMR (500 MHz, DMSO-d6): δ 10.73 (s, br, 1H, NH), 10.49 (s, 1H, OH), 6.48 (s, br, 2H, NH2), 3.39 (td, 2H, J = 7.5 Hz, NCH2), 2.10 (sept, 1H, J1 = 7.5 Hz, J2 = 6.5 Hz, CH), 0.82 (d, 6H, J = 6.5 Hz, CH3). 13C-NMR (500 MHz, DMSO-d6): δ 154.03 (C2), 153.02 (C6), 151.66 (C4), 148.74 (C8), 98.55 (C5), 46.68 (NCH2), 27.63 (CH), 20.22 (2xCH3). m/z (FTMS + ESI) = Found [M+H]+ (C9H13N5O2) 224.1142 requires 223.2360.
2-amino-9-benzyl-6-hydroxy-7,9-dihydro-8H-purin-8-one (13e)
According to the general method with 2-amino-9-benzyl-8-bromo-9H-purin-6-ol (100 mg, 0.31 mmol) and sodium acetate (135 mg, 1.65 mg) to afford a tan solid of yield 40 mg, 50%. 1H-NMR (500 MHz, DMSO-d6): δ 10.80 (s, br 1H, NH), 10.67 (s, 1H, OH), 7.24–7.33 (m, 5H, HBz), 6.52 (s, br, 2H, NH2), 4.80 (s, 2H, NCH2). 13C-NMR (500 MHz, DMSO-d6): δ 154.17 (C2), 152.73 (C4), 148.38 (C6), 137.82 (C8), 128.94 (CBz), 127.67 (CHBz), 127.42 (CHBz), 98.74 (C5), 42.60 (NCH2). m/z (FTMS + ESI) = Found [M+H]+ (C12H11N5O2) 258.0988 requires 257.2530.
2-amino-6-hydroxy-9-phenethyl-7,9-dihydro-8H-purin-8-one (13f)
According to the general method with 2-amino-8-bromo-9-phenethyl-9H-purin-6-ol (100 mg, 0.30 mmol) and sodium acetate (129 mg, 1.57 mg) to afford a tan solid of yield 69 mg, 85%. 1H-NMR (500 MHz, DMSO-d6): δ 10.50 (s, 1H, OH), 7.26–7.30 (m, 2H, HBz), 7.19–7.22 (m, 3H, HBz), 6.93 (s, br, 4H, NH2), 3.81–3.88 (m, 2H, CH2), 2.94–3.00 (m, 2H, CH2). 13C-NMR (500 MHz, DMSO-d6): δ 154.66 (C2), 152.66 (C4), 148.69 (C6), 139.01 (C8), 138.84 (CBz), 129.03 (CHBz), 128.86 (CHBz), 126.80 (CHBz), 98.47 (C5), 34.38 (NCH2), 30.80 (CH2). m/z (FTMS + ESI) = Found [M+H]+ (C13H13N5O2) 272.1146 requires 271.2800.
2-amino-6-hydroxy-9-(3-phenylpropyl)-7,9-dihydro-8H-purin-8-one (13g)
According to the general method with 2-amino-8-bromo-9-(3-phenylpropyl)-9H-purin-6-ol (100 mg, 0.29 mmol) and sodium acetate (125 mg, 1.52 mg) to afford a tan solid of yield 5 mg, 6%. 1H-NMR (500 MHz, DMSO-d6): δ 10.65 (s, br 1H, NH), 10.55 (s, 1H, OH), 7.26–7.32 (m, 2H, HBz), 7.16–7.22 (m, 3H, HBz), 6.49 (s, br, 2H, NH2), 3.64 (t, 2H, J = 7.5 Hz, CH2), 2.57 (t, 2H, J = 7.5 Hz, CH2), 1.91 (quin, 2H, J = 7.5 Hz, CH2). 13C-NMR (500 MHz, DMSO-d6): δ 154.48 (C2), 152.86 (C4), 146.62 (C6), 141.71 (C8), 140.81 (CBz), 131.10 (CHBz), 131.06 (CHBz), 128.78 (CHBz), 128.70 (CHBz), 126.30 (CHBz), 98.42 (C5), 33.21 (NCH2), 30.37 (CH2), 29.06 (CH2). m/z (FTMS + ESI) = Found [M+H]+ (C14H15N5O2) 286.1301 requires 285.3070.
6-(methylthio)-9H-purine (15a)
According to the general alkylation method 3 with 6-mercaptopurine monohydrate (200 mg, 1.18 mmol), DMF (3 ml), DBU (0.18 ml, 1.18 mmol) and iodomethane (0.15 ml, 2.35 mmol) for 3 h. The product extracted with chloroform, and purified by column chromatography with CH2Cl2/MeOH 95:5 as an eluent to afford a white solid of yield 110 mg, 56%. 1H-NMR (500 MHz, DMSO-d6): δ 13.51 (s, br, 1H, NH), 8.71 (s, 1H, H2), 8.44 (s, 1H, H8), 2.67 (s, 3H, CH3). 13C-NMR (500 MHz, DMSO-d6): δ 151.99 (C2), 143.63 (C8), 11.66 (CH3). m/z (FTMS + ESI) = Found [M+H]+ (C6H6N4S) 167.0383 requires 166.2020.
6-(ethylthio)-7H-purine (15b)
According to the general alkylation method 3 with 6-mercaptopurine monohydrate (200 mg, 1.18 mmol), DMF (3 ml), DBU (0.18 ml, 1.18 mmol) and bromoethane (0.18 ml, 2.35 mmol) for 3 h. The product extracted with chloroform, and purified by column chromatography with CH2Cl2/MeOH 95:5 as an eluent to afford a white solid of yield 170 mg, 80%. 1H-NMR (500 MHz, DMSO-d6): δ 13.51 (s, br, 1H, NH), 8.70 (s, 1H, H2), 8.44 (s, 1H, H8), 3.34 (q, 2H, J = 7.0 Hz, SCH2), 1.37 (t, 3H, J = 7.0 Hz, CH3). 13C-NMR (500 MHz, DMSO-d6): δ 151.96 (C2), 147.82 (C8), 22.87 (SCH2), 15.42 (CH3). m/z (FTMS + ESI) = Found [M+H]+ (C7H8N4S) 181.0541 requires 180.2290.
6-(propylthio)-7H-purine (15c)
According to the general alkylation method 3 with 6-mercaptopurine monohydrate (200 mg, 1.18 mmol), DMF (3 ml), DBU (0.18 ml, 1.18 mmol) and 1-bromopropane (0.21 ml, 2.35 mmol) for 3 h. The product extracted with chloroform, and purified by column chromatography with CH2Cl2/MeOH 95:5 as an eluent to afford a white solid of yield 140 mg, 61%. 1H-NMR (500 MHz, DMSO-d6): δ 13.50 (s, br, 1H, NH), 8.69 (s, 1H, H2), 8.43 (s, 1H, H8), 3.33 (t, 2H, J = 7.5 Hz, SCH2), 1.73 (sext, 2H, J = 7.5 Hz, CH2), 1.01 (t, 3H, J = 7.5 Hz, CH3). 13C-NMR (500 MHz, DMSO-d6): δ 151.93 (C2), 143.27 (C8), 30.15 (SCH2), 23.10 (CH2), 13.70 (CH3). m/z (FTMS + ESI) = Found [M+H]+ (C8H10N4S) 195.0696 requires 194.2560.
6-(isobutylthio)-7H-purine (15d)
According to the general alkylation method 3 with 6-mercaptopurine monohydrate (200 mg, 1.18 mmol), DMF (3 ml), DBU (0.18 ml, 1.18 mmol) and 1-bromo-2-methylpropane (0.26 ml, 2.35 mmol) for 3 h. The product extracted with chloroform, and purified by column chromatography with CH2Cl2/MeOH 95:5 as an eluent to afford a white solid of yield 90 mg, 37%. 1H-NMR (500 MHz, DMSO-d6): δ 13.50 (s, br, 1H, NH), 8.68 (s, 1H, H2), 8.44 (s, 1H, H8), 3.29 (d, 2H, J = 6.5 Hz, SCH2), 1.97 (sep, 1H, J1 = 6.5 Hz, J2 = 7.0 Hz, CH), 1.02 (d, 6H, J = 6.5 Hz, 2xCH3). 13C-NMR (500 MHz, DMSO-d6): δ 151.86 (C2), 143.68 (C8), 36.38 (SCH2), 28.68 (CH2), 22.09 (2xCH3). m/z (FTMS + ESI) = Found [M+H]+ (C9H12N4S) 209.1011 requires 208.2830.
6-(benzylthio)-9H-purine (15e)
According to the general alkylation method 3 with 6-mercaptopurine monohydrate (200 mg, 1.18 mmol), DMF (3 ml), DBU (0.18 ml, 1.18 mmol) and benzyl bromide (0.28 ml, 2.35 mmol) for 3 h. The product extracted with chloroform, and purified by column chromatography with CH2Cl2/MeOH 95:5 as an eluent to afford a white solid of yield 140 mg, 49%. 1H-NMR (500 MHz, DMSO-d6): δ 13.55 (s, br, 1H, NH), 8.75 (s, 1H, H2), 8.46 (s, 1H, H8), 7.46 (d, 2H, J = 7.5 Hz, HBz), 7.32 (t, 2H, J = 7.0 Hz, HBz), 7.25 (t, 1H, J1 = 7.0 Hz, J2 = 7.5 Hz, HBz), 4.66 (s, 2H, SCH2). 13C-NMR (500 MHz, DMSO-d6): δ 151.92 (C2), 143.97 (C8), 138.35 (CBz), 129.46 (CHBz), 128.96 (CHBz), 127.64 (CHBz), 32.10 (SCH2). m/z (FTMS + ESI) = Found [M+H]+ (C12H10N4S) 243.0716 requires 242.3000.
6-(phenethylthio)-9H-purine (15f)
According to the general alkylation method 3 with 6-mercaptopurine monohydrate (200 mg, 1.18 mmol), DMF (3 ml), DBU (0.18 ml, 1.18 mmol) and 2-bromoethylbenzene (0.32 ml, 2.35 mmol) for 3 h. The product extracted with chloroform, and purified by column chromatography with CH2Cl2/MeOH 95:5 as an eluent to afford a white solid of yield 160 mg, 53%. 1H-NMR (500 MHz, DMSO-d6): δ 13.52 (s, br, 1H, NH), 8.73 (s, 1H, H2), 8.45 (s, 1H, H8), 7.31–7.32 (m, 4H, HBz), 7.21–7.25 (m, 1H, HBz), 3.61 (t, 2H, J = 7.0 Hz, CH2), 3.03 (t, 2H, J = 7.0 Hz, CH2). 13C-NMR (500 MHz, DMSO-d6): δ 152.01 (C2), 143.33 (C8), 140.58 (CBz), 129.07 (CHBz), 128.85 (CHBz), 126.82 (CHBz), 35.61 (SCH2), 29.72 (CH2). m/z (FTMS + ESI) = Found [M+H]+ (C13H12N4S) 257.0850 requires 256.3270.
6-((3-phenylpropyl)thio)-9H-purine (15g)
According to the general alkylation method 3 with 6-mercaptopurine monohydrate (200 mg, 1.18 mmol), DMF (3 ml), DBU (0.18 ml, 1.18 mmol) and 1-bromo-3-phenylpropane (0.37 ml, 2.35 mmol) for 3 h. The product extracted with chloroform, and purified by column chromatography with CH2Cl2/MeOH 95:5 as an eluent to afford a white solid of yield 200 mg, 63%. 1H-NMR (500 MHz, DMSO-d6): δ 13.52 (s, br, 1H, NH), 8.68 (s, 1H, H2), 8.45 (s, 1H, H8), 7.28–7.31 (m, 2H, HPh), 7.17–7.24 (m, 3H, HPh), 3.35 (t, 2H, J = 7.3 Hz, CH2), 2.76 (t, 2H, J = 7.5 Hz, CH2), 2.03 (quin, 2H, J1 = 7.3, J2 = 7.5 Hz, CH2). 13C-NMR (500 MHz, DMSO-d6): δ 151.94 (C2), 141.61 (CPh), 128.82 (CHPh), 126.36 (CHPh), 39.48 (SCH2), 31.28 (CH2), 27.87 (CH2). m/z (FTMS + ESI) = Found [M+H]+ (C14H14N4S) 271.1011 requires 270.3540.
2,6-bis(methylthio)pyrimidine-4,5-diamine (16a)
According to the general alkylation method 1 with 4,5-diamino-2,6-dimercaptopyrimidine (200 mg, 1.0 mmol), potassium carbonate (150 mg, 1.1 mmol) and iodomethane (0.07 ml, 1.2 mmol) for 3 h. The product was purified by column chromatography with CH2Cl2/MeOH 95:5 as an eluent to afford an off-white solid of yield 112 mg, 49%. 1H-NMR (500 MHz, MeOD): δ 2.57 (s, 3H, SCH3), 2.50 (s, 3H, SCH3). 13C-NMR (500 MHz, MeOD): δ 159.25 (C2), 152.82 (C6), 149.96 (C4), 118.09 (C5), 12.75 (2xSCH3), 11.55 (SCH3). m/z (FTMS + ESI) = Found [M+H]+ (C6H10N4S2) 203.0419 requires 202.2940.
2,6-bis(ethylthio)pyrimidine-4,5-diamine (16b)
According to the general alkylation method 1 with 4,5-diamino-2,6-dimercaptopyrimidine (100 mg, 0.57 mmol), potassium carbonate (150 mg, 1.1 mmol) and bromoethane (0.09 ml, 1.2 mmol) for 3 h. The product was purified by column chromatography with CH2Cl2/MeOH 95:5 as an eluent to afford an off-white solid of yield 99 mg, 76%. 1H-NMR (500 MHz, MeOD): δ 3.20 (q, 2H, J = 7.0 Hz, SCH2), 3.08 (9, 2H, J = 7.0 Hz, SCH2), 1.37 (t, 3H, J = 7.0 Hz, CH3), 1.36 (t, 3H, J = 7.5 Hz, CH3). 13C-NMR (500 MHz, MeOD): δ 158.58 (C2), 152.99 (C6), 148.97 (C4), 118.60 (C5), 24.52 (SCH2), 23.86 (SCH2), 14.49 (CH3), 14.18 (CH3). m/z (FTMS + ESI) = Found [M+H]+ (C8H14N4S2) 230.9 requires 230.3.
2,6-bis(propylthio)pyrimidine-4,5-diamine (16c)
According to the general alkylation method 1 with 4,5-diamino-2,6-dimercaptopyrimidine (200 mg, 1.0 mmol), potassium carbonate (150 mg, 1.1 mmol) and 1-bromopropane (0.06 ml, 1.2 mmol) for 3 h. The product was purified by column chromatography with CH2Cl2/MeOH 95:5 as an eluent to afford an off-white solid of yield 180 mg, 63%. 1H-NMR (500 MHz, MeOD): δ 3.18 (t, 2H, J = 7.5 Hz, SCH2), 3.06 (t, 2H, J = 7.5 Hz, SCH2), 1.73 (sext, 4H, J = 7.5 Hz, CH2), 1.05 (td, 6H, J1 = 7.5 Hz, J2 = 1.0 Hz, CH3). 13C-NMR (500 MHz, MeOD): δ 158.59 (C2), 152.92 (C6), 149.11 (C4), 118.49 (C5), 32.31 (SCH2), 31.40 (SCH2), 23.33 (2xCH2), 23.04 (2xCH2), 12.44 (2xCH3), 12.31 (CH3). m/z (FTMS + ESI) = Found [M+H]+ (C10H18N4S2) 259.1045 requires 258.4020.
2,6-bis(isobutylthio)pyrimidine-4,5-diamine (16d)
According to the general alkylation method 1 with 4,5-diamino-2,6-dimercaptopyrimidine (100 mg, 0.57 mmol), potassium carbonate (150 mg, 1.1 mmol) and 1-bromo-2-methylpropane (0.13 ml, 1.2 mmol) for 3 h. The product was purified by column chromatography with CH2Cl2/MeOH 95:5 as an eluent to afford a light brown solid of yield 90 mg, 50%. 1H-NMR (500 MHz, MeOD): δ 3.14 (d, 2H, J = 6.5 Hz, SCH2), 3.01 (d, 2H, J = 7.0 Hz, SCH2), 1.89–2.00 (m, 2H, CH), 1.06 (d, 3H, J = 2.0 Hz, CH3), 1.05 (d, 3H, J = 2.5, CH3). 13C-NMR (500 MHz, MeOD): δ 158.57 (C2), 152.87 (C6), 149.11 (C4), 118.43 (C5), 38.94 (SCH2), 37.91 (SCH2), 28.81 (CH), 28.47 (CH), 20.93 (4xCH3). m/z (FTMS + ESI) = Found [M+H]+ (C12H22N4S2) 287.0228 requires 286.4560.
2,6-bis(benzylthio)pyrimidine-4,5-diamine (16e)
According to the general alkylation method 1 with 4,5-diamino-2,6-dimercaptopyrimidine (200 mg, 1.0 mmol), potassium carbonate (150 mg, 1.1 mmol) and benzyl bromide (0.14 ml, 1.2 mmol) for 3 h. The product was purified by column chromatography with CH2Cl2/MeOH 95:5 as an eluent to afford an off-white solid of yield 190 mg, 50%. 1H-NMR (500 MHz, MeOD): δ 7.37–7.39 (m, 2H, HBz), 7.19–7.31 (m, 10H, HBz), 4.38 (s, 2H, SCH2), 4.34 (s, 2H, SCH2). 13C-NMR (500 MHz, MeOD): δ 158.08 (C2), 153.16 (C6), 147.77 (C4), 138.50 (CBz), 138.25 (CBz), 128.02 (CHBz), 128.36 (CHBz), 128.02 (CHBz), 127.97 (CHBz), 126.66 (CHBz), 126.48 (CHBz), 119.16 (C5), 34.66 (SCH2), 33.89 (SCH2). m/z (FTMS + ESI) = Found [M+H]+ (C18H18N4S2) 355.1015 requires 354.4900.
2,6-bis(phenethylthio)pyrimidine-4,5-diamine (16f)
According to the general alkylation method 1 with 4,5-diamino-2,6-dimercaptopyrimidine (200 mg, 1.0 mmol), potassium carbonate (150 mg, 1.1 mmol) and 2-bromoethyl benzene (0.16 ml, 1.2 mmol) for 5 h. The product was purified by column chromatography with CH2Cl2/MeOH 95:5 as an eluent to afford an off-white solid of yield 260 mg, 64%. 1H-NMR (500 MHz, MeOD): δ 7.15–7.26 (m, 10H, HBz), 3.45 (t, 2H, J = 7.5 Hz, SCH2), 3.34 (t, 2H, J = 7.5 Hz, SCH2), 2.96 (dt, 4H, J1 = 7.5 Hz, J2 = 3.5 Hz, 2xCH2). 13C-NMR (500 MHz, MeOD): δ 158.39 (C2), 153.07 (C6), 149.11 (C4), 140.64 (CBz), 140.23 (CBz), 128.24 (CHBz), 128.21 (CHBz), 128.03 (CHBz), 125.95 (CHBz), 125.83 (CHBz), 118.95 (C5), 36.18 (CH2), 35.98 (CH2), 31.73 (SCH2), 30.93 (SCH2). m/z (FTMS + ESI) = Found [M+H]+ (C20H22N4S2) 383.1298 requires 382.5440.
2,6-bis((3-phenylpropyl)thio)pyrimidine-4,5-diamine (16g)
According to the general alkylation method 1 with 4,5-diamino-2,6-dimercaptopyrimidine (169 mg, 1.0 mmol), potassium carbonate (128 mg, 1.1 mmol) and 1-bromo-3-phenylpropane (0.15 ml, 1.2 mmol) for 5 h. The product was purified by column chromatography with CH2Cl2/MeOH 95:5 as an eluent to afford an off-white solid of yield 260 mg, 60%. 1H-NMR (500 MHz, MeOD): δ 7.25 (dd, 4H, J1 = 15.5 Hz, J2 = 8.0 Hz, HPh), 7.15 (m, 6H, HPh), 3.14 (dd, 2H, J1 = 13 Hz, J2 = 7.0 Hz, SCH2), 3.03 (t, 2H, J = 7.0 Hz, SCH2), 2.72 (t, 4H, J = 7.0 Hz, CH2), 1.92–2.00 (m, 4H, 2xCH2). 13C-NMR (500 MHz, MeOD): δ 158.43 (C2), 152.96 (C6), 148.84 (C4), 141.53 (CPh), 141.33 (CPh), 128.12 (CHPh), 128.11 (CHPh), 128.03 (CHPh), 128.01 (CHPh), 125.54 (CHPh), 125.50 (CHPh), 118.72 (C5), 34.49 (CPh), 34.32 (CH2), 31.59 (CH2), 31.37 (SCH2), 29.78 (SCH2), 28.95 (SCH2). m/z (FTMS + ESI) = Found [M+H]+ (C22H26N4S2) 411.2652 requires 410.5980.
Biological evaluation
MTT assay
(Mosmann 1983) Breast cancer cell lines MDA-MB-231 and MDA-MB-436 were maintained in Delbecco’s Modified Eagle’s Media (DMEM) supplemented with 10% FCS. Human microvascular endothelial (HMVECs) were maintained in microvascular endothelial cell basal medium 131 with growth supplement (Gibco). MCF10A cells were maintained in mammary epithelial cell growth medium (Lonza) optimized for the growth of mammary epithelial cells in a serum-free environment. All cells were incubated at 37 °C in 5% CO2. Cells were seeded in a 96-well plate at a density of 5 × 103 cells/ml and incubated for 24 h at 37 °C and 5% CO2. The cells were then treated with different concentrations of the test compounds and controls and incubated for 72 h. After this period, 20 µL of sterile MTT solution in PBS (5 mg/mL) was added to each well. The plates were then incubated for 5 h at 37 °C and 5% CO2. Following this, 100 μL of acidified isopropanol was added (0.02% 2M HCl) and the mixture was allowed to stand until the formed formazan crystals had dissolved. The absorbance was measured at 570 nm (BioTeK), and the cell viability was expressed as a percentage of the absorbance of non-treated cells. The experiments were performed in three independent repeats, and the results were expressed as the mean ± the standard error of the mean (SEM).
Cytotoxicity assay
(Essen Bioscience 2017) MDA-MB-231, MDA-MB-436, HMVECs and MCF10A cells were seeded (100 μL per well) at a cell density of 5000 cells/well into a 96-well plate and incubated at 37 °C in 5% CO2 within the IncuCyte ZOOM® system for 24 h. Contrast images were captured every 2 h and analysed using an integrated confluence algorithm. The test compounds and controls were prepared in the media containing the IncuCyte™ Cytotox Reagent at 3x the final concentration, then 50 μL of this solution was added to each well to obtain a dilution of 1:3 with a final assay volume of 150 μL (final assay concentration of Cytotox Reagent 250 nM). Images of the plate were captured at 10x objective, two images per well with phase and fluorescence scanning every 2 h until completion of the assay (72 h). The data were analysed using integrated software.
PathHunterTM cell-based CRYAB/VEGF165 interaction assay
The cells were maintained in a PathHunter AssaycompleteTM culture media and incubated at 37 °C and 5% CO2 until confluent. The U2OS cells were detached with AssaycompleteTM cell detachment reagent, suspended in AssaycompleteTM cell plating reagent and counted. In total, 20 µL of the cell suspension was transferred to each well of a 384-well white-walled, clear bottom plate at the indicated cell densities (5000, 10000, 20000 cells/ml). The seeded cells were incubated for 24 h at 37 °C and 5% CO2, 5 µL of the test compounds (10 μM) dissolved in the vehicle (2% DMSO) were added to the appropriate wells and incubated at room temperature for 90 min. After incubating, 25 µL per well of PathHunter Detection Reagents were added to the plates, incubated for 60 min at room temperature in the dark and the plate read using the POLARstar Omega luminescence reader.
ELISA assay
VEGF165 secreted by MDA-MB-231 cells were assayed using Quantio® ELISA assay kit (R&D Systems). The assay was performed according to the manufacturers’ instructions. MDA-MB-231 cells were grown in a T25 flask at 37 °C and 5% CO2 until 80% confluence. The cells were then treated with the test compounds dissolved in DMSO (0.1%). The cell supernatant was harvested by centrifugation at 2000 × g for 15 min at 6, 12 and 24 h. VEGF165 concentrations were measured in triplicate in each sample. The results are expressed as VEGF165 pg/ml for at least three separate experiments.
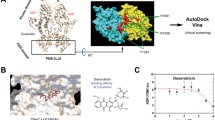
Molecular modelling
All molecular modelling studies were performed with Molecular Operating Environment (MOE) 2015.10 modelling software. The crystal structure of the ACD domain of CRYAB was obtained from the Protein Data Bank (PDB code: 2KLR). Hydrogen atoms were added to the crystal structure minimised with MOE until a gradient of 0.05 Kcal mol−1 Å−1 was reached, using the MMFF94x forcefield. The partial charges were automatically calculated. To simplify calculations and analysis within this hydrophobic active site, no water molecules were considered. The docking site was defined by selecting the important amino acid residues (Tyr122, Arg 123 and Arg 149) and extending to 4.5 Å to include other β7 and β9 amino acid residues. Ligand docking was performed using MOE default settings. After docking, all docked molecules were visually inspected.
Results and discussion
Design of novel analogues and synthetic chemistry
We began our synthetic studies around a dithiopurine-8-one heterocyclic core, proposing the introduction of substituents to improve lipophilicity and enhance cellular penetration. We decided to explore alkylation chemistry to derivatise the core dithiopurine-8-one heterocyclic structure particularly at the sulphur and/or the N7/N9 positions, as well as introducing functional flexibility at position C8. Figure 2 shows the proposed structural changes and exploratory chemistry leading to five small series of new molecules for in vitro anticancer evaluation.
There is clear evidence in the literature that reaction conditions play a crucial role in determining the selectivity of alkylation of purines and pyrimidines. Therefore, various conditions reported in the literature were examined to ascertain the optimal reactions conditions required to obtain selective alkylation at the appropriate positions of the purinone core (Pathak et al. 2004; Cheng and Robins 1958; Robins 1958; Biagi et al. 1996; Corder et al. 2013; Johnston et al. 1958; Robins 1957). Scheme 1 outlines the synthetic route employed in the synthesis of Series 1 analogues 4a–g, characterised by derivatisation of the sulphur groups at positions 2 and 6 with various alkyl groups. The purine core structure 2 was prepared via a thermal cyclisation reaction between the commercially available pyrimidine 1 and urea (Levin et al. 1960). As suggested in the literature, it was expected that under alkaline conditions, alkylation of compound 2 would selectively produce the exclusively S-alkylated product (Joule et al. 1995; Montgomery et al. 1966). In addition, it has been reported that low reaction temperatures favour S- over N-alkylation. However, it was noted that when using K2CO3 or KOH as a base, the tri-substituted product (3) was produced regardless of the stoichiometry of the reagents. The best conditions found to facilitate selective S-alkylation to provide the double alkylation products (4a-g) involved the non-nucleophilic base, 1,8-diazabicyclo[5.4.0]undec-7-ene (DBU) in DMF (Biagi et al. 1996), in yields of 29–74% following purification by column chromatography.
The synthetic route employed in the synthesis of Series 2 analogues 6a–g is outlined in Scheme 2. A thermal cyclisation reaction between pyrimidine (1) and thiourea resulted in the 8-thiopurine (5), which was followed by alkylation reactions with various alkyl halides as described in Scheme 1. Under all the investigated reaction conditions, the products of the alkylation reactions were the tri-substituted purines similar to the products obtained under the previously described alkaline conditions (K2CO3 or KOH), in low-to-moderate yields of 17–54% (Scheme 1). The acidity of the thiopurine NH group adjacent to the pyrimidine N3-atom provides a rationale for alkylation at the N9-position under basic conditions. Although tri-substituted purines have been reported in the literature, they are usually constructed from alkylated pyrimidines cyclised with alkoxides and not in a one-pot synthesis as we achieved (Yang et al. 2005; Villatoro et al. 2015; Ibrahim and Legraverend 2009).
Surprisingly in addition to the tri-substituted thiopurine (6b), methylation of compound 5 also produced the tetra-substituted product 7 (Scheme 3). We postulated that methylation at position N7 is possible since the methyl group is not affected by steric hindrance compared with the bulkier alkyl substituents.
Scheme 4 was employed to give access to selectively N-alkylated purines 13a–g that were deemed necessary for SAR studies. Selective N-alkylation was readily achieved by a reaction between 2-amino-6-chloropurine 8 and various alkyl halides to afford compounds 9a–g/10a–g as a mixture of regioisomers, with the N9-isomer (9a–g) being the major product as previously reported in the literature (Chhabra et al. 2013). Both isomers were successfully separated by silica gel chromatography and fully characterised by NMR. The 1H NMR spectrum showed the alkyl peaks of the N7 product to be shifted downfield due to the deshielding effect of the bulky chloro group. The N9 products were then taken forward in a multi-step synthesis to give the desired N-alkylated purinones 13a–g by introducing a keto-group at position 8. Compounds 9a–g were hydrolysed in aqueous HCl to afford the intermediates 11a–g, which were then brominated to afford the 8-bromopurine intermediates 12a–g (Michael et al. 1993). Finally, the brominated purines were transformed to the desired 8-oxopurines 13a–g through hydrolysis of the bromo group, in low to good yields (6–85%) (Verones et al. 2010).
Lastly, substituted 6-thiopurines 15a–g and 2,6-dithiopyrimindines 16a–g, which represent Series 4 and 5 analogues, respectively, were also synthesised from commercially available precursor compounds 14 and 31, in moderate-to-high yield (37–80%) following the previously described alkylation methods for further SAR studies (Scheme 5).
Biological evaluation—anti-proliferative activity
The analogues were tested for their in vitro inhibitory effect on the viability of the invasive TNBC cell lines MDA-MB-231 and MDA-MB-436, using the MTT endpoint assay (72 h). Additionally, endothelial cell lines, MCF10A and human microvascular endothelial (HMVEC-d) cell lines were used to assess the effects of the analogues on endothelial cells. Table 1 details the result of this in vitro screen expressed as IC50 values from dose-response studies.
In general, several analogues from Series 1 (4a–g), Series 2 (6a–g) and Series 5 (16a–g) showed the most potent anti-proliferative effects, significantly reducing the viability of one or both TNBC cells with IC50 values in the low micromolar range. It should be noted that active analogues were also generally more active as growth inhibitors in the endothelial cell lines HMVEC and MCF10A, perhaps not surprisingly for these early stage lipophilic drug candidates. The selectivity profiles between the different cell types certainly underscore the importance of carrying out more detailed toxicity studies in the future.
Notably, benzyl/phenethyl substituted purinone/thiopurinone analogues 4e, 4f, and 6e, were the most potent in TNBC cell lines, suggesting that this group may be important for biological activity. Across all series, it was also evident that the chain length of the alkyl substituents has an influence on biological activity. For analogues with aliphatic substituents, it was observed that isobutyl > propyl > ethyl > methyl in terms of their anti-proliferative activity within cell lines examined. For example, ethyl substituted analogue 4a showed IC50 values of 97 and 100 μM against MDA-MB-231 and MDA-MB-436 cells, respectively, compared with the propyl substituted analogue 4c, which showed an IC50 value of 29 μM against both TNBC cell lines. However, for analogues with alkyl benzyl substituents, an increase in the chain length of the alkyl benzyl group appears to have a detrimental effect on biological activity. For example, compound 4f with a phenethyl group showed an IC50 value of 3–4 μM in TNBC cell lines, while increasing the chain length to a phenylpropyl group as seen for compound 4g resulted in a complete loss of activity (IC50 > 100 μM).
Overall, analogues from Series 3 and 4 showed no inhibitory effects against either TNBC cell line apart from compound 15f (Series 4). This suggests that the substitution at positions 2 and 6 and the keto group at position-8 may contribute to the observed biological effect. As noted above, the most potent analogues across all series also showed potent growth inhibitory effects on the epithelial and endothelial cell lines MCF10A and HMVEC-d, respectively, without the desired selectivity for TNBC cell lines.
Cytotoxicity evaluation
Compounds that demonstrated the most potent inhibitory effect in the anti-proliferative assay (IC50 ≤ 50 µM) were evaluated for their ability to induce cytotoxicity against the cell lines under investigation. The in vitro cytotoxicity assay was performed using the cyanine nucleic acid dyes developed by Essen BioScience with their IncuCyte™ live-cell imaging system that enables real time detection of cell death. The cells were co-cultured with the IncuCyte™ Cytotox reagent, and test compounds 4c, 4e–f, 6d–e, 15f and 16d were added at different concentrations over a period of 72 h. Images were captured every 2 h for the duration of the assay and the cytotoxicity effect was quantified in real-time with the integrated analysis software. Generally, all the tested compounds showed a significant increase in the cell death characteristics of a cytotoxicity effect compared with the no-drug control, with a few notable exceptions (Fig. 3). All the seven compounds induced a cytotoxic effect at a concentration of 100 μM against the TNBC cells, while cytotoxic effects were detected at a concentration of 50 μM against endothelial cells. These results appear to correlate with the observed anti-proliferative activity and thus, provided confidence for these analogues to undergo further evaluation, notably in our custom cell-based CRYAB/VEGF165 interaction assay to probe their ability to disrupt this interaction.
PathHunterTM cell-based CRYAB/VEGF165 interaction assay
The PathHunterTM cell-based CRYAB/VEGF165 interaction assay is a commercially available, custom cell-based assay designed by DiscoverX. The assay utilizes the enzyme fragment complementation (EFC) technology for studying protein–protein interactions. For the purposes of this research, the assay was designed to monitor the interaction between CRYAB and a cytosolically localised VEGF165. The two candidate proteins were fused to complementation fragments of the β-galactosidase (β-Gal) enzyme. CRYAB was co-expressed with the larger sequence encoding the majority of enzyme, termed enzyme acceptor (EA), while VEGF165 was fused to a small peptide epitope called Prolink (PK). The two separate enzyme fragments are inactive; however, after the constructs were transduced into U2OS cells, the recombined active enzyme is generated as a result of CRYAB/VEGF165 protein–protein interaction. The disruption of the CRYAB/VEGF165 interaction was quantified by measuring the chemiluminescence signal produced as a consequence of the conversion of a non-luminescent substrate to a luminescent product by the active enzyme.
The U2OS cells were seeded at three cell densities (20,000, 10,000 and 5000 cells/well), allowed to adhere to the wells and challenged with different concentrations of the test compounds. Tunicamycin was used as a positive control for this assay because of its ability to elicit the unfolded protein response and thus inhibit angiogenesis (Banerjee et al. 2011). After 90-min incubation period, the non-luminescent substrates were introduced into the cells and luminescent readings taken with a spectrophotometer after 60 min. All experiments were carried out in triplicate. The seven analogues that demonstrated inhibitory and cytotoxicity effects in previous experiments were selected for further evaluation in the cell-based CRYAB/VEGF165 interaction assay. Out of the seven compounds, analogues 4e and 4f were found to significantly attenuate the chemiluminescence signal of U2OS cells at a density of 20,000 cells/well. No significant inhibition effect was observed for the remaining five analogues. The response (RLU) of the analogues at 10 μM and the controls are shown in Fig. 4. Both analogues and tunicamycin demonstrated a significant decrease in chemiluminescence signal in comparison with the vehicle control (DMSO). This inhibitory effect is indicative of the disruption of the interaction between CRYAB and VEGF165.
VEGF165 ELISA measurement
As previously discussed, one of the proposed mechanisms by which CRYAB promotes angiogenesis is by preventing the proteolytic degradation of misfolded or unfolded VEGF resulting in the upregulation of VEGF and maintenance of VEGF signalling (Ruan et al. 2011). Hence, we postulated that disrupting the interaction between CRYAB/VEGF would affect the levels of VEGF produced by breast cancer cells. The concentration of VEGF secreted by MDA-MB-231 cells was measured by the ELISA assay. MDA-MB-231 cells treated with 100 μM of compound 4e showed a significant reduction in VEGF levels compared with the control. A concentration of 160 pg/ml of soluble VEGF was detected by the assay compared with the control (260 pg/ml), representing a 40% decrease in VEGF concentration (Fig. 5). No significant reduction was observed in cells treated with compound 4f. This result supports our hypothesis that the disruption of the interaction between CRYAB/VEGF can lead to inhibition of VEGF production by breast tumour cells.
Molecular modelling studies
Previous work has reported sequence studies for the interaction of important regulatory proteins with CRYAB (Ghosh et al. 2007). Typical of most proteins of the Hsp family, the structure of CRYAB consists of an N-terminal region of ~65 amino acids, preceding a conserved α-crystallin domain (ACD) domain made up of 84 amino acids, and finally a C-terminal region composed of 26 amino acids. The ACD domain consists of eight β strands (β2 to β9) and helical sequences with strong interactions to regulatory proteins. The aforementioned sequence studies demonstrated that the β strands (β3, β7, β8 and β9) play a crucial role in the interactions between many growth factors and CRYAB, including VEGF165. Additionally, an inhibitory competition study with decoy peptides corresponding to CRYAB and VEGF165 confirmed the significance of β7 and β9 in the protein–protein interaction between CRYAB and VEGF165 (Chen et al. 2014). Fortunately from the standpoint of computational drug design, a good crystal structure of the α-crystallin domain (ACD) of human CRYAB obtained from solid-state NMR, small angle X-ray scattering, and computational modelling studies has been published (PDB code: 2KLR) (Jehle et al. 2010).
On this basis, for docking studies a hydrophobic structural pocket was selected that was formed by discontinuous segments of β7 and β9 and containing the amino acids Try122/Arg123 (β7) and Arg149 (β9). In a series of docking simulations carried out with Molecular Operating Environment (MOE 2017), all the analogues and one additional monosubstituted purinone, 6-mercapto-2-(methylthio)purin-8-one, were docked into the selected structural pocket to probe their interaction with the target site.
Figure 6 shows the predicted binding pose of 6-mercapto-2-(methylthio)purin-8-one. The methythio side group spatially occupies the structural pocket, albeit partially. Crucially, one of the nitrogen groups of the imidazolidine ring forms an H-bond with Tyr122. Moreover, there were other hydrophobic interactions between the purine ring and surrounding amino acids, including Asn78 of the β3 strand. Comparatively, 4e (Fig. 7) also retained the key interaction with Tyr122 and an additional interaction with β7 through a H-bond between a thiol group and Ile124. Compound 4f (Fig. 8) interacts with β7 through hydrophobic interaction between the purine core and Arg123. Furthermore, one of the nitrogen groups of the imidazolidine ring of both analogues forms a H-bond with Lys121 also of β7. Finally, it was noted that the benzyl/phenethyl rings penetrates further and occupies more of the binding pocket compared with 6-mercapto-2-(methylthio)purin-8-one. Interestingly, even the aromatic ring of the benzyl/phenethyl groups does not fully occupy this pocket hence the presence of bulky substituents on the ring could promote better insertion, as well as new interactions that could improve on the biological activities of the analogues. Also noticeable is the fact that none of the compounds interact with β9; this may be indicative of its distance (26 amino acids away) from β7. Therefore, introducing bulky substituents to the aromatic ring could bring this group closer to β9 and enhance the interaction between the molecules and CRYAB.
Discussion
The biological and docking studies have highlighted structural features that could be responsible for the observed in vitro activities of the synthesised analogues and will inform future endeavours to design more efficacious CRYAB inhibitors. These observations are summarised below.
Effects of S-alkyl substituents
To improve on the lipophilicities of the purine core structure, various alkyl substituents were introduced to the thio- and/or nitrogen groups. It was observed that across all series, analogues with aromatic phenalkyl substituents demonstrated the most potent biological effects in the various assays. However, the chain length of the phenalkyl group was found to be a limiting factor, that is, an increase in chain length led to a decrease in activity. Furthermore, compounds 4e and 4f, with this benzyl/phenethyl functionality were able to disrupt the interaction between CRYAB/VEGF165 in the protein–protein interaction assay. In addition, compound 4e also decreased the levels of VEGF secreted by MDA-MB-231 cells. In our docking studies, the benzyl/phenethyl group was also found to form key interactions with the selected structural pocket on CRYAB confirming the significance of this group.
Effects of N-substitutions
No improvement in activity was observed for the series of analogues with N-substitutions (Series 3). Additionally, the docking studies showed no interactions between this substitution and CRYAB (data not shown).
Effects of removing the thio group at position-2
Removal of the thio-group at position-2 as seen with Series 3 and 4 analogues could be responsible for the loss of activity in both the various in vitro assays. Crucially, in the docking studies, it appears that this substitution interacts with the structural pocket of CRYAB indicating that this substitution contributes to the biological activities of the analogues.
Functional flexibility at position-8
As Series 4 analogues are effectively non-toxic against the control cell lines, it can be deduced that this substitution may contribute to the observed toxicity of the analogues. However, the toxicity effect may not be limited to this substituent but also the substituted thiol group at position-2, since analogues without this substitution (Series 3 and 4) showed little or no cytotoxic effect.
CRYAB confers protection against apoptosis induced by chemotherapeutic agents by inhibiting caspase-3 activation, sequestration of the anti-apoptotic protein Bcl-2 and prevention of Bcl-2 translocation into the mitochondria. In a study where breast epithelial carcinoma cells were treated with the anticancer agent vinblastine, the phosphorylated form of CRYAB was found to inhibit apoptosis by interacting with Bcl-2, while non-phosphorylated CRYAB induced resistance against vinblastine (Launay et al. 2010). The silencing of CRYAB with antisense or RNAi molecules reversed this effect and sensitised tumour cells to chemotherapeutic and apoptotic agents (Lee et al. 2012). Angiogenesis, a segment of the metastatic cascade and the role of CRYAB as a facilitator of this phenomenon is of particular interest to our group. In the tumour microenvironment, CRYAB modulates angiogenesis through its effect on both cancer and endothelial cells. Studies have shown that CRYAB can prevent proteolytic degradation of VEGF by binding to and correcting unfolded/misfolded VEGF leading to preservation of intracrine VEGF signalling. Inhibition of CRYAB leads to a decrease in endogenous VEGF levels, providing clear evidence that targeting CRYAB may represent a novel approach to suppress angiogenesis. It should be noted that CRYAB is also expressed at high levels in the the heart and cardiomyocytes, conferring protection against stress (such as an ischaemic event) by inhibiting apoptosis (Pereira et al. 2014; Gonçalves et al. 2016; Morrison et al. 2003). Therefore, direct inhibition of CRYAB could lead to unwanted cardiotoxicity. Consequently, we propose a novel strategy of targeting angiogenesis by disrupting the interaction between CRYAB and VEGF with small molecules that competitively bind to the interacting surface. This approach could produce a therapeutic effect while potentially avoiding the unwanted side effects associated with direct inhibition of CRYAB, and serve as a novel anti-angiogenic therapy in the treatment of TNBC.
Conclusions
Even though protein–protein interactions (PPI) are involved in many fundamental biological processes, designing small molecules to target PPI’s has been deemed challenging due to the frequent occurrence of large and featureless protein interfaces. Over the last two decades, as our understanding of the interfaces have progressed, PPI’s have become attractive molecular targets in drug development. Although the interfaces of PPI’s are extensive, studies have shown that not all residues of these interfaces contribute towards the stability of the binding between target- and client proteins, but only the so-called “hotspots” (a number of conserved amino acid residues) are critical for interaction. Thus, most of these “hotspots” have become targets in the drug development process and the last few years have seen a number of small molecule PPI inhibitors enter clinical trials (White et al. 2008; Arkin et al. 2014).
Following this trend, we propose a novel approach of supressing angiogenesis in TNBC with small molecules that target the PPI between CRYAB and VEGF. There is a critical need for targeted therapies in TNBC since the only established treatment for this aggressive disease is chemotherapy, which has been shown to be ineffective in terms of medium- to long-term survival. Angiogenesis is one of the main targets in TNBC due to its role in disease progression, however, most anti-angiogenic therapies have shown mixed and sometimes disappointing results. In the tumour microenvironment, CRYAB is a chaperone protein that has been shown to bind to and correct unfolded/misfolded VEGF promoting angiogenesis. This protein is overexpressed in TNBC and has been found to correlate with poor prognosis in this disease setting, making it an attractive target for the development of new anti-angiogenic therapies. Several studies looking at the interaction between CRYAB and regulator proteins identified a “hotspot” on the surface of CRYAB where VEGF interacts. Aided by molecular docking methods, small molecule hit compounds were identified which bind to this “hotspot” and could potentially inhibit this PPI.
In this paper, we report a proof-of-concept study devised to validate this target. The work started with the synthesis of novel purine-based compounds for structural activity relationship studies. The newly synthesised analogues were then subjected to biological evaluation, firstly, in a cell viability assay where several of the analogues were found to significantly reduce the viability of MDA-MB-231 and MDA-MB-436 TNBC cell lines. The most potent of these analogues were then tested for their ability to disrupt the CRYAB/VEGF interaction in a custom designed cell-based CRYAB/VEGF165 PPI assay. The PPI assay showed that compounds 4e and 4f successfully disrupted the interaction between CRYAB and VEGF165. Additionally compound 4e (100 μM) was found to decrease the amount of VEGF secreted by MDA-MB231 TNBC cells by 40%. Molecular docking studies also suggested that these two analogues interfere with the target “hotspot” through key interactions with specific amino acids. In conclusion, for this work to progress to pre-clinical trials as a viable therapeutic option in TNBC, further design, synthesis and in vitro studies are required to validate this novel concept. Additional studies on lead compound stability and other pharmacokinetic properties will also be required prior to evaluation within relevant in vivo models and identification of a new pre-clinical drug candidate.
References
Arkin MR, Tang YY, Wells JA (2014) Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chem Biol 21:1102–1114
Arrigo AP, Simon S, Gibert B, Kretz-Remy C, Nivon M, Czekalla A, Guillet D, Moulin M, Diaz-Latoud C, Vicart P (2007) Hsp27 (HspB1) and alpha B-crystallin (HspB5) as therapeutic targets. FEBS Lett 581:3665–3674
Banerjee A, Lang JY, Hung MC, Sengupta K, Banerjee SK, Baksi K, Banerjee DK (2011) Unfolded protein response is required in nu/nu mice microvasculature for treating breast tumor with tunicamycin. J Biol Chem 286:29127–29138
Bear HD, Tang G, Rastogi P, Geyer Jr. CE, Robidoux A, Atkins JN, Baez-Diaz L, Brufsky AM, Mehta RS, Fehrenbacher L, Young JA, Senecal FM, Gaur R, Margolese RG, Adams PT, Gross HM, Costantino JP, Swain SM, Mamounas EP, Wolmark N (2012) Bevacizumab added to neoadjuvant chemotherapy for breast cancer. New Eng J Med 366:310–320
Biagi G, Costantini A, Costantino L, Giorgi I, Livi O, Pecorari P, Rinaldi M, Scartoni V (1996) Synthesis and biological evaluation of new imidazole, pyrimidine and purine derivatives and analogs as inhibitors of xanthine oxidase. J Med Chem 39:2529–2535
Chen ZJ, Ruan Q, Han S, Xi L, Jiang WG, Jiang HB, Ostrov DA, Cai J (2014) Discovery of structure-based small molecule inhibitor of alpha B-crystallin against basal-like/triple-negative breast cancer development in vitro and in vivo. Breast Cancer Res Treat 145:45–69
Cheng CC, Robins RK (1958) Potential purine antagonists. 12. Synthesis of 1-alkyl(aryl)-4,6-disubstituted pyrazolo[3,4-d]pyrimidines. J Org Chem 23:852
Chhabra S, Barlow N, Dolezal O, Hattarki MK, Newman J, Peat TS, Graham B, Swarbrick JD (2013) Exploring the chemical space around 8-mercaptoguanine as a route to new inhibitors of the folate biosynthesis enzyme HPPK. PLoS ONE 8:e59535
Corder AL, Subedi BP, Zhang SA, Dark AM, Foss FW, Pierce BS (2013) Peroxide-shunt substrate-specificity for the Salmonella typhimurium O-2-dependent tRNA modifying monooxygenase (MiaE). Biochem 52:6182–6196
De Azevedo WF, Leclerc S, Meijer L, Havlicek L, Strnad M (1997) Inhibition of cyclin-dependent kinases by purine analogues: crystal structure of human cdk2 complexed with roscovitine. FEBS J 243:518–526
Essen Bioscence. IncuCyte™ cytotoxicity assay general protocol. https://www.essenbioscience.com/en/applications/cell-health-viability/cytotoxicity. Accessed 5th December 2017
Fosu-Mensah N, Peris MS, Weeks HP, Cai J, Westwell AD (2015) Advances in small molecule drug discovery for triple-negative breast cancer. Fut Med Chem 7:2019–2039
Garrido C, Paul C, Seigneuric R, Kampinga HH (2012) The small heat shock family proteins family: the long forgotten chaperones. Int J Biochem Cell Bio 44:1588–1592
Ghanem H, Jabbour E, Faderl S, Ghandi V, Plunkett W, Kantarjian H (2010) Clofarabine in leukemia. Exp Rev Hematol 3:15–22
Ghosh JG, Shenoy AK, Clark JI (2007) Interactions between important regulatory proteins and human alpha B crystallin. Biochem 46:6308–6317
Gonçalves DC, Marin TM, Pereira MBM, Santos AM, Leme AFP, Franchini KG (2016) Alpha B-crystallin interacts and attenuates the tyrosine phosphatase activity of Shp2 in cardiomyocytes under mechanical stress. FEBS Lett 590:2232–2240
Goozner M (2011) Avastin hearing leads to more uncertainty over drug’s future. J Natl Cancer Inst 103:1148–1150
Haynes B, Zhang YH, Liu FC, Li J, Petit S, Kothayer H, Bao X, Westwell AD, Mao GZ, Shekhar MPV (2016) Gold nanoparticle conjugated Rad6 inhibitor induces cell death in triple negative breast cancer cells by inducing mitochondrial dysfunction and PARP-1 hyperactivation: synthesis and characterization. Nanomed: Nanotech Biol & Med 12:745–757
Ibrahim N, Legraverend M (2009) High-yielding two-step synthesis of 6,8-disubstituted N-9-unprotected purines. J Comb Chem 11:658–666
Jehle S, Rajagopal P, Bardiaux B, Markovic S, Kühne R, Stout JR, Higman VA, Klevit RE, van Rossum BJ, Oschkinat H (2010) Solid state NMR and SAXS studies provide a structural basis for the activation of alpha B-crystallin oligomers. Nat Struct Mol Biol 17:1037–1042
Johnston TP, Holum LB, Montgomery JA (1958) Synthesis of potential anticancer agents. 16. S-substituted derivatives of 6-mercaptopurine. J Am Chem Soc 80:6265–6271
Joule JA, Mills K, Smith GF (1995) Heterocyclic chemistry, 3rd edn. Wiley-Blackwell, London
Kim LS, Huang S, Lu WX, Lev DC, Price JE (2004) Vascular endothelial growth factor expression promotes the growth of breast cancer brain metastases in nude mice. Clin Exp Metastas- 21:107–118
Launay N, Tarze A, Vicart P, Lilienbaum A (2010) Serine 59 phosphorylation of alpha B-crystallin down-regulates its anti-apoptotic function by binding and sequestering Bcl-2 in breast cancer cells. J Biol Chem 285:37324–37332
Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA (2011) Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 121:2750–2767
Lee JS, Kim HY, Jeong NY, Lee SY, Yoon YG, Choi YH, Yan C, Chu IS, Koh H, Park HT, Yoo YH (2012) Expression of alpha B-crystallin overrides the anti-apoptotic activity of XIAP. Neuro Oncol 14:1332–1345
Levin G, Kalmus A, Bergmann F (1960) Synthesis of 6-thiouric acid and its derivatives. J Org Chem 25:1752–1754
Mayer IA, Abramson VG, Lehmann BD, Pietenpol JA (2014) New strategies for triple-negative breast cancer – deciphering the heterogeneity. Clin Cancer Res 20:782–790
Michael MA, Cottam HB, Smee DF, Robins RK, Kini GD (1993) Alkylpurines as immune potentiating agents – synthesis and antiviral activity of certain alkylguanines. J Med Chem 36:3431–3436
MOE: Molecular Operating Environment - Chemical Computing Group. Molecular Operating Environment user manual. https://www.chemcomp.com/MOE-Molecular_Operating_Environment.htm. Accessed 9th February 2017
Montgomery JA, Hewson K, Clayton SJ, Thomas HJ (1966) Further studies on alkylation of purines. J Org Chem 31:2202
Morrison LE, Hoover HE, Thuerauf DJ, Glembotski CC (2003) Mimicking phosphorylation of alpha B-crystallin on serine-59 is necessary and sufficient to provide maximal protection of cardiac myocytes from apoptosis. Circ Res 92:203–211
Mosmann T (1983) Rapid colorimetric assay for cellular growth and survival – application to proliferation and cytotoxicity assays. J Immunol Methods 65:55–63
O’Reilly EA, Gubbins L, Sharma S, Tully RM, Guang MHZ, Weiner-Gorzel K, McCaffrey J, Harrison M, Furlong F, Kell M, McCann A (2015) The fate of chemoresistance in triple negative breast cancer (TNBC). BBA Clin 3:257–275
O’Shaughnessy J, Dieras V, Glaspy J, Brufsky A, Miller KD, Miles DW, Koralewski P, Phan SC, Bhattacharya S (2009) Comparison of subgroup analyses of PFS from three Phase III studies of bevacizumab in combination with chemotherapy in patients with HER2-negative metastatic breast cancer. Cancer Res 69:512S
Pathak AK, Pathak V, Seitz LE, Suling WJ, Reynolds RC (2004) Antimycobacterial agents. 1. Thio analogues of purine. J Med Chem 47:273–276
Pereira MBM, Santos AM, Gonçalves DC, Cardoso AC, Consonni SR, Gozzo FC, Oliveira PS, Pereira AHM, Figueiredo AR, Tiroli-Cepeda AO, Ramos CHI, de Thomaz AA, Cesar CL, Franchini KG (2014) αB-crystallin interacts with and prevents stress-activated proteolysis of focal adhesion kinase by calpain in cardiomyocytes. Nat Commun 5:5159
Ranpura V, Hapani S, Wu S (2011) Treatment-related mortality with bevacizumab in cancer patients. J Am Med Assoc 305:487–494
Robins RK (1957) Potential purine antagonists. 9. Further studies of some 4,6-disubstituted pyrazolo[3,4-d]pyrimidines. J Am Chem Soc 79:6407–6415
Robins RK (1958) Potential purine antagonists. 15. Preparation of some 6,8-disubstituted purines. J Am Chem Soc 80:6671–6679
Rodriguez G (1994) Fludarabine phosphate – a new anticancer drug with significant activity in patients with chronic lymphocytic leukemia and in patients with lymphoma. Inv New Drugs 12:75–92
Ruan Q, Han S, Jiang WG, Boulton ME, Chen ZJ, Law BK, Cai J (2011) Alpha B-crystallin, an effector of unfolded protein response, confers anti-VEGF resistance to breast cancer via maintenance of intracrine VEGF in endothelial cells. Mol Cancer Res 9:1632–1643
Sahasranaman S, Howard D, Roy S (2008) Clinical pharmacology and pharmacogenetics of thiopurines. Eur J Clin Pharmacol 64:753–767
Steeg PS (2006) Tumor metastasis: mechanistic insights and clinical challenges. Nat Med 12:895–904
Tsang JYS, Lai MWH, Wong KHY, Chan SK, Lam CCF, Tsang AKH, Yu AMC, Tan PH, Tse GM (2012) Alpha-B-crystallin is a useful marker for triple negative and basal breast cancers. Histopathol 61:378–386
Verones V, Flouquet N, Farce A, Carato P, Leonce S, Pfeiffer B, Berthelot P, Lebegue N (2010) Synthesis, biological evaluation and docking studies of 4-amino-tetrahydroquinazolino[3,2-e]purine derivatives. Eur J Med Chem 45:5678–5684
Villatoro MJPDY, Unciti-Broceta JD, Contreras-Montoya R, Garcia-Salcedo JA, Mezo MAG, Unciti-Broceta A, Diaz-Mochon JJ (2015) Amide-controlled, one-pot synthesis of tri-substituted purines generates structural diversity and analogues with trypanocidal activity. Sci Rep 5:9139
von Minckwitz G, Eidtmann H, Rezai M, Fasching PA, Tesch H, Eggemann H, Schrader I, Kittel K, Hanusch C, Kreienberg R, Solbach C, Gerber B, Jackisch C, Kunz G, Blohmer J-U, Huober J, Hauschild M, Fehm T, Müller BM, Denkert C, Loibl S, Nekljudova V, Untch M (2012) Neoadjuvant chemotherapy and bevacizumab for HER2-negative breast cancer. New Eng. J Med 366:299–309
White AW, Westwell AD, Brahemi G (2008) Protein-protein interactions as targets for small-molecule therapeutics in cancer. Expert Rev Mol Med 10:e8
Yang JX, Dang Q, Liu JL, Wei ZL, Wu JC, Bai X (2005) Preparation of a fully substituted purine library. J Comb Chem 7:474–482
Acknowledgements
We thank Cancer Research Wales for a postdoctoral research associate award (to NF-M), and acknowledge the EPSRC UK National Mass Spectrometry Facility at Swansea University for provision of mass spectrometry analysis. We thank Essen BioScience for providing IncuCyte™ live-cell imaging system and reagents for cytotoxicity measurements.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, duplication, adaptation, distribution, and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Fosu-Mensah, N.A., Jiang, W., Brancale, A. et al. The discovery of purine-based agents targeting triple-negative breast cancer and the αB-crystallin/VEGF protein–protein interaction. Med Chem Res 28, 182–202 (2019). https://doi.org/10.1007/s00044-018-2275-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-018-2275-9