Abstract
Mountains are reservoirs for a tremendous biodiversity which was fostered by a suite of factors acting in concert throughout evolutionary times. These factors can be climatic, geological, or biotic, but the way they combine through time to generate diversity remains unknown. Here, we investigate these factors as correlates of diversification of three closely related sections of Gentiana in the European Alpine System. Based upon phylogenetic approaches coupled with divergence dating and ancestral state reconstructions, we attempted to identify the role of bedrock preferences, chromosome numbers coupled with relative genome sizes estimates, as well as morphological features through time. We also investigated extant climatic preferences using a heavily curated set of occurrence records individually selected for superior precision, and quantified rates of climatic niche evolution in each section. We found that a number of phylogenetic incongruences derail the identification of correlates of diversification, yet a number of patterns persist regardless of the topology considered. All the studied correlates are likely to have contributed to the diversification of Gentiana in Europe, however, their respective importance varied through time and across clades. Chromosomal variation and divergence of climatic preferences appear to correlate with diversification throughout the evolution of European Gentiana (Oligocene to present), whereas shifts in bedrock preferences appear to have been more defining during recent diversification (Pliocene). Overall, a complex interaction among climatic, geological and biotic attributes appear to have supported the diversification of Gentiana across the mountains of Europe, which based upon phylogenetic as well as other evidence, was probably also bolstered by hybridization.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mountain systems around the world are more species-rich than their adjacent lowlands, as evident from the global distribution of species richness in plant (e.g., Kier et al. 2005). Higher biodiversity levels in alpine thermal belts were probably established due to a suite of drivers acting in concert locally to foster diversification or even a large number of explosive radiations, while at the same time ensuring their persistence through time (Hoorn et al. 2013). Abiotic drivers, governed by geological and climatic dynamics, are usually considered as prime candidates to explain local diversity in all major mountain systems of the world (reviewed in Muellner-Riehl et al. 2019), yet only a few studies also considered biotic ones (e.g., Lagomarsino et al. 2016; Ebersbach et al. 2017). How these drivers interacted and how this interaction varied through time to generate biodiversity remains poorly understood.
Dating back to the Eocene, the oldest alpine flora in the world most likely originated in the Tibeto-Himalayan Region (THR; Ding et al. 2020), whereas that of the Andes (i.e., the paramo) is notoriously fast evolving (Madriñán et al. 2013). For these two mountain systems, geological processes were overwhelmingly identified as the force activating diversification (e.g., Luebert and Weigend 2014; Favre et al. 2015). Beyond geological dynamics, higher geodiversity appears to be correlated with higher biodiversity levels (Muellner-Riehl et al. 2019). Nevertheless, in the last decade, climatic changes were increasingly viewed as crucial for biodiversity, not only as a potential cause of extinction but also as a powerful force fueling diversification. Unraveling either over millions of years (e.g., Miocene cooling) or via swift cold and warm snaps within a shorter time (i.e., oscillations in the Pleistocene), the evolution of global climate conditions has had a substantial effect on distribution ranges of alpine species, and possibly much faster than geological dynamics ever did. In particular, climate oscillations have been identified as a potent driver for biodiversity in mountain systems (Muellner-Riehl 2019; Muellner-Riehl et al. 2019). For example, in the THR, the “Mountain Geobiodiversity hypothesis” states that climate oscillations fostered a so-called species-pump effect (Hoorn et al. 2013; Sedano and Burns 2010; Mosbrugger et al. 2018), probably at the source of numerous radiations. It is worth noting that whether or not such a species-pump effect occurred within a given mountain system depends on its geographical location and its elevational extent. In the European Alpine System, which is located well beyond the intertropical zone, the overlap between today's and last glacial maximum (LGM) temperature profiles is minimal in comparison to tropical ranges (Muellner-Riehl et al. 2019). This means that suitable habitat for many species was not to be found within European mountain ranges when the temperature plummeted in the past. As a result, the survival of most of the European alpine flora during the LGM occurred ex-situ at the periphery of the Alps and beyond (e.g., Schönswetter et al. 2002; Kropf et al. 2003; Windmaißer et al. 2016) except for a few nunatak refugia (Schönswetter et al. 2005; Kosiński et al. 2019; Schönswetter and Schneeweiss 2019; Pan et al. 2020). These drastic displacements of distribution ranges have certainly contributed to prolonged isolation of lineages in geographically distant glacial refugia, thus also contributing to a considerable degree to divergence and possibly diversification (Kadereit et al. 2004; Nieto Feliner 2011; Paule et al. 2018), although this might not constitute a species-pump effect per se.
In addition to geological and climatic processes, a series of biotic drivers contributed to current biodiversity levels in mountain systems of the world. Although these are less frequently investigated (Muellner-Riehl et al. 2019), several studies have identified morphological traits associated with increased diversification rates (e.g., Matuszak et al. 2016). In most cases, these key innovations were clearly advantageous in harsh alpine conditions. For example, the cushion habit in Saxifraga section Porphyrion (Ebersbach et al. 2017) and Androsace (Roquet et al. 2013) are believed to have enhanced diversification rates in alpine regions throughout Eurasian mountain systems. Interestingly, the effect of key innovations (or their loss) on diversification rates may vary temporally throughout the evolution of a genus, as was the case of heterostyly in Primula (deVos et al. 2014). Besides, even in the absence of increased diversification rates, key innovation may still help lineages to seize new ecological opportunities and thus foster divergence, as was the case in Alchemilla lineages of the Afroalpine (Gehrke et al. 2016). Other biological modifications may allow lineages to colonize new habitats and embark on their own evolutionary track, thus contributing to biodiversity. For instance, it has been shown that polyploids appear to spread more than diploids into new environments (te Beest et al. 2012), for example, those rendered available after glacial retreat (e.g., Burnier et al. 2009). As a result, polyploids are thought to be more abundant in both arctic and alpine floras (e.g., Brochmann et al. 2004). Furthermore, hybrid speciation may follow polyploidization, thus generating new lineages, as for example in Saxifraga (reviewed in Ebersbach et al. 2020). Any chromosomal rearrangement may contribute to divergence and diversification, sometimes in correlation with climatic modifications (e.g., Carex; Escudero et al. 2012). Finally, the evolution of environmental preferences (abiotic niche space, e.g., climate and soil) has only rarely been investigated as a driver of diversification within a more comprehensive context, taking modifications of the abiotic background through time into account. Such a complex approach requires assembling a vast array of geological, climatic and biological data, as well as a robust phylogenetic background. The European Alpine System appears thus as prime mountain system to investigate this topic because owing to a long research tradition, a plethora of data are available, including precise geological maps, numerous georeferenced occurrence data, not to mention solid taxonomic treatments.
Although biotic and abiotic drivers have been investigated in depth in few taxa such as Saxifraga and the Andean bellflowers (several genera, Campanulaceae: Lobelioideae), other taxa appear promising because of their ubiquitous strong alpine affiliation. This is the case of Gentiana L., a species-rich genus (ca. 360 spp.) of predominantly alpine plants that is distributed in many major mountain systems in the world, as e.g. the THR, the European Alps and the Andes. Gentiana is composed of 13 sections, of which delineation and phylogenetic relationships have been clarified molecularly in the last decades (Yuan and Kuepfer 1995; Favre et al. 2010, 2014, 2020). Biogeographic analyses have shown that the genus originated in the THR probably by the end of the Eocene (Favre et al. 2016), which is the estimated time of the earliest occurrence of alpine vegetation there, based on multiple taxa (Ding et al. 2020). With over 260 species, the THR is currently the main center of biodiversity for Gentiana (Ho and Liu 1990). After an early diversification in situ, Gentiana repeatedly dispersed out of Tibet and independently colonized Europe, at least six to eight times. Except in one case, the colonization of Europe occurred during and after Late Miocene (Gentiana section Pneumonanthe Gaudin, G. sect Cruciata Gaudin and G. sect. Chondrophyllae Bunge). The single early account of dispersal to Europe dates back to the Oligocene and was followed by steady diversification up to today (Favre et al. 2016). Three closely related lineages are attributed to this early diversification, namely sections Calathianae Froel. (ca. 10 spp.), Ciminalis (Adans.) Dumort (7 spp.) and Gentiana (7 spp.). These monophyletic sections (Favre et al. 2020) are endemic or near-endemic to the European Alpine System (e.g., Alps, Pyrenees, Dinarides, and Carpathians), with only two species occurring as far east as Lake Baikal. These, however, represent only very recent expansions (Favre et al. 2016). Morphologically, the three European sections are easy to distinguish. Only G. sect. Calathianae displays funnelform corollas, whereas those of the two other sections are campanulate, urceolate, or widely obconical. The only exception is G. lutea, which displays a deeply divided corolla resembling that of Swertia L. All species of G. sect. Gentiana display multi-flowered, often clustered inflorescences on tall stems, whereas the flowers in the two other sections are single and terminal. With their distribution centered in the European Alpine System, the three sections G. sect. Calathianae, G. sect. Ciminalis, and G. sect. Gentiana represent an ideal model to investigate potential factors that contributed to their diversification and how these combined through time in the European Alpine System.
It is worth mentioning that in the literature, authors often refer to “drivers” by default, without providing statistical support for a causal relationship between environmental conditions and diversification. Typically, a plethora of case studies claims that a geological process (e.g., uplift) acted as a driver, whereas they only show a temporal overlap between the two. In this study, we will thus refer to “correlates” of diversification instead of drivers, indeed because equaling a correlate with a driver is not straightforward. Using an integrative approach, we will investigate their relative contributions through time in the European Alpine System, with a near-complete species sampling of Gentiana and two phylogenetic hypotheses.
Materials and methods
Samples, DNA extraction and sequencing
Our sampling design included almost all (22/24) species of the closely related sections Calathianae, Ciminalis and Gentiana, excluding G. orbicularis Schur, and G. ligustica R.Vilm. & Chopinet. Most species (n = 19) were collected in the wild in Switzerland, Italy and Austria in 2018, 2019 and 2020. Voucher specimens were deposited at FR. Fresh leaf samples were dried and preserved in silica gel. We complemented our field sampling by retrieving material from herbarium specimens (M) and from specimens cultivated in the Jardin Gentiana (Leysin, Switzerland). Species were identified using Flora Helvetica (Lauber et al. 2012), Flora Alpina (Aeschimann et al. 2004) and Flora Europaea (Tutin et al. 1964–1980). We also included species of almost all accepted sections of Gentiana as outgroups (Favre et al. 2020). We excluded the monotypic G. sect. Phyllocalyx (Kusn.) T.N.Ho and G. sect Chondrophyllae because of their particularly derived sequences (Fu et al. 2021a). Accession numbers are provided in Appendix I [accessions to be provided upon acceptance].
For DNA extractions, we used the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany), following the manufacturer’s protocol with a longer incubation time in the lysis step (ca. two hours). Whole genome library preparation (insert size 350 bp) and sequencing were performed by Novogene (Novogene BioTech, Inc. Beijing, China), using the NovaSeq 6000 PE 150 (Illumina, San Diego, USA) sequencing system. Two Gb of raw data were generated per sample. Some additional data produced via anchored hybrid enrichment were available from a previous study (Favre et al. 2020; see Appendix I). We imported raw data in Geneious Prime® 2019.0.4 (Kearse et al. 2012) (Biomatters Ltd, Auckland, New Zealand), where raw reads were trimmed with BBDuk and mapped (Geneious mapper) against the entire plastid and rDNA-cistron (incl. ETS, 18S, ITS 1, 5.8S, ITS2, and 26S) sequences available for closely related species in earlier studies (plastids from Fu et al. 2021a; rDNA-cistron from Favre et al. 2020).
Phylogenetic analyses and molecular dating
Sequences were aligned in Geneious Prime using MAFFT v.7 (Katoh and Standley 2013). The resulting alignments were inspected and manually edited, and finally reduced to informative sites. We ran Bayesian inference (BI) and Maximum likelihood (ML) for both alignments separately (rDNA, plastid). For the ML analysis, we used raxmlGUI v.2.0 beta (Edler et al. 2019) for RAxML v.8 (Stamatakis 2014), with GTRGAMMA as the model of evolution. We obtained statistical support for nodes via bootstrap analysis including 1000 replicates (Felsenstein 1985). We performed BI analyses with MrBayes v.3.2.6 (Ronquist et al. 2012). Four runs were started from random trees, with 4 coupled incrementally heated Monte Carlo Markov Chains each (MCMC; 1 cold and 3 heated), for 10 million generations sampling every 1000th. The potential scale reduction factor (PSRF) values, effective sample sizes (ESS), and average standard deviations of split frequencies (asdsf) were well within acceptable values (ESS ≫ 200, PSRF between 0.999 and 1.001, asdsf < 0.01). We discarded the first quarter (25%) of the sampled trees as burn-in, then a majority rule consensus tree and posterior probabilities (PP) were computed. Trees were visualized in Figtree v.1.4.0 (Rambaut 2010). Complete phylogenetic trees with node support for BI and ML analyses are presented in Appendix II.
To perform molecular dating, we ran the analyses separately for the plastid and rDNA datasets, using the HKY substitution model, the Yule model and the relaxed clock model. We used a calibration strategy identical to Fu et al. (2021a): The split between G. sect. Cruciata and G. sect. Pneumonanthe was constrained with taxonomically unambiguous seed fossils from the Early Miocene (Mai and Walther 1988; Mai 2000) using a lognormal node age prior with an offset at 16 Ma, a mean of 1 and a standard deviation of 1. Seed fossils were attributed to a species belonging to the G. sect. Cruciata, of which seed morphology is rather uniform and known to differ from those of G. sect. Pneumonanthe (Davitashvili and Karrer 2010, and references therein). Hence, the age of the seed fossils represent a minimum age for the split between these two sections. Given the scarcity of fossil evidence in gentians, we also constrained the crown age of the genus using the date calculated in a broad-scale phylogenetic study of angiosperms (Janssens et al. 2020). We used uniform priors (Schenk and Axel 2016) for the 21.25–38.21 Ma age range, covering the entire 95% highest posterior density (HPD) interval from Janssens et al. (2020). The MCMC had 10 million generations, which were sampled every 1000th generation, discarding the initial 25% as burn-in. Convergence was assessed in TRACER v.1.5 (http://tree.bio.ed.ac.uk/software/tracer/) and deemed suitable given ESS values (> 200). Trees were summarized in TreeAnnotator v.1.7.5 using the mean height option (Drummond et al. 2007). Analyses were run on CIPRES (Miller et al. 2010). The plastid and rDNA-cistron dated trees are presented and compared in Fig. 1.
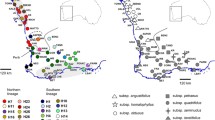
Dated phylogenetic trees of three studied European sections of Gentiana based on plastid and rDNA data. The grey bars show the 95% highest posterior density intervals. All nodes are fully supported (1.00 posterior probability) unless otherwise stated. Plotted on the tree are shifts or expansions of four traits (bedrock preferences, corolla type, life cycle and inflorescence type) based on ancestral states reconstruction using Mesquite. Extant trait states are indicated to the right of each species and are color coded according to the legends to the left. Plastid and rDNA phylogenetic trees produced by Bayesian and Maximum Likelihood analyses are available in Appendix II. The crown node of G. sect. Ciminalis is not supported (ns)
Georeferencing and evaluation of available locality data
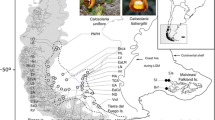
Our georeferenced occurrence point dataset matched the DNA sampling with the addition of G. ligustica. For topographically complex areas, we applied the strongest control possible on locality data retrieved from databases (i.e., location verified within < 300 m). In contrast, for areas characterized by only minor elevational variation and by uniform climatic conditions (e.g., vast plains and plateaus), some degree of uncertainty was allowed (up to 3 km), particularly in areas for which only a few data points were available. Locality data were gathered from several sources. First of all, high-accuracy occurrence records for Switzerland were obtained from Infoflora (https://www.infoflora.ch), and from fieldwork campaigns lead by the authors or collaborators in Switzerland, Austria, Italy, Bulgaria and Romania (e.g., see Bartók et al. 2015). These datasets guarantee a < 50 m accuracy. Additional data were downloaded from GBIF (Last accessed: December 2019), and for these, strict data cleaning was applied. For example, occurrences associated with pre-defined quadrants across a country (with the center of the quadrant as corresponding GPS point) or gardens were discarded altogether. GPS points provided without associated locality descriptions were discarded unless they originated from specialized platforms such as iNaturalist (https://www.inaturalist.org/). We evaluated all remaining GBIF data points individually in Google Earth (Gorelick et al. 2017), carefully verifying that given locality descriptions and elevations (whenever the latter was available) matched the GPS coordinates provided. Whenever possible, imprecise GPS coordinates were refined to increase precision, but only when locality descriptions included unmistakable landmarks (e.g., mountain tops, lake shores). Occurrence data for which the precise locality could not be identified with certainty were discarded. In a few cases, we had to take taxonomic synonymy into account, which led to the re-assignment of occurrences of some taxa (e.g., Gentiana terglouensis subsp. schleicheri (Vacc.) Tutin) to others (e.g., Gentiana schleicheri (Vacc.) Kunz). Finally, we double-checked that occurrences matched the known distribution of each species, and eliminated all points outside of their respective ranges. For this, we used Flora Helvetica (Lauber et al. 2012), Flora Alpina (Aeschimann et al. 2004) and Flora Europaea (Tutin et al. 1964–1980). When the remaining number of occurrences was particularly low (overall or within a part of their distribution range), we searched additional literature to recover additional locations and georeferenced them. This was for example the case for all species occurring in the Carpathians and the Dinarides (e.g., Christe et al. 2014; Bartók et al. 2015). In the case of G. froelichii Jan ex Rchb., six additional points were generated in similar habitats near known occurrences to overcome severe data limitations for this narrow endemic species (these additional occurrences are listed in Appendix V and VI).
Chromosome numbers and relative genome size
We systematically reviewed the literature and compiled an extensive list of previously published chromosome counts for the species included in the molecular dataset. Since we were also interested in the spatial distribution of cytotypes, we only included counts that were accompanied by detailed locality information. Furthermore, taking into account the synonymy, records for which the determination was doubtful, or records that fell outside of the distribution range of a species were discarded. The list of reviewed chromosome numbers is given in Appendix IIIb.
To asses the variability in chromosome numbers, relative genome size (RGS) of plants collected in the scope of this project was estimated by flow cytometric analyses of silica gel-dried leaves using the Partec CyFlow space (Partec, Münster, Germany) fitted with a high power UV LED (365 nm). Leaf tissues of the analyzed sample and internal standard Glycine max cv. Polanka (2C = 2.50 pg) or Pisum sativum cv. Ctirad (2C = 9.09 pg) (Doležel et al. 1994, 1998) were homogenized using a razor blade in a plastic Petri-dish containing 1 ml of ice-cold Otto I buffer (0.1 M citric acid, 0.5% Tween 20; Otto 1990). The suspension was filtered through Partec CellTrics® 30 µm to remove tissue debris and incubated for at least 10 min at room temperature. Isolated nuclei in filtered suspension were stained with 1 ml of Otto II buffer (0.4 M Na2HPO4 × 12H2O) containing the AT-specific fluorochrome 4′,6-diamidino-2-phenylindole (DAPI; 4 µg ml–1) and β-mercaptoethanol (2 µg ml–1). The relative fluorescence intensity was recorded for 4000 particles with one to three replicates per accession. RGS was calculated from the means of fluorescence histograms visualized using the FloMax v.2.4d software (Partec, Münster, Germany) as a sample/standard ratio. Only histograms with coefficients of variation (CVs) < 5% for the G0/G1 peak of the sample were considered. In the case of CVs exceeding this threshold, the measurement was discarded and the sample was reanalyzed. Pisum sativum was used as a primary standard and Glycine max was used for samples of which signal overlapped with that of P. sativum. The sample-standard ratios estimated using the internal standard G. max were adjusted to those using P. sativum by multiplying the values by the coefficient of 3.204 which was based on 11 repeats of ratios among the 2 standards. The list of the studied samples with a distribution map is provided in Appendix IV.
Ancestral states reconstructions
For a series of ecological and morphological traits, we reconstructed ancestral states using Mesquite v. 2.75 (Maddison and Maddison 2011) using a parsimony model, with categorical characters treated as unordered. Ancestral state reconstructions were performed on both the plastid and the rDNA trees. For each species, we identified bedrock preference as calcareous, indifferent or siliceous referring mostly to Flora Alpina (Aeschimann et al. 2004). Other traits we investigated were the shape of the corolla (campanulate, funnelform or deeply lobed), the type of inflorescence (terminal flowers, cyme), as well as the life cycle (annual, perennial).
In addition, ancestral chromosome number was reconstructed in ChromEvol v.2.0 (Glick and Mayrose 2014) also along both plastid and rDNA trees. The input file was based on the reviewed majority haploid chromosome numbers. We used 100,000 simulations, default settings and a rooted phylogenetic tree with G. pneumonanthe as an outgroup resulting from trimming to studied sections of either the plastid or the rDNA tree. Gentiana pneumonanthe was selected as an outgroup because its chromosome number (2n = 26) is stable for all species of G. sect. Pneumonanthe and very common in G. sect. Cruciata, whereas chromosome numbers are unknown for the closest relative of European gentians (i.e., the monotypic G. sect. Phyllocalyx) and mostly unknown and highly variable in the mostly annual G. sect. Chondrophyllae. Moreover, Yuan et al. (1996) did suggest 2n = 26 as plesiomorphic for this entire clade, however with a different sectional delineation as in our study. As a comparison, we also ran the analyses without any outgroup species. ChromEvol implements a series of likelihood models for the evolution of chromosome numbers. By comparing the fit to data of different models parametrizing different chromosomal processes (e.g., loss/gain of single chromosomes, whole whole-genome duplications), it provides insights into the chromosome number evolution. For each model, the program estimated the rates for the possible transitions, inferred the set of ancestral states, and estimated the location along the tree for which chromosome number alteration occurred. The best fitting models were chosen based on Akaike information criterion (AIC).
Climatic preferences and geographic overlap
To quantify climatic preferences among the European species of Gentiana, we downloaded the WorldClim dataset v.2.1 for the highest possible spatial resolution of 30 s (~ 1 km2, Fick and Hijmans 2017). The dataset comprises 19 bioclimatic variables (e.g., annual mean temperature, min temperature of the coldest month) derived from monthly temperature and rainfall values for the years 1970–2000 as well as the underlying elevation data derived from the Shuttle Radar Topography Mission. Bioclimatic variable and elevation values were extracted for all occurrence data points in R (R Core Team 2020) and RStudio (RStudio Team 2020) using the raster package (Hijmans and van Etten 2012). Since we were mainly interested in the climatic niche differences of European mountain species, we restricted the queried occurrence data points to those located in continental Europe (map extent visible in Fig. 3) as the occurrence of some species in distant areas such as Iceland (G. nivalis) are likely to be recent expansions of distribution ranges. Elevational ranges of each species were then quantified based on the retained occurrence points using the above mentioned elevation raster layer.
Correlations between variables were then assessed in base R and visualized using the corrplot R package (Wei and Simko 2017), and highly correlated variables (Pearson’s r > 0.9) were removed from the dataset resulting in the following variable set: Annual mean temperature (Bio1), mean diurnal range (Bio2), isothermality (Bio3), temperature seasonality (Bio4), mean temperature of the wettest quarter (Bio8), mean temperature of the driest quarter (Bio9), annual precipitation (Bio12), precipitation seasonality (coefficient of variation, Bio15), precipitation of the warmest quarter (Bio18), and precipitation of the coldest quarter (Bio19). Climatic and elevation preferences of each species were assessed using principal component analysis (PCA) using the factoextra R package (Kassambara and Mundt 2017) and compared among sections, major clades and sister species using analysis of variance (ANOVA) in base R. Results were visualized using the ggplot2 package (Wickham 2016). Rates of climatic niche evolution were calculated based on the means of the selected bioclimatic variables and the first two principal components. To account for temporal and topological uncertainty, we used a subset of 500 trees (for both the plastid and the rDNA trees), randomly chosen from the posterior distribution from our BEAST analyses and pruned to contain only ingroup species. Climatic niche evolution rates (σ2) were then modeled separately for each section under a Brownian motion model using the fitContinuous() function of the R package geiger (Harmon et al. 2008). Higher values of σ2 (rate of variance accumulation per unit of branch length) are generally interpreted as signals of faster niche evolution (Felsenstein 1985; O'Meara et al. 2006; Saupe et al. 2018). Finally, raw σ2 values of each variable and dataset were rescaled to values between 0 and 1 for visualization purposes.
Geographic overlap between sister species distributions was assessed at res = 0.2 using the “Chesser&Zink” statistic of the lets.overlap() function in the letsR package (Vilela and Villalobos 2015). This statistic quantifies the degree of overlap between two distribution ranges as the proportion of the smaller range that is contained in the larger range (Chesser and Zink 1994).
Results
Phylogenetic relationships and molecular dating
The full plastid alignment comprised 155,347 bp, with sequences ranging from 146,687 bp to 148,976 bp for species sequenced via genome skimming. For species sequenced via anchored hybrid enrichment (Lemmon et al. 2012; see Favre et al. 2020), sequences were incomplete (as expected with this approach), ranging between 115,679 bp and 120,632 bp. Sequences of the rDNA were all complete, with a length ranging from 6059 to 6187 bp, producing together an alignment of 6227 bp. Sites with missing data and ambiguous positions occurred particularly in non-coding regions and were removed. Finally, reduced alignments (informative sites only) were 15,614 (plastid) and 677 (rDNA) characters, respectively.
The topologies based on the plastid and rDNA datasets were incongruent (Fig. 1, Appendix II), although the monophyly of the sections as well as several other clades (incl. the G. verna clade, the G. lutea clade, and the G. nivalis clade) were relatively well attested. Phylogenetic relationships among sections, and the relative position of G. terglouensis and G. froelichii to their congeners differed between the trees. Gentiana sect. Ciminalis was strongly supported only in the rDNA dataset (see Fig. 1; Appendix II). In was not in the plastid phylogeny, where G. sect. Ciminalis was deeply divided in two well-supported clades (Fig. 1), one including G. clusii Perrier & Song, G. alpina Villars and G. dinarica Beck, the other clade comprising the remaining species. Within G. sect. Calathianae, two distinct clades were well defined. The first one included G. nivalis L., G. pumila Jacq. and G. utriculosa L., whereas the second clade included the remaining species (e.g., G. verna), of which phylogenetic relationships were well supported in the plastid tree, and less so in the rDNA tree. Finally, G. sect. Gentiana was partially resolved in the plastid tree, with a clade formed by the blue flowering species (G. froelichii, G. asclepiadea L.) which was sister to a clade encompassing only yellow or red flowering species (e.g., G. lutea L.). In contrast, G. froelichii and G. asclepiadea did not cluster together in the rDNA tree, and the phylogenetic relationships among red and yellow flowering gentians differed but in both cases with relatively low support. In the outgroup, all sections were well supported, and their phylogenetic relationships were as in earlier studies (e.g., Favre et al. 2020).
Divergence time estimates were usually similar for comparable nodes of the plastid and rDNA dated trees (Fig. 1), and in the range of earlier studies although we recovered broader 95% HPD intervals than some (compare with Fu et al. 2021a), and slightly older ages than others (e.g., Favre et al. 2016). We found that the crown age of Gentiana was ca. 36 (39–31) Ma in the plastid tree, and ca. 34 (38–28) in the rDNA tree. The three sections started to diverge from each other at ca. 24 (31–16) Ma in the plastid tree, and 27 (35–20) Ma in the rDNA tree. The crown ages of G. sect. Gentiana and G. sect. Calathianae strongly overlapped between the plastid and rDNA (respectively), with ca. 15 (23–7) Ma and ca. 15 (24–6) Ma for the former and ca. 12 (18–6) Ma ca. 15 (22–9) Ma for the latter. In contrast, the crown ages of G. sect. Ciminalis differed strongly between the two trees with 23 (31–12) Ma and 10 (19–3) Ma for the plastid and rDNA trees, respectively. Indeed, in the plastid tree, a deep split was observed in G. sect. Ciminalis, and the crowns of these two clades were of contrasting ages, with ca. 1.5 (0.5–3.5) Ma and ca. 9 (16–3) Ma for the clade containing G. acaulis and the clade containing G. clusii, respectively. This deep split is not present in the rDNA tree, and since phylogenetic relationships differ among species, further age comparisons in this section are not possible. In the plastid tree of G. sect. Gentiana, the two blue-flowered species of (G. asclepiadea and G. froelichii) diverged from each other at ca. 12 (20–2.5) Ma, whereas the yellow or red flowering species started to diversify ca. 2.7 (4.9–1.3) Ma of the yellow or red flowering species was older: 6 (11–2) Ma. In G. sect. Calathianae, the ages recovered for clade formed by G. nivalis, G. pumila and G. utriculosa were almost identical, with ca. 10 (15–5) Ma in the plastid tree, and ca. 10 (15–4) Ma in the rDNA tree. The other clade of this section (the G. verna clade) was ca. 8 (12–3.6) Ma in the plastid tree, and 12 (18–7) Ma in the rDNA one.
Occurrence dataset
To date, our dataset (see Appendix VI) includes the most accurate selection of georeferenced occurrences for European species of Gentiana (20,841), with all GPS points attested within a maximum of 300 m within topographically complex areas. Overall, we found that ca. 14% out of 74,436 occurrences originally downloaded from GBIF were precise enough to be included in this study (10,484 occurrences). Some of them could be included after being refined based upon precise locality descriptions (1480). Including the data provided by Infoflora for Switzerland (9309), as well as additional literature and fieldwork information (1048), the total per section was as follows: G. sect. Calathianae (9373), G. section Ciminalis (4288), G. sect. Gentiana (7180). Over 40 occurrences were available for most species, with particularly abundant records for G. nivalis (4435), G. verna (3148), G. asclepiadea (2379) and G. lutea (2356). The data were critically limited only for three rather narrowly distributed species, including G. terglouensis Hacq. (24), G. dinarica (22) and G. froelichii (23). For these three species, some additional occurrences were mined in publications, and additional points were generated nearby existing records (6 for G. froelichii), listed in Appendix V and VI). The data available from GBIF and Infoflora were sufficient to represent the distribution range of all species in the European Alpine System, although in the Alps we observed a slightly lower density of points in Tyrol (Austria), for which we could not obtain further data. After cropping the dataset to include only these mountain ranges, our final dataset included 17,143 occurrences. Table 1 summarizes the origin of occurrence data, as well as their distribution among species.
Chromosome numbers and relative genome size
Altogether, we identified 305 original chromosome counts for all studied ingroup species (Table 2, Appendix IIIb), with widespread taxa being counted more often than rare ones, as expected. Chromosome numbers appear to be stable within 15 of the 22 species that we investigated. For example, for all six species of G. sect. Ciminalis n = 18 or 2n = 36 chromosomes were reported consistently. In contrast, some variation was discovered among species of the two other sections. Chromosome numbers were highest in G. sect. Gentiana, (2n = 40, 42 or 44), and counts in the clade formed by G. burseri Lapeyr., G. lutea, G. pannonica Scop., G. punctata L. and G. purpurea L. were uniformly 2n = 40. However, in the same section, the species G. froelichii and G. asclepiadea had 2n = 42 and 2n = 36 or 44 chromosomes, respectively. The latter species is widespread (from the Alps to the Caucasus) and counts are available throughout its distribution range, but 2n = 36 was only counted in some samples from Slovakia. Since 2n = 36 is otherwise found only in G. sect. Ciminalis, it is possible that local G. clusii were mistaken for depauperate G. asclepiadea. The section with the most chromosomal variation was G. sect. Calathianae (2n = 14–42). Although chromosome number seems stable in G. bavarica L. (2n = 30), G. brachyphylla Vill. (2n = 28) and G. nivalis (2n = 14), the records for the six other species each contained at least one divergent count. The closely related species G. nivalis, G. pumila (2n = 20), and G. utriculosa (2n = 22), had the lowest chromosome counts overall. However, for the two latter species, it appears that either polyploidy may exist (G. utriculosa, 2n = 33), or some chromosomal aberrations such as aneuploidy and the presence of B chromosomes (e.g., 2n = 21 in G. pumila, 2n = 28 + 0-6B in G. utriculosa). Such anomalies were also discovered in several other species of the G. sect. Calathianae. In this lineage, aberrant chromosome numbers were detected in G. rostanii Reut. ex Verl. (2n = 31, 28 + 0-2B), G. schleicheri (Vacc.) Kunz (2n = 31, 33, 30, 30 + 1B), G. verna (2n = 30, 28 + 0–2/3B) and G. terglouensis (2n = 30, 40 or 42). Finally, in G. lutea and G. bavarica, subspecies did not differ from each other in terms of chromosome numbers (see Appendix IIIb). In Gentiana verna, 91 of the 100 records overwhelmingly indicated the chromosome number to be 2n = 28, however, in some subspecies only aberrant counts were recorded, as for example G. verna ssp. tergestina (Beck) Hayek and G. verna ssp. pontica (Soltok.) Hayek (both 2n = 28 + 0-2B).
A total of 102 individuals of Gentiana were analyzed by flow cytometry (Table 2, Appendix IV), representing all species included in the phylogeny, except G. rostanii. Coefficients of variation for the G0/G1 peak of the analyzed samples ranged from 0.79 to 4.92 (mean = 1.78). In line with previously published chromosome numbers, the variation of RGS among species as well as within the whole section Ciminalis was low, supporting the conserved chromosome number 2n = 36. Similarly, in G. sect. Gentiana the RGS from 0.70 to 0.74 showed a close relationship to 2n = 40. Two exceptions were G. froelichii (2n = 42) and G. asclepiadea (2n = 44) for which RGS of 0.53 and 0.68 were recovered, respectively. Even though chromosomal variation was reported in G. sect. Calathianae, RGS of studied accessions revealed almost no intraspecific variation. Hence, we assume that recovered RGS represent dominant cytotypes for particular species. Interestingly, the RGS of both G. bavarica and G. verna subspecies were slightly differentiated, when compared to the nominal subspecies.
Ancestral state reconstructions
Based on both the plastid and rDNA trees, our reconstructions showed that the common ancestor of G. sect. Calathianae, G. sect. Ciminalis and G. sect. Gentiana was most likely a perennial plant with campanulate flowers arranged in cymes, preferring calcareous bedrock (Fig. 1). During their evolution, floral differentiation occurred twice, first with a shift to funnelform flowers dating back to the Mid Miocene at the stem of G. sect. Calathianae, and more recently in the Pliocene for G. lutea (deeply lobed flowers). However, the number and timing of shifts from cymes to single terminal flowers differed whether the reconstructions were performed with the plastid or the rDNA tree. Using the former, three shifts from cymes to single terminal flowers were detected. The first one occurred during the divergence of the common ancestor of all species G. sect. Ciminalis (between Eocene and the first half of the Miocene), the second was associated with the stem of the G. verna clade in G. sect. Calathianae (second half of the Miocene and Pliocene), and the last one for G. pumila (around the Miocene/Pliocene boundary). With the latter, only one clear shift was recovered shortly after the Oligocene–Miocene boundary, probably before the divergence of G. sect. Calathianae and G. sect. Ciminalis (which cluster together in the rDNA tree), whereas the sequence of shifts for the clade containing G. nivalis remains unclear.
In contrast to morphological traits, shifts (or expansions) from calcareous to siliceous bedrocks were reconstructed in all three sections and appeared to have been concentrated to relatively recent times (almost always since the Pliocene). For example, two shifts were detected in G. sect. Ciminalis, namely for G. acaulis and G. alpina, regardless of the phylogenetic tree used. However, reconstructions based on the plastid tree and the rDNA tree differed for G. section Calathianae and G. sect. Gentiana. In the latter, the reconstruction using the rDNA tree is straightforward because only a single shift occurred in the Late Pliocene, which was followed by the divergence of three silicolous species: G. purpurea, G. punctata and G. burseri. In contrast, the reconstruction with the plastid tree was inconclusive: we could not identify the preference of the common ancestor of the clade composed of the yellow and red flowering species, although some differentiation in bedrock preferences clearly occurred. Finally, in G. section Calathianae, there were three expansions and one shift (G. brachyphylla) detected in both trees.
To infer the transitions of the haploid chromosome numbers along the branches all eight models implemented in ChromEvol were tested along the plastid and the rDNA tree. Analyses based on both trees recovered the same model assuming a constant rate of change with no duplication (CONST_RATE_NO_DUPL, Table 2) as best-fitting. However different ancestral states (Fig. 2, Appendix IIIa) were recovered for both trees. The ancestral state of the haploid chromosome number for all three studied sections based on the rDNA tree was n = 23. The overall calculated loss rate of chromosomes (61.03) was about six times higher than the gain rate (9.17). Thus, reductions in haploid chromosome number occurred more frequently than increases and were observed in all three sections (Fig. 2). Interestingly, reconstructions based on the plastid tree revealed n = 18 for the common ancestor and the overall calculated loss rate of chromosomes (55.39) was only slightly higher than the gain rate (41.59). Hence, reductions in haploid chromosome number also occurred more frequently but was suggested only in G. sect. Calathianae (Fig. 2). In G. sect. Ciminalis the chromosome number remained stable as the ancestral state and in G. sect. Gentiana a gain of two chromosomes was proposed. When reconstructing the ancestral states along mid-point rooted phylogenies (without outgroup) the results were consistent with minor differences in the reconstructions along the plastid phylogeny (Appendix IIIa).
ChromEvol inferences for three studied European sections of Gentiana based on the respective haploid chromosome numbers and/or their frequencies, using the plastid and rDNA tree without outgroup (Appendix II). ChromEvol inferences using trees including outgroups are presented in Appendix IIIa. Haploid chromosome numbers and their respective frequency (when several values were found) are indicated after each species name. The ML ancestral chromosome numbers estimated are given next to nodes. Pie charts on nodes indicate the probability of different haploid numbers, which are color coded as indicated. A review of available chromosome counts, as well as ChromEvol inferences are available in Appendix IIIb
Geographic overlap
To better understand the role of allopatric speciation for the species considered here, we quantified the geographic overlap between closely related species. Values of geographic overlap varied widely, from 0 for example for comparisons involving G. dinarica which only occurs in a limited distribution area in the Dinarides and the Apennines, to 1.0 between G. verna and G. schleicheri, indicating that the restricted distribution of G. schleicheri is completely engulfed in the distribution area of G. verna (Table 4, Appendix V). Mean overlap was relatively similar among the three sections, with overlap between closely related species of G. sect. Ciminalis being the lowest (0.29 and 0.23 according to cp and rDNA topologies, respectively) compared to G. sect. Gentiana (0.31, 0.34) and G. sect. Calathianae (0.45, 0.52). We present a map of species richness (total overlap of all species in our dataset) in Fig. 3.
Climate preferences and climatic niche evolution
Despite substantial geographic overlap (0.65–0.72, Table 4), Gentiana sect. Calathianae, G. sect. Gentiana and G. sect. Ciminalis displayed significantly different elevational ranges, with species of G. sect. Gentiana recorded from the lowest mean elevations (1387 m asl) compared to the other two sections (mean G. sect. Calathianae: 1860 m asl, mean G. sect. Ciminalis: 1720 m asl). These elevational differences were also mirrored in significant differentiation in some extant section-specific climatic preferences, even though overall climatic niche spaces were broadly overlapping between the three sections (Appendix V). Gentiana sect. Calathianae was significantly differentiated from the other two sections along PC1, which was mostly driven by seasonality-dependent variables (Bio 4, 8, 15; Table 4, Appendix V) as well as PC2, which was predominantly driven by Bio1 (annual mean temperature) and Bio9 (mean temperature of the driest quarter). Gentiana sections Gentiana and Ciminalis were separated from each other by subtle but significant differentiation along PC1 (Table 4, Appendix V).
When considering differentiation within each section, we found that main clades were almost always accompanied by significant differentiation in terms of both elevational ranges and climatic preferences as summarized by PC1 and PC2 (Table 4). More recent splits between closely related species were also often characterized by significant levels of differentiation in both elevation and climatic preferences, however, this was commonly caused by one sister species (or closely related species) occupying a relatively large niche space and the other one being restricted to a subset of that space (e.g., G. rostanii and G. brachyphylla, G. froelichii and G. asclepiadea), a pattern also mirrored by the geographic distributions of these species (Appendix V).
Finally, the three sections differed strongly in their rates of climatic niche parameter evolution. Despite incongruences between plastid and rDNA data, patterns of evolutionary rates (σ2) were broadly consistent: Rates were comparatively low for all assessed climatic variables in G. sect. Gentiana and G. sect. Calathianae with the notable exception of elevated niche evolution rate of some precipitation-related variables (Bio8: Mean temperature of the wettest quarter, Bio12: Annual precipitation, Bio15: Precipitation seasonality) in G. sect. Calathianae. Gentiana sect. Ciminalis displayed comparatively fast rates of evolution for all remaining bioclimatic variables, resulting in overall high evolution rates along PC2 (Fig. 4, Appendix V).
Maximum a posteriori density values for estimated evolutionary rates (σ2) for nine selected WorldClim variables and the first two principal components of each of the three studied European sections of Gentiana. Based on the rDNA and plastid topologies, rates were modeled separately for each section under a Brownian motion model in R. Tiles are color coded (lower rates are indicated by darker colors, higher rates are indicated by lighter colors). Full table available in Appendix V
Discussion
By combining molecular data with morphological and ecological information, as well as occurrence, climate and karyological data, we attempted to assess the importance of various correlates of diversification in three closely related European sections of Gentiana. Overall, we find that the importance of these correlates may vary through time and among sections, clades and closely related species. However, incongruences between the plastid and rDNA trees weaken our ability to identify precise sequences of events in the diversification of European gentians. Nevertheless, some of the recovered patterns were congruent between analyses, allowing us to draw some general conclusions, and ultimately the occurrence of incongruences suggests new lines of investigation.
Phylogenetic incongruences
Incongruences between rDNA and plastid phylogenies have been recurrent in studies tackling European gentians since the early time of sequencing. Of course, many of these incongruences were not detected as such in some earlier studies because only one genetic compartment was considered (e.g., Yuan et al. 1996; Gielly and Taberlet 1996; Mishiba et al. 2009), or because of the relatively low support recovered for crucial nodes (e.g., Rossi 2012). This latter aspect allowed several studies to concatenate plastid and nuclear sequence data, which now, in the light of considerably larger datasets, appears to have been of little justification (e.g., Favre et al. 2010). For example, we find that the relationships among the three sections vary depending on whether the plastid or the rDNA is considered (Figs. 1, 2 and Appendix II). Although containing only about half of the European species, phylogenetic relationships based on 294 single-copy nuclear genes (Favre et al. 2020) and the rDNA were identical among European sections. Even older phylogenetic reconstruction based only on ITS1 and ITS2 (Yuan et al. 1996) suggested the same topology, yet with relatively low support. Hence, it appears that regardless of the amount of data and the concerted evolution of this repetitive nuclear region, rDNA proved to accurately mirror nuclear evolution.
Furthermore, incongruences were observed within each of the three sections, and again, some of them were already suggested in earlier studies, however with lower or no support. For example in G. sect. Calathianae, phylogenetic relationships varied due to the different placement of G. terglouensis. Our results are thus identical to those of Haemmerli (2007), with G. terglouensis being the sister species of G. bavarica when plastid data are used, and sister to the entire G. verna clade with nuclear markers. In addition, in G. sect. Ciminalis, two contrasting topologies were found, including a split deeply dividing the section in two clades in the plastid phylogeny which is absent from the rDNA phylogeny. In the plastid tree, we recover a clustering of species similar to Christe et al. (2014) and Hungerer and Kadereit (1998), and although RAPD and ITS phylogenies in Hungerer and Kadereit (1998) differ slightly from ours, comparisons are rendered difficult due to a lack of support in this earlier study. Finally, G. sect. Gentiana was only poorly investigated so far (but see Rossi 2012), and our study does provide a new account of phylogenetic relationships among its species, including incongruences as found in other sections. In a nutshell, we find that G. asclepiadea and G. froelichii cluster together only in the plastid tree, and that the phylogenetic relationships among the other (yellow or red flowering) species vary between the trees, although with relatively low support. In summary, rDNA and plastid DNA tell slightly different stories about the evolution of European gentians, rendering the identification of correlates of drivers of diversification more challenging. Nevertheless, in the following, we will show that some patterns persist independently from the topological differences, or discuss how these incongruences may affect our interpretations about correlates of diversification.
Colonization of Europe and early divergence among European sections
Regardless of the topology, we found that the common ancestor of the three sections colonized Europe sometime between the Late Oligocene and the Early Miocene (as in Favre et al. 2016), and most likely, this perennial plant was calcicolous and characterized by campanulate flowers arranged in cymes. At this point in time, the nascent Pyrenees and European Alps underwent rapid exhumation (Curry et al. 2019; Steck 2008), the latter attaining ca. 2500 m by the Early Miocene in their central part (Kuhlemann et al. 2007). This is consistent with the study of Schlunegger and Kissling (2007) who inferred high alpine elevations already at 25 Ma, and others (e.g., van Hinsbergen et al. 2020). As for all mountain systems of the world, some contradicting scenarios for the timing of the uplift exist for European mountains (e.g., Botsyun et al. 2020 and references therein). Yet, despite geological uncertainty, the immigration of pre-adapted lineages such as Gentiana may indicate the existence of high alpine vegetation early on in their uplift history. The establishment of alpine vegetation in the Tibeto-Himalayan region, where Gentiana originated (Favre et al. 2016), was dated to the Early Oligocene (Ding et al. 2020), indicating that the evolution of this genus almost completely overlaps with the uplift of major European and Asian mountain systems. Whether or not geological processes had an active effect on diversification in Europe is debatable because the Alps represent a small orogen in comparison to the much larger and more topographically complex THR and Andes. Besides, because of their more northern location, recent climate oscillations had a stronger impact in European mountain ranges (Muellner-Riehl et al. 2019), likely causing more extinction.
After the colonization of Europe, the three sections diverged from each other (Fig. 1) and relatively rapidly evolved into morphologically distinct entities. Indeed, each section was already characterized by a unique combination of reproductive traits in the Mid Miocene. The sequence of morphological changes slightly differs whether the rDNA or the plastid tree is used for reconstructions. For example, one single shift from multi-flowered cymes to single terminal flowers was detected for the common ancestor of G. sect. Calathianae and G. sect. Ciminalis when these are depicted as sister sections in the rDNA phylogeny, whereas two shifts were recovered (for G. sect. Ciminalis and the G. verna clade of G. sect. Calathianae, respectively) with the plastid tree. With both trees though, a shift from campanulate to funnelform flowers was recovered for G. sect. Calathianae. This shift is significant, as it was probably correlated with a switch in pollinators (bombusophilous to lepidophilous), which may have acted as a driver. After the Mid Miocene, only small morphological modifications occurred, except for the evolution of deeply lobed corollas in G. lutea. Even then, this morphology is not unique to G. lutea, and might constitute a reversal when the broader evolution of Gentianaceae is taken into account. Indeed, a large number of distantly related species do present this type of corollas (e.g., in Swertia).
Chromosome and genome size evolution
Polyploids are known to be more frequent in alpine and Arctic areas (Brochmann et al. 2004). Although Gentiana is markedly alpine, we only detected a single polyploid count in our chromosome number review (G. utriculosa, Table 3, Appendix IIIb), and we also did not detect any polyploid individuals based on flow cytometric screening. This indicates that polyploidization was probably uncommon during the evolution of European gentians, and thus did not act as a correlate or driver of their diversification. However, for their early evolution, we found slightly contrasting results in ancestral reconstructions of chromosome numbers, depending on the phylogeny used. For rDNA tree-based reconstruction (Fig. 2, Appendix IIIa), high haploid numbers (n = 22, 23) were recovered for the common ancestor, whereas smaller ones (n = 18, 19) were reconstructed along the plastid tree. Taking into account the phylogenetic distribution of chromosome numbers in extant species, the former result would inevitably indicate a trend to chromosomal reduction, descending dysploidy and/or aneuploidy (Fig. 2, Appendix IIIa). In contrast, the plastid tree reconstruction suggests a mix of chromosome loss, gains and stasis throughout the evolution of European gentians. If the ancestral reconstruction based on the rDNA tree was to be preferred, our results would allow further speculating that descending dysploidy followed an event of whole genome duplication (WGD) before the divergence of the three sections. An ancient polyploid origin of the entire clade is also supported by early karyological studies (reviewed in Meľnyk et al. 2014) suggesting that several species are in fact natural tetraploids (e.g., G. verna, 2n = 4x = 28; Favarger 1952; Mueller 1982) or even octaploids in G. sect. Gentiana (2n = 8x = 40; Favarger 1949; Skalinska 1951). Some authors even tentatively recognized that most Gentianaceae are natural polyploids, although based only upon a small fraction of species (Rork 1949). Ancient polyploidization events (Oligocene) could have provided the ecological flexibility necessary for the lineage to escape the THR and colonize Europe. Also, dysploidy is known to have followed pulses of polyploidization (Levin 2020), which occurred 60 and 65 million years ago (e.g., van de Peer et al. 2017) and in more recent periods, including the Paleocene–Eocene (e.g., Cai et al. 2019) and the Miocene (e.g., Estep et al. 2014). Nevertheless, it is difficult to attest whether the timing of hypothetical polyploidization in this Gentiana lineage corresponds to either of these waves since 95% HPD intervals span over millions of years (Fig. 1), but ours and previous (Favre et al. 2016) molecular dating indicate the branch along which polyploidization may have occurred could be dated from Oligocene to middle Miocene. Investigating the Asian lineages most closely related to the European gentians could allow selecting one of these two scenarios as the most likely. Unfortunately, no chromosome counts or genomes sizes are yet available for G. phyllocalyx, the only member of the sister lineage of European gentians (e.g., see Favre et al. 2020). Alternatively, an in-depth investigation of paralogous genes may allow identifying and dating ancient polyploidization events, similarly to those detected at the Cretaceous–Tertiary boundary (Fawcett et al. 2009).
In any case, all our results suggest that the differentiation among sections was accompanied by chromosomal processes. As in several other alpine taxa such as Phyteuma (Schneeweiss et al. 2013), and Saxifraga (Mas de Xaxars et al. 2015; Ebersbach et al. 2020), dysploidy and aneuploidy have most likely contributed to the diversification of Gentiana. Indeed, chromosomal variation was observed in G. sect. Gentiana and G. sect. Calathianae, but not in G. sect. Ciminalis (Fig. 2). In this section, however, RGS varied considerably among species (between 0.85 in G. occidentalis to 1.18 in G. clusii, Appendix IV) despite an overall stable chromosome number (2n = 36). Surprisingly, the reconstruction for the crown node of G. sect. Ciminalis was not unequivocally n = 18 because n = 19 was only slightly less probable regardless of the tree used for reconstruction. At least for the plastid tree, we could argue that this result is due to the ChromEvol constraints towards branch length combined with the presence of the deep split in the topology of G. sect. Ciminalis. In contrast, both plastid and rDNA trees indicate a descending dysploidy or aneuploidy in G. sect. Gentiana. This echoes the result of an early karyological study in the related section Chondrophyllae, where rapid karyotypic evolution probably happened by a combination of polyploidy followed by descending dysploidy (Kuepfer and Yuan 1996; Yuan et al. 1998), and was proposed for G. terglouensis as well (Mueller 1982). Early diverging G. asclepiadea (2n = 44) and G. froelichii (2n = 42) possess more chromosomes than other species of G. sect. Gentiana (2n = 40), but lower RGS (0.53–0.67 vs 0.71–0.74) suggesting ascending aneuploidy as a mechanism. This was not the case of the yellow or red flowering species of G. sect. Gentiana (2n = 20), of which RGS is conserved (0.70–0.74, Appendix IV). Finally in G. sect. Calathianae, the clade formed by G. nivalis (2n = 14), G. pumila (2n = 20), and G. utriculosa (2n = 22) generally had less chromosomes than its sister clade (the G. verna group: ca. 2n = 28–42), but comparable RGS (0.39–0.53 vs 0.41–0.42, respectively). This result would further support descending dysploidy as a general mechanism in the section, although for G. nivalis (2n = 14, RGS 0.28), descending aneuploidy and/or significant deletions seems to be more probable. However, descending dysploidy or aneuploidy is only clearly suggested by the rDNA-cistron tree in G. sect. Calathianae, the plastid tree supporting a relative stasis from the sectional crown node throughout the clade containing G. verna. Thus, conflicting phylogenetic relationships (and a lack of RGS data for G. rostanii) render the interpretation of chromosomal evolution within the section more challenging, but the presence of several counts indicating aneuploidy or the presence of B chromosomes (Appendix IIIb) might indicate active chromosomal rearrangements or recent hybrid origin (Jones et al. 2008). For example, in G. verna ssp. tergestina, additional B chromosomes were reported (Appendix IIIb), and a higher RGS (0.53) was found in comparison to nominal subspecies (0.47 ± 0.01), similarly as for G. verna AFAT19003b (RGS 0.51) for which additional chromosomes might be assumed. However, to avoid the bias resulting from AT-specific staining using DAPI, these issues need to be addressed further by absolute genome size using intercalating fluorochromes in follow-up studies.
Climate
We also found that the three sections differ in terms of their climatic niche preferences, with all sections exhibiting subtle but significant differentiation along the first principal component in addition to differentiation of G. sect. Calathianae along PC2. However, the overall climatic niche space occupied by each section was very similar (Appendix V). Since the three sections do not show major differences in their distributions among the assessed mountain systems, this suggests that all three sections were able to effectively colonize the available climatic niche space in these mountains. In parallel, we uncovered that the three Gentiana sections are characterized by divergent niche evolution rates (Fig. 4), with G. sect. Ciminalis displaying relatively high rates for almost all bioclimatic variables. This all perennial section being the most uniform morphologically and karyologically, we expected some degree of climatic differentiation, on top of shifts in bedrock preferences (see below; Christe et al. 2014).
Regardless of topological incongruences, each section revealed at least one Miocene divergence event, and these events almost always seem to have been associated with strong differentiation of climatic niche preferences and elevational distribution ranges (Table 4, Appendix V). For example, according to the plastid tree, two clades of G. sect. Calathianae (clade 1: G. bavarica, G. terglouensis, G. brachyphylla, G. rostanii; clade 2: G. verna, G. schleicheri) were well-separated climatically and in terms of elevation (mean difference: 419.2 m asl) and these two clades were in turn slightly but significantly differentiated from clade 3 (G. nivalis, G. pumila, G. utriculosa). Even though the rDNA topology is slightly different for this section, Miocene divergence events still show the same pattern. One could speculate that during the Miocene cooling, the treeline would have progressively dropped, expanding the original suitable area for alpine plants, and possibly producing more extreme alpine habitats. Lineages in each section could have possibly taken advantage of this situation by expanding their climatic niche space towards more extreme conditions, although we did not formally test this scenario. Similarly, colonization of new habitats or areas could have been followed by climatic niche evolution in the newly emerging species. In any case, these divergence events likely laid the foundation for later niche differentiation and also underscore the crucial role of Miocene in evolution of European Gentiana. As mentioned above, the three sections also differed widely in their overall rates of niche evolution. The ability to adapt more quickly to varying climatic conditions via fast niche evolution would have been most advantageous during periods of rapid climatic changes, such as Pleistocene climatic oscillations which imposed particularly strong selective pressures in the European Alpine System. Following this line of arguments, climatic niche evolution would not only have been an important correlate during the early and intermediate evolution of the studied Gentiana sections, but also played a particular role during more recent evolutionary processes. Moreover, judging by their extant distribution ranges, most of these intrasectional clades show substantial geographic overlap (values ranging from 0.58 to 0.79, except for G. section. Gentiana: 0.49), suggesting that Miocene divergence was more strongly driven by niche evolution and other factors such as shifts in life cycles in G. sect. Calathianae (Fig. 1, Appendix V) than by vicariance. However, extant clade-wise distribution ranges might not reflect the past distribution. Indeed, intense displacement and strong floristic exchange among European mountain ranges have occurred. Also, our comparison of distribution range does not consider extinction events likely, making it most relevant only for more recent times.
Recent diversification was accompanied by climatic and correspondingly, elevational differentiation in all three studied sections (Table 4). At the same time, closely related species and species groups often contained one species with relatively broad climatic tolerances and one with relatively restricted ones. Even though mean climatic preferences showed significant differences in these comparisons, overall niche occupancies were often broadly overlapping, with the more restricted niche tolerances often being a subset of the broader niche (e.g., G. burseri and G. lutea, G. brachyphylla and G. rostanii, Appendix V). Although differences in the degrees of range overlap were small overall, the closely related species of G. sect. Ciminalis which displayed the highest rates of climatic niche evolution were characterized by the lowest degree of geographic overlap (Table 3). Taken together, these results indicate that fast niche evolution was associated with geographical vicariance, at least during the more recent evolution of this section. However, climatic preferences and niche evolution rates presented here are all based on the global Bioclim dataset of ca. 1 km2 resolution. At high elevation, alpine environments are known for their heterogeneity in microclimatic conditions (Körner 2011), thus higher resolution rasters of relevant climatic and environmental conditions, as well as formal species distribution modeling will be needed to fully unravel the role of niche divergence within the sections.
Evolution of soil preferences
Starting from this calcicolous common ancestor (regardless of the tree topology), we found several shifts or expansions from calcareous to siliceous bedrock preferences in every section. Overall, shifts of soil preferences do not appear to be associated with other attributes we investigated, including chromosome numbers, morphology or life cycle. Repeated patterns of vicariant speciation on adjacent substrate types may result in clades characterized by a high substrate diversity, as for example in the Amazonia (Fine et al. 2005) and the Cape (Cowling et al. 2009; Schnitzler et al. 2011). This phenomenon is also well known in the Alps for a long time (e.g., Unger 1836; Landolt 1960; Aeschimann et al. 2004), although it was rarely shown within a phylogenetic context (but see e.g., Moore and Kadereit 2013). This is surprising since repeated adaptation to contrasting soil conditions is likely to have been the driver for the obvious divide between the calcicolous and silicolous flora in the European Alpine System.
Most of these shifts were dated within the last few million years (Pliocene to present), regardless of phylogenetic trees used for reconstructions (Fig. 1). This may appear surprising because in the presence of both types of bedrocks, shifts could have also occurred early in the evolution of European gentians. First of all, shifts to silicolous ecology may have been rare in the past simply because of uplift dynamics in the European Alpine System. Indeed, it is to be expected that prior to the uplift, siliceous rocks were buried below thick sedimentary layers (mostly calcareous), and were exposed only as the uplift and erosion progressed. This aspect is however not treated in the geological literature. Another explanation is that extinction was more severe for old silicolous lineages through time than for calcicolous ones. Although speciation and diversification based on substrate specialization have been detected for several taxa in the Alps, it does not mean that differentiation occurred within the range, as shown in Minuartia (Moore and Kadereit 2013). Indeed, the survival of silicolous species through cold periods and their current occupation of the Alps and the Pyrenees is not trivial. Siliceous bedrocks being concentrated within the mountain ranges, which were under the ice during the LGM, silicolous species had much fewer opportunities than calcicolous ones to survive on nunataks in situ (Stehlik 2000; Parisod and Besnard 2007; Schneeweiss and Schönswetter 2011; Pan et al. 2020) or in the nearby foreland (e.g., Schönswetter et al. 2002; Kropf et al. 2003; Windmaißer et al. 2016). As shown by Christe et al. (2014), G. alpina and G. acaulis (silicolous, G. sect. Ciminalis) bear all the marks of glacial survival in distant refugia (i.e., Iberian Peninsula; Christe et al. 2014) followed by a rapid expansion, whereas calicolous species (e.g., G. clusii) show stronger patterns of local differentiation suggesting survival in a series of isolated but closeby refugia during the LGM. This pattern is not only observed in Gentiana (see Smyčka et al. 2018 and references therein), so the likelihood to successfully disperse back and forth from glacial refugia to mountain ranges may thus have been asymmetrically dependent on soil preferences, an aspect still relatively poorly understood in all mountain systems of the world. Such asymmetry in dispersal potential may have caused disproportionate extinction in silicolous through time, resulting in the apparent domination of limestone species throughout the evolution of European gentians.
As for other correlates of diversification, phylogenetic incongruences partly complicate describing the patterns of ecological shifts in European gentians. Nevertheless, it is clear that the majority of shifts to silicolous ecology occurred at terminal branches in both plastid and rDNA trees (e.g., for G. acaulis and G. alpina), meaning there was only one case of speciation within siliceous bedrocks, in G. sect. Gentiana. In this section, and only visible from the rDNA tree, three species evolved after a shift to silicolous ecology, namely G. punctata, G. purpurea and G. burseri. In comparison, these three species do not cluster together in the plastid phylogeny, but it is worth mentioning that several nodes in this part of the phylogeny are not unequivocally supported. In this case, not only the rDNA tree appears slightly more reliable, but also known genes responsible for adaptation to different soils and bedrocks are nuclear. For instance, ion transport SULTR1;1 and GLR2.5 genes are involved in the divergence of calcicolous and silicolous ecotypes in Arabidopsis lyrata (Guggisberg et al. 2018), or MRS2/MGT gene family probably allowed Microthlaspi erraticum to cope with highly calcareous soils (Mishra et al. 2020). How many genes are actually involved in bedrock-related shifts is unknown, as well as whether the genetic background of adaptation differs among shifts in closely related species. In addition, whether these adaptive genes may cross species borders via hybridization and adaptive introgression remains also unknown.
The ghost of hybridization
It is clear that hybridization has been a major player in the diversification of several species-rich mountain taxa in Europe, as for example in Saxifraga (Ebersbach et al. 2020) or Potentilla (Paule et al. 2012). However, at present, little is known about the history and frequency of hybridization in Gentiana and how it relates to adaptation to different bedrocks and soils. The incongruences highlighted in our study may in fact derive from hybridization and gene flow, and if so, then hybridization occurred in most lineages of European gentians, starting as early as the divergence of the three sections. Further evidence supports the occurrence of hybridization and/or gene flow in Gentiana. For example, in G. sect. Ciminalis, Christe et al. (2014) interpreted the lesser power of plastid markers in resolving species delineation as a sign of chloroplast capture. In addition, based on both biparentally and maternally inherited markers, Haemmerli (2007) suggested that the high chromosome number of G. terglouensis may be of a hybrid origin between the clades named subsect. Calathianae (incl. G. nivalis) and Vernae (incl. G. verna). This would explain the different phylogenetic position of this species in the plastid and rDNA trees (see Appendix II), and is supported by rather variable chromosome numbers and RGS in G. sect. Calathianae. Furthermore, several natural hybrids have been detected in the wild in all sections, as for example in G. sect. Gentiana between G. lutea and other species such as G. burseri, G. punctata and G. purpurea (Anchisi et al. 2010). Also, beyond Europe, some studies have revealed rare cases of hybrid speciation (Fu et al. 2021b). Therefore, to complete the picture of correlates of diversification, additional investigations on the role of hybridization in European Gentiana need to be performed, for example using population genomic approaches and demographic reconstructions.
Conclusions
Using an integrative approach and despite phylogenetic incongruences, our study provides an overview of potential correlates likely involved in the diversification of three closely related sections of the alpine genus Gentiana in Europe. Once early morphological differentiation was established, some correlates appear to have been pivotal throughout the evolution of this group (e.g., the evolution of climate preferences, dysploidy), the effect of others appearing to have been more restricted in time (e.g., bedrock preferences, shifts in the life cycle). By partially unfolding this sequence of events in Gentiana, our study sets the base for investigations on the genomic mechanisms supporting soil adaptation and climatic niche evolution and their contribution to speciation. Due to multiple phylogenetic incongruences throughout the topology, as well as other lines of evidence, investigating genomic mechanisms of adaptation will most likely have to be set within a context of recurrent hybridization. Finally, because of the northern location of the European Alpine System and their relatively small size, geological processes may be less relevant for diversification than they are in other highly diverse mountain systems such as the THR or the Andes. Nevertheless, we argue that the relative importance of biotic factors or drivers (e.g., bedrock preferences) are widely underestimated in major tropical and subtropical mountain systems of the world, and should be investigated in more detail.
Availability of data and materials
Molecular data are available online, and occurrence data are provided as appendix.
Code availability
Not applicable.
References
Aeschimann D, Lauber K, Moser DM, Theurillat JP (2004) Flora alpina, vol 2. Haupt Verlag, Bern
Anchisi E, Bernini A, Cartasegna N, Polani F (2010) Genziane d’Europa. Verba & Scripta, Pavia
Bartók A, Hurdu BI, Szatmari PM (2015) Distribution of endangered Gentiana clusii EM Perrier & Songeon in the Romanian Carpathians—a critical overview. Contrib Bot 50:15–32
Botsyun S, Ehlers TA, Mutz SG, Methner K, Krsnik E, Mulch A (2020) Opportunities and challenges for paleoaltimetry in “small” orogens: Insights from the European Alps. Geophys Res Lett 47:e2019GL086046
Brochmann C, Brysting A, Alsos IG, Borgen L, Grundt HH, Scheen AC, Elven R (2004) Polyploidy in arctic plants. Biol J Linn Soc 82:521–536
Burnier J, Buerki S, Arrigo N, Kuepfer P, Alvarez N (2009) Genetic structure and evolution of alpine polyploid complexes: Ranunculus kuepferi (Ranunculaceae) as a case study. Mol Ecol 18:3730–3744
Cai L, Xi Z, Amorim AM, Sugumaran M, Rest JS, Liu L, Davis C (2019) Widespread ancient whole-genome duplications in Malpighiales coincide with Eocene global climatic upheaval. New Phytol 221:565–576
Chesser RT, Zink RM (1994) Modes of speciation in birds: a test of Lynch’s method. Evolution 48:490–497
Christe C, Caetano S, Aeschimann D, Kropf M, Diadema K, Naciri Y (2014) The intraspecific genetic variability of siliceous and calcareous Gentiana species is shaped by contrasting demographic and re-colonization processes. Mol Phylogenet Evol 70:323–336
Cowling RM, Procheş S, Partridge TC (2009) Explaining the uniqueness of the Cape Flora: incorporating geomorphic evolution as a factor for explaining its diversification. Mol Phylogenet Evol 51:64–74
Curry ME, van der Beek P, Huismans RS, Wolf SG, Muñoz JA (2019) Evolving paleotopography and lithospheric flexure of the Pyrenean Orogen from 3D flexural modeling and basin analysis. Earth Planet Sci Lett 515:26–37
Davitashvili N, Karrer G (2010) Taxonomic importance of seed morphology in Gentiana (Gentianaceae). Bot J Linn Soc 162:101–115
deVos JM, Hughes CE, Schneeweiss GM, Moore BR, Conti E (2014) Heterostyly accelerates diversification via reduced extinction in primroses. Proc R Soc B 281:20140075
Ding WN, Ree RH, Spicer RA, Xing YW (2020) Ancient orogenic and monsoon-driven assembly of the world’s richest temperate alpine flora. Science 369:578–581
Doležel J, Doleželová M, Novák FJ (1994) Flow cytometric estimation of nuclear DNA amount in diploid bananas (Musa acuminata and M. balbisiana). Biol Plant 36:351–357
Doležel J, Greilhuber J, Lucretti S, Meister A, Lysák MA, Nardi L, Obermayer R (1998) Plant genome size estimation by flow cytometry: Inter-laboratory comparison. Ann Bot 82:17–26
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214
Ebersbach J, Schnitzler J, Favre A, Muellner-Riehl AN (2017) Evolutionary radiations of the species-rich mountain genus Saxifraga L. BMC Evol Biol 17:119
Ebersbach J, Tkach N, Röser M, Favre A (2020) The role of hybridisation in the making of the species-rich arctic-alpine genus Saxifraga (Saxifragaceae). Diversity 12:440
Edler D, Klein J, Antonelli A, Silvestro D (2019) raxmlGUI 2.0 beta: a graphical interface and toolkit for phylogenetic analyses using RAxML. bioRxiv. https://doi.org/10.1101/800912
Escudero M, Hipp AL, Waterway MJ, Valente LM (2012) Diversification rates and chromosome evolution in the most diverse angiosperm genus of the temperate zone (Carex, Cyperaceae). Mol Phylogenet Evol 63:650–655
Estep MC, McKain MR, Diaz DV, Zhong JS et al (2014) Allopolyploidy, diversification, and the Miocene grassland expansion. Proc Natl Acad Sci USA 111:15149–15154
Favarger C (1949) Contribution à l’étude caryologique et biologique des Gentianacées. Bull Soc Bot Suisse 59:62–86
Favarger C (1952) Contribution à l’étude caryologique et biologique des Gentianacées. II. Bull Soc Bot Suisse 62:244–257
Favre A, Yuan YM, Kuepfer P, Alvarez N (2010) Phylogeny of subtribe Gentianinae (Gentianaceae): biogeographic inferences despite limitations in temporal calibration points. Taxon 59:1701–1711
Favre A, Matuszak S, Sun H, Liu E, Yuan YM, Muellner-Riehl AN (2014) Two new genera of Gentianinae (Gentianaceae): Sinogentiana and Kuepferia supported by molecular phylogenetic evidence. Taxon 63:342–354
Favre A, Päckert M, Pauls SU, Jaehnig SC, Uhl D, Michalak I, Muellner-Riehl AN (2015) The role of the uplift of the Qinghai-Tibetan Plateau for the evolution of Tibetan biotas. Biol Rev 90:236–253
Favre A, Michalak I, Chen CH, Wang JC, Pringle JS, Matuszak S, Sun H, Yuan YM, Struwe L, Muellner-Riehl AN (2016) Out-of-Tibet: the spatio-temporal evolution of Gentiana (Gentianaceae). J Biogeogr 43:1967–1978
Favre A, Pringle JS, Heckenhauer J, Kozuharova E, Gao QB, Moriarty Lemmon E, Lemmon AR, Sun H, Tkach N, Gebauer S, Sun SS, Fu PC (2020) Phylogenetic relationships and sectional delineation within Gentiana (Gentianaceae). Taxon 69:1221–1238
Fawcett JA, Maere S, Van de Peer Y (2009) Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. PNAS 106:5737–5742
Felsenstein J (1985) Phylogenies and the comparative method. Am Nat 125:1–15
Fick SE, Hijmans RJ (2017) WorldClim 2: new 1km spatial resolution climate surfaces for global land areas. Int J Climatol 37:4302–4315
Fine PVA, Daly DC, Villa Muñoz G, Mesones I, Cameron KM (2005) The contribution of edaphic heterogeneity to the evolution and diversity of Burseraceae trees in the Western Amazon. Evolution 59:1464–1478
Fu PC, Sun SS, Twyford AD, Li BB, Zhou RQ, Chen SL, Gao QB, Favre A (2021a) Lineage-specific plastid degradation in subtribe Gentianinae (Gentianaceae). Ecol Evol. https://doi.org/10.1002/ece3.7281
Fu PC, Twyford AD, Sun SS, Wang HY, Xia MZ, Tan CX, Zhou XJ, Chen SL (2021b) Recurrent hybridization underlies the evolution of novelty in Gentiana (Gentianaceae) in the Qinghai-Tibetan Plateau. AoB Plants 13:plaa068
Gehrke B, Kandziora M, Pirie MD (2016) The evolution of dwarf shrubs in alpine environments: a case study of Alchemilla in Africa. Ann Bot 117:121–131
Gielly L, Taberlet P (1996) A phylogeny of the European gentians inferred from chloroplast trnL (UAA) intron sequences. Bot J Linn Soc 120:57–75
Glick L, Mayrose I (2014) ChromEvol: assessing the pattern of chromosome number evolution and the inference of polyploidy along a phylogeny. Mol Biol Evol 31:1914–1922
Gorelick N, Hancher M, Dixon M, Ilyushchenko S, Thau D, Moore R (2017) Google earth engine: planetary-scale geospatial analysis for everyone. Remote Sens Environ 202:18–27
Guggisberg A, Liu X, Suter L, Mansion G, Fischer MC, Fior S, Roumet M, Kretzschmar R, Koch MA, Widmer A (2018) The genomic basis of adaptation to calcareous and siliceous soils in Arabidopsis lyrata. Mol Ecol 27:5088–5103
Haemmerli M (2007) Molecular aspects in systematics of Gentiana sect. Calathianae Froel. Université de Neuchâtel, Th. no 1969
Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W (2008) GEIGER: investigating evolutionary radiations. Bioinformatics 24:129–131. https://doi.org/10.1093/bioinformatics/btm538
Hijmans RJ, van Etten J (2012) raster: Geographic analysis and modeling with raster data. R package version 2.0-12. http://CRAN.R-project.org/package=raster
Ho TN, Liu SW (1990) The infrageneric classification of Gentiana (Gentianaceae). Bull Br Mus Nat Hist Bot 20:169–192
Hoorn C, Mosbrugger V, Mulch A, Antonelli A (2013) Biodiversity from mountain building. Nat Geosci 6:154
Hungerer KB, Kadereit JW (1998) The phylogeny and biogeography of Gentiana L. sect. Ciminalis (Adans.) Dumort.: a historical interpretation of distribution ranges in the European high mountains. Perspect Plant Ecol Evol Syst 1:121–135
Janssens SB, Couvreur TLP, Mertens A, Dauby G, Dagallier LMJ, Abeele SV et al (2020) A large-scale species level dated angiosperm phylogeny for evolutionary and ecological analyses. Biodivers Data J 8:e39677
Jones RN, Viegas W, Houben A (2008) A century of B chromosomes in plants: so what? Ann Bot 101:767–775
Kadereit JW, Griebeler EM, Comes HP (2004) Quaternary diversification in European alpine plants: pattern and process. Phil Trans R Soc Lond B 359:265–274
Kassambara A, Mund F (2017) Package ‘factoextra’. Extract and visualize the results of multivariate data analyses. R package version 1.0.7. http://CRAN.R-project.org/package=factoextra
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S et al (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Kier G, Mutke J, Dinerstein E, Ricketts TH, Küper W, Kreft H, Barthlott W (2005) Global patterns of plant diversity and floristic knowledge. J Biogeogr 32:1107–1116
Körner C (2011) Coldest places on earth with angiosperm plant life. Alp Bot 121:11–22
Kosiński P, Sękiewicz K, Walas Ł, Boratyński A, Dering M (2019) Spatial genetic structure of the endemic alpine plant Salix serpillifolia: Genetic swamping on nunataks due to secondary colonization? Alp Bot 129:107–121
Kropf M, Kadereit JW, Comes HP (2003) Differential cycles of range contraction and expansion in European high mountain plants during the Late Quaternary: insights from Pritzelago alpina (L.) O. Kunthe (Brassicaceae). Mol Ecol 12:931–949
Kuepfer P, Yuan YM (1996) Karyological studies on Gentiana sect.Chondrophyllae (Gentianaceae) from China. Plant Syst Evol 200:161–176
Kuhlemann J (2007) Paleogeographic and paleotopographic evolution of the Swiss and Eastern Alps since the Oligocene. Glob Planet Change 58:224–236
Lagomarsino LP, Condamine FL, Antonelli A, Mulch A, Davis CC (2016) The abiotic and biotic drivers of rapid diversification in Andean bellflowers (Campanulaceae). New Phytol 210:1430–1442
Landolt E (1960) Unsere Alpenflora. Schweizer Alpen Club, Zurich
Lauber K, Wagner G, Gygax A (2012) Flora Helvetica. Haupt Verlag, Bern
Lemmon AR, Emme SA, Lemmon EM (2012) Anchored hybrid enrichment for massively high-throughput phylogenomics. Syst Biol 61:727–744
Levin DA (2020) Did dysploid waves follow the pulses of whole genome duplications? Plant Syst Evol 306:75
Luebert F, Weigend M (2014) Phylogenetic insights into Andean plant diversification. Front Ecol Evol 2:27
Maddison WP, Maddison DR (2011) Mesquite: a modular system for evolutionary analysis version 2.75. http://www.mesquiteproject.org
Madriñán S, Cortés AJ, Richardson JE (2013) Páramo is the world’s fastest evolving and coolest biodiversity hotspot. Front Genet 4:192
Mai DH (2000) Die mittelmiozänen und obermiozänen Floren aus der Meuroer und Raunoer Folge in der Lausitz. Teil I: Farnpflanzen, Koniferen und Monokotyledonen. Palaeontogr Abt B,pälaophyto 256:1–68
Mai DH, Walther H (1988) Die pliozänen Floren von Thüringen, Deutsche Demokratische Republik. Quartärpaläontologie 7:55–297
Mas De Xaxars G, García-Fernánder A, Barnola P, Martín J, Mercandé A, Vallès J, Vargas P, Vigo J, Garnatje T (2015) Phylogenetic and cytogenetic studies reveal hybrid speciation in Saxifraga subsect. Triplinervium (Saxifragaceae). J Syst Evol 53:53–62
Matuszak S, Favre A, Schnitzler J, Muellner-Riehl AN (2016) Key innovations and climatic niche divergence as drivers of diversification in subtropical Gentianinae (Gentianaceae) in the region of the Qinghai-Tibetan Plateau. Am J Bot 103:899–911
Mel’nyk VM, Drobyk NM, Twardovska MO, Kunakh VA (2014) Karyology of European Species of Genus Gentiana L. In: Rybczyński J, Davey M, Mikuła A (eds) The Gentianaceae—Volume 1: Characterization and Ecology. Springer, Berlin, Heidelberg
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov 2010, New Orleans, pp 1–8
Mishiba K, Yamane K, Nakatsuka T, Nakano Y, Yamamura S, Abe J, Kawamura H, Takahata Y, Nishihara M (2009) Genetic relationships in the genus Gentiana based on chloroplast DNA sequence data and nuclear DNA content. Breed Sci 59:119–127
Mishra B, Ploch S, Runge F, Schmuker A, Xia X, Gupta DK, Sharma R, Thines M (2020) The Genome of Microthlaspi erraticum (Brassicaceae) Provides insights into the adaptation to highly calcareous soils. Front Plant Sci 11:943
Moore JM, Kadereit JW (2013) The evolution of substrate differentiation in Minuartia series Laricifoliae (Caryophyllaceae) in the European Alps: In situ origin or repeated colonization? Am J Bot 100:2412–2425
Mosbrugger V, Favre A, Muellner-Riehl AN, Päckert M, Mulch A (2018) Cenozoic evolution of geo-biodiversity in the Tibeto-Himalayan Region. Hoorn C, Perrigio A, Antonelli A (ds), Mountains, climate, and biodiversity. Wiley-Blackwell, Oxford, pp 429–448
Mueller G (1982) Contribution à la cytotaxonomie de la section Cyclostigma Griseb. du genre Gentiana. Feddes Repert 93:625–722
Muellner-Riehl AN (2019) Mountains as evolutionary arenas: patterns, emerging approaches, paradigm shifts, and their implications for plant phylogeographic research in the Tibeto-Himalayan Region. Front Plant Sci 10:195
Muellner-Riehl AN, Schnitzler J, Mosbrugger V, Favre A (2019) Origins of global mountain plant biodiversity: testing the ‘mountain-geobiodiversity hypothesis. J Biogeogr 46:2826–2838
Nieto Feliner G (2011) Southern European glacial refugia: a tale of tales. Taxon 60:365–372
O'Meara BC, Ané C, Sanderson MJ, Wainwright PC (2006) Testing for different rates of continuous trait evolution using likelihood. Evolution 60:922–933
Otto F (1990) DAPI staining of fixed cells for high-resolution flow cytometry of nuclear DNA. Methods Cell Biol 33:105–110
Pan D, Hülber K, Willner W, Schneeweiss GM (2020) An explicit test of Pleistocene survival in peripheral versus nunatak refugia in two high mountain plant species. Mol Ecol 29:172–183
Parisod C, Besnard G (2007) Glacial in situ survival in the Western Alps and polytopic autopolyploidy in Biscutella laevigata L. (Brassicaceae). Mol Ecol 16:2755–2767
Paule J, Scherbantin A, Dobeš C (2012) Implications of hybridsation and cytotypic differentiation in speciation assessed by AFLP and plastid haplotypes—a case study of Potentilla alpicola La Soie. BMC Evol Biol 12:132
Paule J, Dunkel FG, Schmidt M, Gregor T (2018) Climatic differentiation in polyploid apomictic Ranunculus auricomus complex in Europe. BMC Ecol 18:16
R Core Team (2020) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/
Rambaut A (2010) FigTree v1.4.0, available at http://tree.bio.ed.ac.uk/software/figtree/
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542
Roquet C, Boucher FC, Thuiller W, Lavergne S (2013) Replicated radiations of the alpine genus Androsace (Primulaceae) driven by range expansion and convergent key innovations. J Biogeogr 40:1874–1886
Rork CL (1949) Cytological studies in the Gentianaceae. Am J Bot 36:687–701
Rossi M (2012) Taxonomy, phylogeny and reproductive ecology of Gentiana lutea L. [Ph.D. thesis]. Bologna (IT): Alma Mater Studiorum—University of Bologna. http://amsdottorato.cib.unibo.it/4740/
RStudio Team (2020) RStudio: Integrated Development for R. RStudio, PBC, Boston. http://www.rstudio.com/
Saupe EE, Barve N, Owens HL, Cooper JC, Hosner PA, Peterson AT (2018) Reconstructing ecological niche evolution when niches are incompletely characterized. Syst Biol 67:428–438
Schenk JJ, Axel J (2016) Consequences of secondary calibrations on divergence time estimates. PLoS ONE 11:e0148228
Schlunegger F, Rieke-Zapp D, Ramseyer K (2007) Possible environmental effects on the evolution of the Alps-Molasse Basin system. Swiss J Geosci 100:383–405
Schneeweiss GM, Schönswetter P (2011) A re-appraisal of nunatak survival in artic-alpine phylogeography. Mol Ecol 20:190–192
Schneeweiss GM, Pachschwöll C, Tribsch A, Schönswetter P, Barfuss MH, Esfeld K, Weiss-Schneeweiss H, Thiv M (2013) Molecular phylogenetic analyses identify alpine differentiation and dysploid chromosome number changes as major forces for the evolution of the European endemic Phyteuma (Campanulaceae). Mol Phylogenet Evol 69:634–652
Schnitzler J, Barraclough TG, Boatwright JS, Goldblatt P, Manning JC, Powell MP, Rebelo T, Savolainen V (2011) Causes of plant diversification in the Cape biodiversity hotspot of South Africa. Syst Biol 60:343–357
Schönswetter P, Schneeweiss GM (2019) Is the incidence of survival in interior Pleistocene refugia (nunataks) underestimated? Phylogeography of the high mountain plant Androsace alpina (Primulaceae) in the European Alps revisited. Ecol Evol 9:4078–4086
Schönswetter P, Tribsch A, Barfuss M, Niklfeld H (2002) Several Pleistocene refugia detected in the high alpine plant Phyteuma globulariifolium Sternb. & Hoppe (Campanulaceae) in the European Alps. Mol Ecol 11:2637–2647
Schönswetter P, Stehlik I, Holderegger R, Tribsch A (2005) Molecular evidence for glacial refugia of mountain plants in the European Alps. Mol Ecol 14:3547–3555
Sedano RE, Burns KJJ (2010) Are the Northern Andes a species pump for Neotropical birds? Phylogenetics and biogeography of a clade of Neotropical tanagers (Aves: Thraupini). J Biogeogr 37:325–343
Skalińska M (1951) Cytological studies in Gentiana-species from the Tatra and Pieniny Mts. Bull Acad Polon Sci Ser Sci Biol 1:119–137
Smyčka J, Roquet C, Renaud J, Thuiller W, Zimmermann NE, Lavergne S (2018) Disentangling drivers of plant endemism and diversification in the European Alps—a phylogenetic and spatially explicit approach. Perspect Plant Ecol Evol Syst 30:31–40
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Steck A (2008) Tectonics of the Simplon massif and Lepontine gneiss dome: deformation structures due to collision between the underthrusting European plate and the Adriatic indenter. Swiss J Geosci 101:515–546
Stehlik I (2000) Nunataks and peripheral refugia for alpine plants during quaternary glaciation in the middle part of the Alps. Bot Helv 110:25–30
te Beest M, Le Roux JJ, Richardson DM, Brysting AK, Suda J, Kubešová M, Pyšek P (2012) The more the better? The role of polyploidy in facilitating plant invasions. Ann Bot 109:19–45
Tutin TG, Heywood VH, Burges NA, Moore DM, Valentine DH, Walters SM, Webb DA (1964–1980) Flora Europaea 1–5. Cambridge University Press, Cambridge
Unger F (1836) Über den Einfluss des Bodens auf die Vertheilung der Gewächse, nachgewiesen in der Vegetation des nordöstlichen Tirol's. Rohrmann und Schweigerd, Wien
Van de Peer Y, Mizrachi E, Marchal K (2017) The evolutionary significance of polyploidy. Nat Rev Genet 18:411–424
Van Hinsbergen DJJ, Torsvik TH, Schmid SM, Maţenco LC, Maffione M, Vissers RLM, Gürer D, Spakman W (2020) Orogenic architecture of the Mediterranean region and kinematic reconstruction of its tectonic evolution since the Triassic. Gondwana Res 81:79–229
Vilela B, Villalobos F (2015) letsR: a new R package for data handling and analysis in macroecology. Methods Ecol Evol 6:1229–1234. https://besjournals.onlinelibrary.wiley.com/doi/abs/10.1111/2041-210X.12401
Wei T, Simko V (2017) R package "corrplot": Visualization of a correlation matrix. R package version 0.84. https://github.com/taiyun/corrplot
Wickham H (2016) ggplot2: Elegant graphics for data analysis. Springer-Verlag, New York. https://ggplot2.tidyverse.org
Windmaißer T, Kattari S, Heubl G, Reisch C (2016) Glacial refugia and postglacial expansion of the alpine–prealpine plant species Polygala chamaebuxus. Ecol Evol 6:7809–7819
Yuan YM, Kuepfer P (1995) Molecular phylogenetics of the subtribe Gentianinae (Gentianaceae) inferred from the sequences of internal transcribed spacers (ITS) of nuclear ribosomal DNA. Plant Syst Evol 196:207–226
Yuan YM, Kuepfer P, Doyle JJ (1996) Infrageneric phylogeny of the genus Gentiana (Gentianaceae) inferred from nucleotide sequences of the internal transcribed spacers (ITS) of nuclear ribosomal DNA. Am J B 83:641–652
Yuan YM, Kuepfer P, Zeltner L (1998) Chromosomal evolution of Gentiana and Jaeschkea (Gentianaceae), with further documentation of chromosome data for 35 species from western China. Plant Syst Evol 210:231–247
Acknowledgements
We would like to thank Stefan Dressler (FR) and Andreas Fleischmann (M), for allowing access to the herbarium specimens and granting permission to retrieve samples for DNA analysis. We are thankful to the Jardin Gentiana (Leysin, Switzerland) for generously providing samples. Many thanks to Bernhard Pospichal, Susanne Fritz, and Charlène Heiniger for their support during field campaigns. DNA extractions were performed in the Grunelius-Möllgaard Laboratory (GML), Senckenberg Research Institute (Frankfurt am Main), with the motivating support of Heike Kappes. We are thankful to Carola Greve and Damian Baranski at the LOEWE-TBG for their constant help dealing with sequencing and genomic data management. We are thankful to Raluca Enescu, Attila Bartók and Ekaterina Kozuharova who provided additional occurrences for Eastern Europe. Many thanks to Guillaume Dupont-Nivet for supporting us with the geological history of the European mountains. We would like to thank Info Flora—The National Data and Information Center of the Swiss Flora—infoflora.ch, for providing occurrence data. Financial support for this study was provided by the German Science Foundation (Deutsche Forschungsgemeinschaft), project no. FA1117/1-2 (to AF), and internal funds of Senckenberg Research Institute (to JP).
Funding
Open Access funding enabled and organized by Projekt DEAL. Financial support for this study was provided by: (1) German Science Foundation (Deutsche Forschungsgemeinschaft) project no. FA1117/1–2 (to AF). (2) Internal funds of Senckenberg Research Institute (to JP).
Author information
Authors and Affiliations
Contributions
AF, JP and JE have contributed equally in the concept and realization of the study, as well as in writing the manuscript. With the help of all authors, AF took the lead for molecular work and associated analyses, as well as for compiling occurrence data, JP led the flow cytometry experiment and associated analyses, JE led the review of chromosome counts as well as the analyses of climate data.
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Favre, A., Paule, J. & Ebersbach, J. Incongruences between nuclear and plastid phylogenies challenge the identification of correlates of diversification in Gentiana in the European Alpine System. Alp Botany 132, 29–50 (2022). https://doi.org/10.1007/s00035-021-00267-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00035-021-00267-6