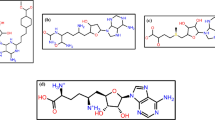
Abstract
NAD+-dependent DNA ligases (LigA) are essential enzymes found only in bacteria and some virus species. This makes them attractive drug targets. Based on the crystal structure of the NAD+ binding domain of the M. tuberculosis enzyme (MtuLigA) and virtual screening, we have earlier identified several novel classes of inhibitors for this enzyme. These inhibitors bind to the adenylation domain and compete with the cofactor NAD+. Recently, we identified that the BRCT domain is essential for the enzyme activity of MtuLigA. We have used virtual screening to identify compounds from the CAP database that should potentially bind to the BRCT domain. These will now be evaluated as inhibitors of the enzyme with a novel mechanism of action. Challenges faced in designing specific and potent inhibitors of the enzyme which can distinguish between the human adenosine triphosphate (ATP)-dependent ligase and MtuLigA are additionally discussed in this report. Proposed strategies for the design of potent inhibitors with desired properties are also outlined.





Similar content being viewed by others
References
Benarroch D, Shuman S (2006) Characterization of mimivirus NAD+-dependent DNA ligase. Virology 353:135–43
Ciarrocchi G, MacPhee DG, Deady LW, Tilley L (1999) Specific inhibition of the eubacterial DNA ligase by arylamino compounds. Antimicrob Agents Chemother 43:2766–2772
Doherty AJ, Suh SW (2000) Structural and mechanistic conservation in DNA ligases. Nucleic Acids Res 28:4051–58
Duncan K (1998) The impact of genomics on the search for novel tuberculosis drugs. Novartis Found Symp 217:228–37
Engler MJ, Richardson CC (1982) In: Boyer PD (ed) The Enzymes. Academic, New York, pp 15:3–29
Feng H, Parker JM, Lu J, Cao W (2004) Effects of deletion and site-directed mutations on ligation steps of NAD+-dependent DNA ligase: A biochemical analysis of BRCA1 C-terminal domain. Biochemistry 43:12648–12659
Jeon HJ, Shin HJ, Choi JJ, Hoe HS, Kim HK, Suh SW, Kwon ST (2004) Mutational analyses of the thermostable NAD+-dependent DNA ligase from Thermus filiformis. FEMS Microbiol Lett 237:111–118
Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8:477–86
Lehman IR (1974) DNA ligase: Structure, mechanism, and function. Science 186:790–797
Lu J, Tong J, Feng H, Huang J, Afonso CL, Rock DL, Barany F, Cao W (2004) Unique ligation properties of eukaryotic NAD+-dependent DNA ligase from Melanoplus sanguinipes entomopoxvirus. Biochim Biophys Acta 1701:37–48
Marti-Renom MA, Stuart A, Fiser A, Sánchez R, Melo F, Sali A (2000) Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct 29:291–325
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 9:1639–662
Ravishankar R, Ravindran M, Suguna K, Surolia A, Vijayan M (1997) Crystal structure of the peanut lectin-T-antigen complex. Carbohydrate specificity generated by water bridges. Curr Sci 72:855–861 (also, Curr Sci 1999;76:1393)
Ravishankar R, Suguna K, Surolia A, Vijayan M (1999) The crystal structures of the complexes of peanut lectin with methyl-β-galactose and N-acetyllactosamine and a comparative study of carbohydrate binding in Gal/GalNAc specific legume lectins. Acta Crystallogr D 55:1375–1382
Sriskanda V, Moyer RW, Shuman S (2001) NAD+-dependent DNA ligase encoded by a eukaryotic virus. J Biol Chem 276:36100–36109
Srivastava SK, Tripathi RP, Ravishankar R (2005) NAD+-dependent ligase (Rv3014c) from M. tuberculosis H37Rv: Crystal structure of the adenylation domain and identification of novel inhibitors. J Biol Chem 280:30273–30281
Srivastava SK, Dube D, Tewari N, Dwivedi N, Tripathi RP, Ravishankar R (2005) Mycobacterium tuberculosis NAD+-dependent DNA ligase is selectively inhibited by glycosylamines compared with human DNA ligase I. Nucleic Acids Res 33:7090–7101
Srivastava SK, Dube D, Vandna K, Jha AK, Hajela K, Ravishankar R (2007) NAD+ -dependent DNA ligase (Rv3014c) from Mycobacterium tuberculosis: Novel structure-function relationship and identification of novel inhibitors. Proteins 69:97–111
Takeuchi T, Ishidoh T, Iijima H, Kuriyama I, Shimazaki N, Koiwai O, Kuramochi K, Kobayashi S, Sugawara F, Sakaguchi K, Yoshida H, Mizushina Y (2006) Structural relationship of curcumin derivatives binding to the BRCT domain of human DNA polymerase lambda. Genes Cells 11:223–35
Vriend G (1990) WHAT IF: a molecular modeling and drug design program. J Mol Graph 8:52–56
Wilkinson A, Smith A, Bullard D, Lavesa-Curto M, Sayer H, Bonner A, Hemmings A, Bowater R (2005) Analysis of ligation and DNA binding by Escherichia coli DNA ligase (LigA). Biochim Biophys Acta 1749:113–122
Acknowledgements
The work was funded by the Council of Scientific and Industrial Research, India grant CMM0017 and SMM0003. DD, VK, and SKS acknowledge fellowships from CSIR. This is communication number 7135 from CDRI.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dube, D., Kukshal, V., Srivastava, S.K. et al. NAD+-dependent DNA ligase (Rv3014c) from M. tuberculosis: Strategies for inhibitor design. Med Chem Res 17, 189–198 (2008). https://doi.org/10.1007/s00044-007-9052-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-007-9052-5