Abstract
The endocytic adaptor protein 2 (AP-2) complex binds dynactin as part of its noncanonical function, which is necessary for dynein-driven autophagosome transport along microtubules in neuronal axons. The absence of this AP-2-dependent transport causes neuronal morphology simplification and neurodegeneration. The mechanisms that lead to formation of the AP-2-dynactin complex have not been studied to date. However, the inhibition of mammalian/mechanistic target of rapamycin complex 1 (mTORC1) enhances the transport of newly formed autophagosomes by influencing the biogenesis and protein interactions of Rab-interacting lysosomal protein (RILP), another dynein cargo adaptor. We tested effects of mTORC1 inhibition on interactions between the AP-2 and dynactin complexes, with a focus on their two essential subunits, AP-2β and p150Glued. We found that the mTORC1 inhibitor rapamycin enhanced p150Glued–AP-2β complex formation in both neurons and non-neuronal cells. Additional analysis revealed that the p150Glued–AP-2β interaction was indirect and required integrity of the dynactin complex. In non-neuronal cells rapamycin-driven enhancement of the p150Glued–AP-2β interaction also required the presence of cytoplasmic linker protein 170 (CLIP-170), the activation of autophagy, and an undisturbed endolysosomal system. The rapamycin-dependent p150Glued–AP-2β interaction occurred on lysosomal-associated membrane protein 1 (Lamp-1)-positive organelles but without the need for autolysosome formation. Rapamycin treatment also increased the acidification and number of acidic organelles and increased speed of the long-distance retrograde movement of Lamp-1-positive organelles. Altogether, our results indicate that autophagy regulates the p150Glued–AP-2β interaction, possibly to coordinate sufficient motor-adaptor complex availability for effective lysosome transport.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The effective cooperation of endomembrane components and the cytoskeleton is necessary for efficient intracellular communication and cell contacts with the extracellular environment. Microtubules are cytoskeleton elements that are essential for both the integrity of membrane compartments and their long-distance movement [1, 2]. Microtubules are dynamic and polarized, meaning that their ends (referred to as plus and minus) can undergo dynamic changes and are not identical [3]. This polarization determines the rules of directed cargo transport along microtubules by molecular motors, e.g., kinesins and dynein [4, 5]. The latter transports cellular cargo from the plus end to the minus end of microtubules [5,6,7]. Dynein does not act alone; it requires additional protein complexes to efficiently hold cargo and move along microtubules. One of these complexes is dynactin, a large multiprotein complex that initiates dynein movement, increases its processivity, and supports cargo attachment [5, 8, 9]. Dynactin consists of two major parts: sidearm and actin-related protein 1 (Arp-1) rod [5, 7, 10, 11]. The sidearm binds microtubules and dynein [5, 7, 10, 11]. The Arp-1 rod, in cooperation with dynein activators and adaptors, is responsible for cargo binding [5, 7, 10,11,12,13,14,15]. p150Glued is part of the sidearm, the largest dynactin subunit, and a member of the microtubule plus-end tracking protein (+ TIP) family [16]. Its binding to microtubule plus ends and its plus-end tracking behavior require the presence of cytoplasmic linker protein 170 (CLIP-170) [17, 18]. In some model systems (e.g., neuronal axons), this is essential for the initiation of dynein-dynactin-bound cargo transport along tyrosinated microtubules [8, 19].
The adaptor protein 2 (AP-2) complex consists of two large subunits (α, β), one medium subunit (µ), and one small subunit (σ) [20]. All four subunits contribute to the trunk of the AP-2 complex, but α and β2 C-termini project outside the trunk as α and β2 appendages (i.e., ears), respectively [20]. Canonically, AP-2 serves as a cargo adaptor complex in clathrin-mediated endocytosis [21]. However, evidence supports AP-2 functions outside the initiation of clathrin-mediated endocytosis, particularly in macroautophagy (hereafter called autophagy), lysosome tubulation, and microtubular transport [22,23,24,25,26,27]. The latter function was first discovered in neurons, in which AP-2 was found to be central to the retrograde transport of neuronal amphisomes that are produced by autophagosome-late endosome fusion and in axons act as signaling organelles that carry activated receptors for neurotrophins, such as tropomyosin receptor kinase B (TrkB) [24, 28] to the cell soma. The lack of this AP-2-dependent transport in axons resulted in disturbances in the morphology of dendrites and neurodegeneration [22, 24]. For dynein cargo adaptor function, AP-2 binds microtubule-associated protein 1A/1B-light chain 3 (LC3) on the amphisome surface via its AP-2µ subunit, whereas the AP-2β ear was shown to co-immunoprecipitate with p150Glued [24].
Autophagy is a cellular process during which cells trap proteins or organelles (e.g., mitochondria) that are designated for degradation in double-membrane structures, called autophagosomes, and deliver them to lysosomes [29,30,31,32]. Autophagosome formation is a multistep process. Mammalian/mechanistic target of rapamycin complex 1 (mTORC1) is among its best-known regulators [33, 34]. Low mTORC1 activity allows autophagy initiation, but also autophagosome maturation, microtubular transport, and fusion with the lysosome [35,36,37,38,39,40]. Notably, in the case of neurons, the role of mTOR inhibition in autophagy initiation, particularly in axons, is still debated because of conflicting findings on whether rapamycin potentiates this process [41,42,43,44].
The effective termination of autophagy requires the fusion of autophagosomes or amphisomes with lysosomes, which contain degradative enzymes that are needed for autophagosome cargo destruction. It heavily relies on autophagosome and lysosome transport along microtubules. Dynein-dynactin transports autophagosomes retrogradely for fusion with lysosomes [45,46,47]. To meet autophagosomes, lysosomes may use both kinesins and dynein-dynactin [48, 49]. Lysosomes are dispersed through the cytoplasm with two distinguishable pools: perinuclear and peripheral [49]. The peripheral pool serves additional purposes (e.g., exocytosis [50]); when the demand for lysosomes greatly increases, however, such as during nutrient starvation that initiates autophagy, they move via dynein-dynactin transport toward autophagosomes that are already positioned in the cell center. To date, only two adaptors (ALG2 and JIP4) have been shown to recruit dynein-dynactin to lysosomes on demand upon nutrient starvation [51, 52].
Although AP-2-dynactin was shown to transport TrkB-positive amphisomes in neurons, unclear is whether the AP-2-dynactin complex also forms naturally in non-neuronal cells, in which amphisomes are considered very transient. Further details of the AP-2-dynactin interaction and its potential regulation are lacking. Our unpublished preliminary mass spectrometry data suggested a potential role for mTOR in the regulation of the p150Glued–AP-2β interaction. This is particularly intriguing when considering the important role of kinases in the regulation of microtubular transport [39, 53] and a recent finding that dynein can be recruited to autophagosomes by LC3 and Rab-interacting lysosomal protein (RILP) when mTORC1 activity is low [39]. Therefore, the present study investigated whether mTORC1 controls the p150Glued–AP-2 interaction and, if so, how and for what purpose. We found that mTORC1 inhibition enhanced the p150Glued–AP-2β interaction in both neurons and non-neuronal cells. We also found that p150Glued–AP-2β complex formation, boosted by mTORC1 inhibition, in non-neuronal cells required an intact dynactin complex and the undisturbed initiation of autophagy and endolysosomal pathway. We also found that the autophagy-induced p150Glued–AP-2β interaction occurred on lysosomes, which accelerated their retrograde motility. Thus, we revealed a novel mechanism whereby functions of essential components of cellular transport machinery are regulated at the level of autophagy initiation.
Materials and methods
Plasmids and siRNA
The following plasmids were commercially available or described previously: pEGFPC1 (Clontech), β-actin-GFP and β-actin-tdTomato [54], HA-BirA [55], pAvi-tag-thrombin-HA (also known as Bio-Thrombin-HA; [56], pEGFPC1-Ap2b1 [57], pEGFPC2-Avi-tag-p150Glued [24], pEGFP-TrkB (kind gift from Volker Haucke) [24], pEGFPC2-Avi-tag-βGal (also known as bioβ-Gal) [58], EB3-GFP [59], pEGFP-CLIP-170 and pEGFPC1-CLIP-170-Δhead [60] (gift from Anna Akhmanova), pEGFPC1-p50 [61] (gift from Casper Hoogenraad), pEGFP-N1-Lamp1-GFP [62] (gift from Juan Bonifacino), pET-28-His6-AP-2β appendage domain [24] (gift from Volker Haucke), pGEX-4T1 (Merck, catalog no. GE28-9545-49), pGEX-4T1-GST-Eps15 (aa 541–790; gift from Mark McNiven) [63], and pGFP-LC3 [64] (gift from Iwona Ciechomska). Additional plasmids generated for this study are described in Supplementary Materials and Methods in Supplementary Information. The following siRNAs were purchased from Invitrogen: Select Negative Control No. 1 siRNA (catalog no. 4390843; siCtrl), Silencer Select siRNA rat Clip1#1 (catalog no. 4390771, ID: s134775; siCLIP-70), Silencer Select siRNA human Clip1#1 (catalog no. 4392420, ID: s12372; siCLIP-170), Silencer Select siRNA rat Atg5 (catalog no. 4390771, ID: s172246), and Silencer Select siRNA rat Snap29 (catalog no. 4390771, ID: s138000).
Antibodies
Commercially available primary antibodies that were used for this study are listed in Table S1. Rabbit anti-pan CLIP antibody (clone 2221; Western blot, 1:500) that recognizes both cytoplasmic linker protein 115 (CLIP-115) and CLIP-170 was a kind gift from Casper Hoogenraad [65]. Alexa Fluor 488-, 568-, 594-, and 647-conjugated secondary antibodies (anti-mouse, anti-goat, and anti-rabbit) were obtained from Thermo Fisher. Horseradish peroxidase (HRP)-conjugated secondary antibodies were obtained from Jackson ImmunoResearch. Anti-mouse/anti-rabbit IRDye 680RD and IRDye 800CW were purchased from LI-COR Biosciences.
Cell line and primary neuronal cultures and transfection
Rat2, HEK293, and primary hippocampal neurons cultures and transfections were performed using standard previously published protocols. For details, please refer to Supplementary Information.
Drugs and drug treatment
All drugs, unless indicated otherwise, were dissolved in dimethylsulfoxide (DMSO), the final concentrations of which in the culture medium did not exceed 0.1%. For mTOR inhibition, Rat2 and HEK293T cells were treated with rapamycin (100 nM, Calbiochem, catalog no. 553210) or AZD-8055 (100 nM, Cayman Chemical, catalog no. 16978-5) for 2 h before the experiment (see figure legends for detailed descriptions). For translation inhibition, cells were treated for 2 h with 35 µM cycloheximide (Calbiochem, catalog no. 239763). For mTOR inhibition-dependent autophagy arrest at the initiation step, 25 µM SBI-0206965 (Merck, catalog no. SML1540) was used for 2.5 h (see figure legends for detailed descriptions). When combined with rapamycin, SBI-0206965 was added 30 min before the addition of rapamycin. To inhibit autophagic flux, cells were treated for 2 h with 60 µM 1-adamantyl(5-bromo-2-methoxybenzyl)amine (ABMA; Medchemexpress, catalog no. HY-124801) or 50 µM chloroquine (dissolved in water; Sigma-Aldrich, catalog no. C6628). For lysosomal vacuolar (H +)-adenosine triphosphatase (vATPase) inhibition, cells were treated for 2 h with bafilomycin A1 (Baf A1; 100 nM; Bioaustralis, catalog no. 88899-55-2). When cells were treated with an inhibitor and rapamycin, these compounds were administered at the same time and incubated for 2 h. For the alkalization of the cellular environment, 20 mM NH4Cl (dissolved in water; Sigma-Aldrich, catalog no. 213330) was added for 3 h. When cells were treated with both NH4Cl and rapamycin, NH4Cl was added to the cells 1 h prior to rapamycin, and the cells were then incubated with both drugs for an additional 2 h. Ciliobrevin D (30 µm; Sigma-Aldrich, catalog no. 250401) was added 30 min prior to imaging and 1.5 h after rapamycin treatment. To induce autophagy independently from mTORC1 inhibition, 100 μM L-690330 (Tocris, catalog no. 0681) was added to the cells for 3 h. Nocodazole (100 nM; Sigma-Aldrich, catalog no. M1404) was used to inhibit microtubule dynamics. For live-cell imaging experiments, the drug was added to Rat2 cells 1 h before imaging. For the PLA experiments, in which nocodazole was added to Rat2 cells alone or combined with rapamycin, it was added 2 h 15 min or 1 h before fixation, depending on whether it was used before or after rapamycin treatment. Before live imaging or fixation, neurons were treated with either vehicle (0.1% DMSO, 2 h) or rapamycin (100 nM, 2 h).
Animals and rapamycin treatment
Rapamycin treatment and brain protein lysate isolation were performed according to a protocol that was approved by the 1st Ethical Committee in Warsaw (Poland; decision no. 843/2008 and 288/2012). Mature (3-month-old) male Wistar rats were used for the experiments. Rapamycin was initially dissolved in 100% ethanol at a 0.1 mg/ml concentration and stored at −20 °C. Immediately before the injection, rapamycin was diluted in a vehicle solution that contained 5% Tween 80 and 5% PEG 400 (low-molecular-weight grade of polyethylene glycol; Sigma) and injected intraperitoneally (i.p.; 10 mg/kg) three times per week for 1 week. A control group of rats was injected with a vehicle solution that contained 5% Tween 80, 5% PEG 400, and 4% ethanol. Protein extraction from adult rat brains is described in detail in Supplementary Information.
Proximity ligation assay and PLA-EM
Standard PLA procedures were performed as described previously [58] and detailed procedure is described in Supplementary Information. For PLA-EM, Rat2 cells were grown for 24 h and fixed for 15 min in 4% PFA with the addition of 0.1% glutaraldehyde in PBS. The cells were then washed three times with PBS. Afterward, cells in PBS were permeabilized by three cycles of freezing in liquid nitrogen, thawed, incubated for 20 min with 1% sodium borohydride in PBS, washed three times with PBS, incubated for 20 min at room temperature with 3% hydrogen peroxide in PBS/ethanol (1:1), and washed again three times in PBS. Next, fixed and permeabilized cells were incubated for 1 h in 5% bovine serum albumin (BSA) in PBS at room temperature. Afterward, the cells were incubated for 48 h at 4 °C with primary mouse anti-p150Glued and rabbit anti-AP-2β antibodies that were diluted in 0.1% donkey serum/PBS and washed three times in PBS at room temperature. The cells were then incubated for 60 min at 37 °C with PLA probes (Sigma-Aldrich, catalog no. DUO92002 and DUO92004) and washed twice for 5 min with buffer A (Sigma-Aldrich, catalog no. DUO82046). Ligation and amplification were performed according to the manufacturer’s protocol using DuoLink In Situ Detection Reagents Brightfield (Sigma-Aldrich, catalog no. DUO92012). After the PLA reaction, the cells were additionally fixed with 2.5% glutaraldehyde in PBS for 2 h at 4 °C and washed twice with PBS and once with deionized water. Next, the cells were incubated with 3% hexamethylenetetramine, 5% silver nitrate, and 2.5% disodium tetraborate for 10 min at 60 °C, washed three times with water, once with 0.05% tetrachloroauric acid, once with 2.5% sodium thiosulfate, and finally three times with water (all at room temperature). As the last step, the cells were postfixed with 1% osmium tetroxide for 1 h at room temperature, washed with water, incubated in 1% aqueous uranyl acetate for 1 h, dehydrated with increasing dilutions of ethanol, and infiltrated with epoxy resin (Sigma-Aldrich, catalog no. 45-359-1EA-F). After resin polymerization at 60 °C, fragments of coverslips with embedded cells were cut out with scissors and glued to the resin blocks. The blocks were then trimmed and cut with a Leica ultramicrotome (Ultracut R) to obtain ultrathin Sects. (70 nm thick) and collected on 100 mesh copper grids (Agar Scientific, catalog no. AGS138-1). Specimen grids were examined with a Tecnai T12 BioTwin transmission electron microscope (FEI) that was equipped with a 16 megapixel TemCam-F416 (R) camera (Tietz Video and Imaging Processing Systems).
Immunofluorescence and fixed cell image acquisition and analysis
Procedures used for immunofluorescence and fixed cell image acquisition and analysis are described in detail in Supplementary Information.
Live imaging of microtubule dynamics
Procedures used for live imaging and analysis of microtubule dynamics are described in detail in Supplementary Information.
Live imaging of Lamp-1-GFP objects
For the lysosomal-associated membrane protein 1 (Lamp-1) object motility analysis, Rat2 cells were electroporated with Lamp-1-GFP plasmid and imaged approximately 22 h later with an Andor Revolutions XD spinning disc microscope, with the same setup and settings as described above. Time-lapse movies were collected over 3 min at 0.3 s intervals, resulting in 600 frames. Movies were analyzed using the ImageJ “TrackMate” plugin [66]. Only objects that were visible for more than 4 consecutive frames were considered. Objects with movement lengths shorter than 6.8 µm were considered not motile. The number of motile and non-motile objects divided by cell area and their ratio were measured. Other calculated values that were used for the analysis included the following: length of the Lamp-1 object run (Track Displacement), time during which the objects were visible (Track Duration), and speed with which the object moved (Track Mean Speed). For the analysis of Lamp-1-GFP objects in the cell center and in the periphery, Lamp-1-GFP objects were assigned manually to these compartments based on maximum projections.
Live imaging of neurons
An Andor Revolutions XD confocal spinning disc microscope and a Chamlide magnetic chamber were used for the in vivo imaging of cells. Cell recording was performed at 37 °C with 5% CO2 in a thermostat-controlled incubator. Series of images were acquired at 502 × 501 pixel resolution, with 1 s interval between frames. The total imaging time was 3 min. The images were collected using the 63 × objective and 1 × optovar. Neuronal processes were manually tracked with ImageJ. Based on these tracks, kymographs were created in ImageJ with the “Kymograph Clear” plugin [67]. By tracing lines on the kymographs, the number of particles that moved at a given time and distance was determined, and their velocity was calculated by determining the difference between the height of the starting point and end point of a given particle.
Biochemical procedures
All standard biochemical procedures including protein production, pull-down assays, immunoprecipitation, Western blot, kinase assays and RNA isolation and Quantitative Real-Time PCR are described in details in Supplementary Information.
Statistical analysis
The exact numbers of cells (n) that were examined for the respective experiments and number of repetitions of each experiment (N) are provided in the figure legends. The statistical analyses were performed using GraphPad Prism 9 software or Rstudio. The Shapiro–Wilk test was used to assess whether the data distribution met the assumptions of a normal distribution. For comparisons between two groups, the t-test (in the case of a normal distribution) or Mann–Whitney test (in the case of a non-normal distribution) was used to verify statistical significance. For comparisons between more than two groups, the data were analyzed using one-way analysis of variance (ANOVA), followed by the Bonferroni multiple-comparison post hoc test (in the case of a normal distribution) or Kruskal–Wallis test and Dunn’s multiple-comparison post hoc test (in the case of a non-normal distribution). For comparisons between two factors, the data were analyzed using two-way ANOVA, followed by Tukey’s multiple-comparison post hoc test. The statistical significance of qRT-PCR data was assessed with the one-sample Student’s t-test.
Results
mTORC1 inhibition increases p150 Glued –AP-2β interaction in neurons and non-neuronal cells
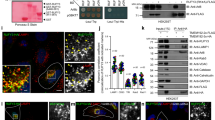
Recent work shows that mTOR inhibition increases the biosynthesis of RILP and enhances its recruitment to autophagosomes, potentiating their transport in different cell types, including neurons [39]. This finding prompted us to investigate whether rapamycin, an inhibitor of mTORC1, also affects the AP-2–dynactin interaction that is responsible for the axonal transport of amphisomes [24]. We performed the IP of endogenous AP-2β from brain lysates from control rats and rats that were treated with rapamycin for 8 days. Rapamycin treatment effectively decreased the phosphorylation of ribosomal protein S6 (P-S6) at Ser235/236, confirming efficient mTORC1 inhibition, and increased the co-IP of p150Glued with AP-2β. We observed no noticeable difference in overall levels of p150Glued or AP-2β in corresponding input fractions (Fig. 1A).
mTORC1 inhibition increases p150Glued–AP-2β interaction. A Western blot analysis of levels of endogenous p150Glued, AP-2β, and P-S6 (Ser235/236) and co-immunoprecipitation of endogenous AP-2β with p150Glued from brain lysates from rats that were treated with rapamycin (RAPA +) or vehicle (RAPA-). INPUT, 10% of lysate used for immunoprecipitation. Shown is a representative example from N = 3 independent experiments. B–E Dynamics of p150Glued-tdTomato and AP-2β-GFP co-transport in axons of neurons that were treated for 2 h with 0.1% DMSO or 100 nM rapamycin (RAPA). B Representative snapshots of 60 μm segment of axon and C corresponding kymographs of p150Glued-tdTomato- and AP-2β-GFP-positive objects. See also Supplementary Fig. S1 and Movies 1–2. Scale bar = 10 μm. D Number of all p150Glued-tdTomato/AP-2β-GFP objects in axons per 60 μm. The data are expressed as the mean ± SEM. N = 4 independent experiments. n = 15 cells per variant. *p < 0.05 (Mann–Whitney test). E Number of mobile p150Glued-tdTomato/AP-2β-GFP-positive objects in axons per 60 μm. The data are expressed as the mean ± SEM. N = 4 independent experiments. n = 15 cells per variant. ns nonsignificant (Mann–Whitney test). F Western blot analysis of levels of endogenous p150Glued, AP-2β, and P-S6 (Ser235/236) and co-immunoprecipitation of endogenous p150Glued with AP-2β from HEK293T cells that were treated for 2 h with 0.1% DMSO (RAPA-) or 100 nM rapamycin (RAPA +). Input, 10% of lysate used for immunoprecipitation. Shown is a representative example from N = 5 independent experiments. G Representative images of Rat2 fibroblasts that were treated for 2 h with 0.1% DMSO or 100 nM rapamycin (RAPA) with p150Glued–AP-2β PLA signals (magenta), immunofluorescently labeled tubulin (green), and DAPI-stained nuclei (blue). Scale bar = 10 μm. H Quantification of the number of p150Glued–AP-2β PLA puncta in cells that were treated as in G. The data are expressed as the mean number of PLA puncta per cell, normalized to the control variant (DMSO) ± SEM. N = 5 independent experiments. n = 201 cells for each experimental variant. ***p < 0.001 (Student’s t-test). (I) Representative images of Rat2 fibroblasts that were treated for 2 h with 0.1% DMSO or 100 nM AZD-8055 with p150Glued-AP-2β PLA signals (magenta), immunofluorescently labeled tubulin (green), and DAPI-stained nuclei (blue). Scale bar = 10 μm. J Quantification of the number of p150Glued-AP-2β PLA puncta in cells that were treated as in I. The data are expressed as the mean number of PLA puncta per cell, normalized to the control variant (DMSO) ± SEM. N = 5 independent experiments. n = 211 cells (DMSO), 188 cells (AZD-8055). *p < 0.05 (Student’s t-test)
mTORC1 inhibition potentiates the AP-2–dynactin interaction in the brain. Therefore, we next tested effects of rapamycin on the AP-2β and p150Glued interaction in axons of hippocampal neurons, in which its functional significance was demonstrated [24]. However, in view of the above-mentioned studies on mTOR-dependent RILP recruitment to autophagosomes performed on week-old neurons in culture [39] we decided to use less mature cells than in our previous work on the interaction of AP-2β and p150Glued. Of note, however, is that in such young neurons axonal and synaptic developmental processes are very active [62], while in DIV14 neurons, used by Kononenko et al. [24] these processes are already completed and transport was studied in more mature axons, which already formed synapses. Therefore, it should be kept in mind that results presented below cannot be directly compared to our previous work. We transfected neurons that were cultured in vitro (DIV5) with plasmids that encoded tdTomato-tagged p150Glued and green fluorescent protein (GFP)-tagged AP-2β. Two days later, we imaged the behavior of fluorescently tagged proteins in axons in DMSO-treated (control) cells and cells that were treated with 100 nM rapamycin for 2 h (Fig. 1B–E, Movie 1–4, Fig. S1A [Supplementary Figures in Supplementary Information]). Similar to brain tissues, rapamycin decreased P-S6 levels (Fig. S1B, C). At the same time total S6 levels remained unchanged (Fig. S1D, E). We also observed a significant increase in the number of p150Glued–AP-2β-positive objects (Fig. 1D), suggesting that mTORC1 inhibition boosted the p150Glued–AP-2β interaction in neurons, but these objects were largely immobile. The difference between fractions of mobile AP-2β/p150Glued-positive objects was also statistically nonsignificant under the tested conditions (Fig. 1E). To further test whether mTORC1 inhibition would similarly affect the behavior of cargo transported by p150Glued–AP-2β, we checked whether the colocalization of TrkB with AP-2β also changes after rapamycin treatment. To this end, we repeated the experiment as described above, but this time using neurons cotransfected with plasmids encoding TrkB-GFP and mCherry-AP-2β. In the rapamycin-treated neurons, the number of TrkB/AP-2β positive objects in the axons indeed increased, but the observed difference did not reach statistical significance (Fig. S1F–I, Movies 5–8). However, these results confirm that TrkB coexists with the AP2 complex in axons as in mature neurons [24]. In summary, we concluded that mTORC1 inhibition in neurons enhanced the p150Glued–AP-2β interaction in axons and that this carrier likely also transports TrkB as previously described [24].
We next investigated whether mTORC1 inhibition has a similar effect on the co-occurrence of p150Glued and AP-2β in non-neuronal cells using HEK293T and Rat2 cell lines. HEK293T cells were selected because they are frequently used for biochemical experiments that require large amounts of material. On the other hand, Rat2 cells were used because they originate from the same species as our neurons to avoid potential species-specific differences in molecular mechanisms. Furthermore, Rat2 cells are not cancer cells, which in our opinion makes them suitable for the study of physiological mechanisms. Under basal culture conditions, there was some evidence of an AP-2–dynactin interaction, demonstrated by IP (HEK293T), immunofluorescence co-localization (Rat2), and the PLA (Rat2; Fig. 1F–J; Fig. S2). The treatment of HEK293T cells with 100 nM rapamycin for 2 h decreased P-S6 levels and increased AP-2β–p150Glued co-IP (Fig. 1F). In Rat2 cells, rapamycin decreased S6 phosphorylation without changing abundance of S6 (Fig. S2A–C) and enhanced the p150Glued–AP-2β interaction, measured by immunofluorescence signal co-localization analysis and PLA using antibodies against endogenous proteins (Figs. 1G, H, S2D–I). Co-localization analysis showed relatively low co-localization under basal conditions and an increase in rapamycin-treated cells (Fig. S2D–H). The PLA results additionally confirmed the biochemical and immunofluorescence evidence that is described above (Figs. 1G, H, S2I). AZD-8055, an ATP-competitive inhibitor of mTOR (2 h, 100 nM), also effectively decreased P-S6 immunofluorescence (Fig. S2A–C) and increased the p150Glued–AP-2β PLA signal in Rat2 cells (Fig. 1I, J), supporting our hypothesis that mTOR inhibition enhances the studied interaction.
The best-known function of mTORC1 is the positive regulation of protein synthesis. Therefore, we treated Rat2 cells for 2 h with cycloheximide (CHX) (35 µM), a widely used protein synthesis inhibitor, to test whether the increase in the PLA signal was attributable to a decrease in translation. However, we did not detect a significant difference in the PLA signal in CHX-treated cells compared with control cells (Fig. S3). Overall, our results suggest that mTORC1 inhibition enhances the interaction between p150Glued and AP-2β in different cell types but not because of the canonical function of mTORC1 as a translational enhancer.
Excluding the possibility that protein synthesis inhibition was a main driver of an increase in the p150Glued–AP-2β interaction prompted us to test whether p150Glued or AP-2β are substrates of mTOR. In vitro kinase assays, using GFP-AP-2β or GFP-p150Glued and the mTOR active fragment, excluded such a possibility (Fig. S4A). Furthermore, inspection of the available datasets of mTOR-dependent phosphoproteomes ([e.g., 68, 69]) did not support the mTOR-dependent phosphorylation of other dynactin or AP-2 subunits. mTOR inhibition in HEK293T cells also did not affect the connection between the dynactin sidearm and its Arp1 rod, indicated by the lack of differences in the co-IP of p150Glued with p62 and Arp1 between analyzed conditions (Fig. S4B). Thus, we concluded that the observed effects of mTORC1 inhibition on the p150Glued–AP-2β interaction were not driven by direct actions of mTOR on the dynactin complex or AP-2β.
p150Glued interaction with AP-2β is indirect and requires dynactin integrity
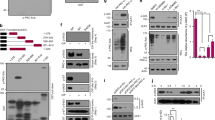
Our previous study [24] and the data described above show that AP-2 and p150Glued can form a complex, but further characterization is needed. Therefore, in the following experiments, we first focused on the biochemical characterization of this interaction. Using an Avi-tag pull-down assay, we previously demonstrated that full-length p150Glued that is produced in HEK293T cells can effectively bind the E. coli-produced β2 ear of AP-2β [24]. Therefore, we used this system to characterize the p150Glued–β2-ear interaction further and clarified which p150Glued domains are required. We first compared the ability of the N-terminal (1–490 aa; N) and C-terminal (490-end; C) parts of p150Glued and the full-length protein (Fig. 2A) that is produced in HEK293T cells to bind the His-tagged β2 ear. The C-terminal part of p150Glued was as effective as the full-length protein, whereas its N-terminus did not bind the AP-2 fragment, exactly like β-galactosidase that served as the negative control (Fig. 2B). Moreover, a shorter fragment of the C-terminus (1049-end; C2; Fig. 2A), which is known for its contribution to the dynactin interaction with cargo adaptors [13, 70, 71], also bound the β2 ear (Fig. 2C). Notably, however, the newest structural and biochemical data raise the issue of whether the C-terminus of p150Glued binds cargo adaptors directly [10, 11]. Indeed, when both C-terminal fragments of p150Glued were produced in E. coli, no interaction with the AP-2β fragment was observed (Fig. 2D, E). But, the β2 ear interacted with Eps15 protein (541–790 aa fragment fused to GST; [63, 72]), which was used as a positive control. The most C-terminal part of p150Glued is known for its role in connecting the dynactin sidearm with the Arp-1 rod that binds cargo [73]. Indeed, Western blot indicated that during the p150Glued Avi-tag pull-down, regardless of the harsh washing conditions, the dynactin Arp1-rod proteins (Arp1 and p62) also co-purified from HEK293T cells (Fig. 2F). This suggests that intact dynactin might be involved in formation of the multi-protein complex that contains p150Glued and AP-2β. Indeed, overexpression of the p50 subunit of dynactin, which is routinely used to disrupt dynactin complex integrity by dissociation of the sidearm and Arp1 rod [74, 75]; Fig. 2G), completely blocked the rapamycin-driven increase in the p150Glued-AP-2β PLA signal in Rat2 cells (Fig. 2H, I). Thus, although p150Glued and AP-2β are unlikely to bind each other directly, their interaction (evident upon mTORC1 inhibition) requires the C-terminus of p150Glued and an undisturbed interaction between the dynactin sidearm and Arp-1 rod.
AP-2β interaction with p150Glued is indirect and requires intact dynactin-dynein complex. A Diagram of the full p150Glued and N, C, and C2 fragments that were used in the study. B Western blot analysis of E. coli-produced His-AP-2β-ear binding to in vivo biotinylated AviHA-tagged p150Glued N or C fragments or AviGFP-tagged p150Glued or AviGFP-tagged β-galactosidase that was pulled down from HEK293T cells using Avi-tag pull down. Input, 10% of lysate added to the assay. Shown is a representative example from N = 3 independent experiments. C Western blot analysis of E. coli-produced His-AP-2β-ear binding to in vivo biotinylated AviHA-tagged p150Glued C or C2 fragments or AviGFP-tagged p150Glued or AviGFP-tagged β-galactosidase that was pulled down from HEK293T cells using Avi-tag pull down. Input, 10% of lysate added to the assay. Shown is a representative example from N = 2 independent experiments. D, E Western blot analysis of pull down of E. coli-purified recombinant GST- p150Glued C, GST- p150Glued C2, GST-Eps15, or GST (negative control) with recombinant His-AP-2β ear. Shown is a representative example from N = 2 independent experiments. F Western blot analysis of endogenous p62 and Arp1 binding to in vivo biotinylated AviHA-tagged p150Glued N or C2 fragments or full AviGFP-tagged p150Glued or AviGFP-tagged β-galactosidase that was pulled down from HEK293T cells using Avi-tag pull down. Shown is a representative example from N = 3 independent experiments. G Western blot analysis of the co-immunoprecipitation of endogenous p150Glued with other subunits of dynactin-dynein complex (DIC1/2, p62, and Arp1) from HEK293T cells that were transfected with pEGFPC1 or pEGFPC1-p50 plasmids. Shown is a representative example from N = 3 independent experiments. H Representative images of Rat2 fibroblasts that were transfected with pEGFPC1 or pEGFPC1-p50 (green) and treated for 2 h with 0.1% DMSO or 100 nM rapamycin (RAPA) with p150Glued–AP-2β PLA signals (magenta) and DAPI-stained nuclei (blue). Scale bar = 10 μm. I Quantification of the number of p150Glued–AP-2β PLA puncta in cells that were treated as in H. The data are expressed as the mean number of PLA puncta per cell, normalized to the control variant (GFP + DMSO) ± SEM. N = 4 independent experiments. n = 96 cells (GFP + DMSO), 81 cells (GFP + RAPA), 91 cells (GFP-p50 + DMSO), 92 cells (GFP-p50 + RAPA). **p < 0.01, ***p < 0.001, ns, nonsignificant (two-way ANOVA followed by Tukey’s multiple-comparison post hoc test)
CLIP-170 is needed for p150 Glued –AP-2β interaction
p150Glued is a + TIP. Therefore, we tested whether the p150Glued–AP-2β interaction requires its ability to target dynamic microtubules. p150Glued microtubule plus-end targeting requires the presence of CLIP-170, which is also an mTOR substrate [17, 18, 58]. Thus, we tested whether CLIP-170 knockdown impacts the p150Glued–AP-2β interaction. We knocked down CLIP-170 in Rat2 and HEK293T cells using rat- and human-specific siRNAs, respectively, and performed the PLA and IP 72 h later. Both siRNAs against CLIP-170 effectively reduced CLIP-170 levels compared with cells that were transfected with control siRNAs (Fig. 3A–D). The loss of CLIP-170 also potently prevented the rapamycin-induced p150Glued–AP-2β interaction (Fig. 3A, E, F). Notably, CLIP-170 knockdown did not affect the overall distribution of AP-2β in either control or rapamycin-treated cells (Fig. S5A).
CLIP-170 but not microtubule dynamics is needed for p150Glued–AP-2β interaction. A Western blot analysis of endogenous CLIP-170, p150Glued, AP-2β, S6, and P-S6 (Ser235/236) levels and co-immunoprecipitation of endogenous AP-2β with p150Glued in HEK293T cells that were transfected with control siRNA (siCtrl) or siRNA against human CLIP-170 (siCLIP-170) and treated for 2 h with 0.1% DMSO or 100 nM rapamycin (RAPA). Input, 10% of lysate added for immunoprecipitation. Shown is a representative example from N = 3 independent experiments. B Western blot analysis of endogenous CLIP-170 and CLIP-115 levels in non-transfected (NT) Rat2 cells or Rat2 cells that were transfected with siCtrl or rat siCLIP-170. C Representative images of Rat2 cells that were transfected with siCtrl or rat siCLIP-170, with immunofluorescently labeled endogenous CLIP-170 (green) and nucleus stained with Hoechst 33258 (blue). Scale bar = 10 µm. D Quantitative analysis of CLIP-170 immunofluorescence in Rat2 cells that were treated as in C. The results are presented as the average intensity of CLIP-170 immunofluorescence in the cell ± SEM. N = 5 independent experiments. **p < 0.01 (Student’s t-test). E Representative images of Rat2 fibroblasts that were transfected with siCtrl or rat siCLIP-170, and treated for 2 h with 0.1% DMSO or 100 nM rapamycin (RAPA), with p150Glued–AP-2β PLA signals (magenta), immunofluorescently labeled tubulin (green), and DAPI-stained nuclei (blue). Scale bar = 10 μm. F Quantification of the number of p150Glued–AP-2β PLA puncta in cells that were treated as in E. The data are expressed as the mean number of PLA puncta per cell, normalized to the control variant (siCtrl + DMSO) ± SEM. N = 4 independent experiments. n = 149 cells (siCtrl + DMSO), 130 cells (siCtrl + RAPA), 161 cells (siCLIP-170 + DMSO), 142 cells (siCLIP-170 + RAPA). **p < 0.01, ns, nonsignificant (two-way ANOVA followed by Tukey’s multiple-comparison post hoc test). G Representative images of Rat2 fibroblasts with p150Glued–AP-2β PLA signals (magenta), immunofluorescently labeled tubulin (green), and DAPI-stained nuclei (blue). Cells were treated for 2 h with 0.1% DMSO or 100 nM rapamycin (RAPA) or treated for 2 h 15 min with 100 nM nocodazole alone or in combination with 100 nM rapamycin that was added 15 min after nocodazole (PRIOR nocodazole + RAPA). Scale bar = 10 μm. H Quantification of the number of p150Glued–AP-2β PLA puncta in cells that were treated as in G. The data are expressed as the mean number of PLA puncta per cell, normalized to the control variant (DMSO) ± SEM. N = 3 independent experiments. n = 126 cells (DMSO), 125 cells (RAPA), 133 cells (nocodazole), 139 cells (PRIOR nocodazole + RAPA). *p < 0.05, **p < 0.01, ns, nonsignificant (one-way ANOVA followed by Bonferroni multiple-comparison post hoc test). I Representative images of Rat2 fibroblasts with p150Glued–AP-2β PLA signals (magenta), immunofluorescently labeled tubulin (green), and DAPI-stained nuclei (blue). Cells were treated for 2 h with 0.1% DMSO or 100 nM rapamycin (RAPA) or treated for 1 h with 100 nM nocodazole alone or added in the middle of 2 h of 100 nM rapamycin incubation (RAPA + POST nocodazole). Scale bar = 10 μm. J Quantification of the number of p150Glued–AP-2β PLA puncta in cells that were treated as in I. The data are expressed as the mean number of PLA puncta per cell, normalized to the control variant (DMSO) ± SEM. N = 3 independent experiments. n = 128 cells (DMSO), 118 cells (RAPA), 136 cells (nocodazole), 121 cells (RAPA + POST nocodazole). **p < 0.01, ***p < 0.001, ns, nonsignificant (one-way ANOVA followed by Bonferroni multiple-comparison post hoc test)
CLIP-170 knockdown may result, in addition to p150Glued displacement, in substantial changes in microtubule dynamics, especially in cells that do not express CLIP-115 [17]. CLIP-115 is highly homologous to CLIP-170 but its C-terminus is shorter and lacks some residues potentially responsible for binding metal ions [65, 76]. Nevertheless, CLIP-170 and CLIP-115 shares several microtubule-related functions [65]. Although Rat2 cells express both CLIPs and our siRNAs did not target CLIP-115 (Fig. 3B), we directly tested effects of CLIP-170 knockdown on microtubule dynamics and the role of microtubule dynamics in the AP-2–dynactin interaction. Indeed, CLIP-170 knockdown had no effect on EB3-GFP mobility that highlights dynamic plus ends of microtubules (Fig. S5B–F, Movies 9–10). Next, we performed a p150Glued–AP-2β PLA in Rat2 cells that were treated with 100 nM nocodazole that was added either 15 min before or 1 h after rapamycin treatment. At such low concentrations, nocodazole blocks plus-end microtubule dynamics instead of depolymerizing microtubules [59]. Indeed, 1 h of the nocodazole treatment of Rat2 cells resulted in the loss of EB3-GFP and CLIP-170 comets, confirming the inhibition of microtubule dynamics (Fig. S6, Movies 11–14). Such treatment did not affect the p150Glued–AP-2β PLA signal under basal conditions or in response to rapamycin treatment (Fig. 3G–J). In contrast to nocodazole, rapamycin treatment did not affect EB3-GFP microtubule plus-end tracking behavior (Fig. S7A–D, Movie 15, 16). Notably, rapamycin did not change the plus-end tracking behavior of CLIP-170–GFP that was overexpressed in Rat2 cells (Fig. S7E–H, Movie 17, 18). This observation is consistent with our previous data on the lack of effect of rapamycin on endogenous CLIP-170 microtubule binding in neurons and HeLa cells [58].
Overall, the data suggest that microtubule dynamics, at least in the short term, is not needed for the p150Glued–AP-2β interaction. Low-dose nocodazole treatment should also result in p150Glued displacement from dynamic microtubule plus ends, like CLIP-170, but we did not observe any impact of nocodazole on p150Glued–AP-2β complex formation. Thus, p150Glued displacement from microtubules unlikely explains the effects of CLIP-170 knockdown on the p150Glued–AP-2β PLA interaction. We confirmed this hypothesis using a dominant-negative CLIP-170 mutant that lacked the N-terminal part of the protein and was previously shown to severely affect microtubule dynamics and displace p150Glued from microtubule plus ends [60]. The 48 h overexpression of this protein in Rat2 cells did not prevent p150Glued–AP-2β interaction measured with PLA in the rapamycin-treated cells (Fig. S8). In summary, CLIP-170 is needed for the p150Glued–AP-2β interaction, but two of its canonical functions (i.e., the regulation of microtubule dynamics and targeting p150Glued to microtubule plus ends) do not appear to be directly or immediately involved.
mTORC1-dependent autophagy triggers p150Glued–AP-2β interaction
Like mTOR, AP-2 was postulated to be essential for autophagy initiation [25]. Thus, we investigated whether mTORC1-controlled autophagy under the conditions that were used in the present study is induced and required for the rapamycin-driven p150Glued–AP-2β interaction. Treatment with rapamycin decreased P-S6 level, demonstrating mTORC1 inhibition and resulting in an increase in beclin-1 phosphorylation at Ser30, which is an Ulk-1 target and considered an early marker of autophagy (Fig. S9A). Furthermore, after 2 h, rapamycin also increased the ratio of the lipidated form of LC3 (LC3B II) to non-lipidated LC3B I, which is routinely used to assess autophagy (Fig. S9A, B). Furthermore, rapamycin also increased the formation of large LC3B foci, further confirming autophagy induction (Fig. S9C). To verify whether autophagy induction is required for the mTORC1 inhibition-driven AP-2–dynactin interaction, we investigated whether pretreatment with the autophagy initiation inhibitor SBI-0206965 (25 µM; 30 min before rapamycin administration) counteracts the effects of rapamycin. As expected, pretreatment with SBI-0206965 was sufficient to decrease rapamycin-induced beclin-1 phosphorylation at Ser30, LC3 lipidation, and the formation of endogenous large LC3 foci (Fig. S9A–C). Notably, blocking autophagy initiation completely abolished the rapamycin-induced increase in the AP-2β- p150Glued interaction, measured by IP and the PLA in HEK293T and Rat2 cells, respectively (Figs. 4A–D, S9D). To further confirm that early steps of autophagy are required for the rapamycin-induced interaction of AP-2β and p150Glued, we tested whether the knockdown of Atg5, a key protein for this process, exerts an identical effect as SBI-0206965. Rat2 cells were transfected with siAtg5 and siCtrl for 72 h. Western blot showed that siAtg5 effectively reduced Atg5 levels in transfected cells compared with the control and simultaneously inhibited the autophagy process, indicated by a decrease in the LC3B II/LC3B I ratio (Fig. S9E–G). The silencing of Atg5 also effectively counteracted the rapamycin-induced increase in the p150Glued–AP-2β PLA signal (Fig. 4E, F). Thus, the induction of autophagy is needed for p150Glued–AP-2β protein complex formation upon mTORC1 inhibition.
Autophagy induction upon mTORC1 inhibition is needed for p150Glued–AP-2β interaction. A Western blot analysis of endogenous p150Glued, AP-2β, S6, and P-S6 (Ser235/236) levels and co-immunoprecipitation of endogenous AP-2β with p150Glued in HEK293T cells that were treated with 0.1% DMSO for 2 h, 100 nM rapamycin (RAPA) for 2 h, 25 μM SBI-0206965 for 2 h 30 min, or 25 μM SBI-0206965 for 30 min and 100 nM rapamycin for 2 h (RAPA + SBI-0206965). Input, 10% of lysate used for immunoprecipitation. Shown is a representative example from N = 2 independent experiments. B Results of quantitative analysis of co-immunoprecipitation of experiments performed is as in A. C Representative images of Rat2 fibroblasts with p150Glued–AP-2β PLA signals (magenta), immunofluorescently labeled tubulin (green), and DAPI-stained nuclei (blue). Cells were treated with 0.1% DMSO for 2 h, 100 nM rapamycin (RAPA) for 2 h, 25 μM SBI-0206965 for 2 h 30 min, or 25 μM SBI-0206965 for 30 min and 100 nM rapamycin for 2 h (RAPA + SBI-0206965). Scale bar = 10 μm. D Quantification of the number of p150Glued-AP-2β PLA puncta in cells that were treated as in B. The data are expressed as the mean number of PLA puncta per cell, normalized to the control variant (DMSO) ± SEM. N = 5 independent experiments. n = 184 cells (DMSO), 189 cells (RAPA), 169 cells (SBI-0206965), 174 cells (RAPA + SBI-0206965). *p < 0.05, ns, nonsignificant (one-way ANOVA followed by Bonferroni multiple-comparison post hoc test). E Representative images of Rat2 fibroblasts that were transfected with siCtrl or rat siAtg5 for 72 h and then treated for 2 h with 0.1% DMSO or 100 nM rapamycin (RAPA), with PLA p150Glued–AP-2β signals (magenta), immunofluorescently labeled tubulin (green), and DAPI-stained nuclei (blue). Scale bar = 10 μm. F Quantification of the number of p150Glued–AP-2β PLA puncta in cells that were treated as in D. The data are expressed as the mean number of PLA puncta per cell, normalized to the control variant (siCtrl + DMSO) ± SEM. N = 4 independent experiments. n = 199 cells (siCtrl + DMSO), 178 cells (siCtrl + RAPA), 195 cells (siAtg5 + DMSO), 211 cells (siAtg5 + RAPA). ***p < 0.001, **p < 0.01, ns, nonsignificant (two-way ANOVA followed by Tukey’s multiple-comparison post hoc test)
Autophagy initiation is sufficient for p150 Glued –AP-2β interaction
Our results above show that the initiation of autophagy is essential for effects of mTORC1 inhibition on p150Glued–AP-2β complex formation. However, a key issue is whether autophagy initiation, even when mTORC1 is active, is sufficient to induce a similar effect. To investigate this possibility, we treated Rat2 cells with L-690330, an inhibitor of inositol monophosphatase and mTOR-independent activator of autophagy [77], and performed an p150Glued–AP-2β PLA. After 3 h of L-690330 (100 µM) treatment, autophagy levels increased, indicated by the LC3B II/LC3B I ratio and formation of large LC3B foci, whereas the level of phosphorylated S6 at Ser235/236 did not decrease as expected (Fig. 5A–C). This treatment also increased the p150Glued–AP-2β PLA signal similarly to rapamycin treatment (Fig. 5D, E). Altogether, our results show that autophagy induction alone is sufficient to induce AP-2–dynactin complex formation. Additionally, as in the case of rapamycin treatment, CLIP-170 knockdown blocked the L-690330-induced increase in the p150Glued–AP-2β interaction (Fig. 5F–H). This observation suggests potential novel autophagy-related activities of CLIP-170. Thus, we tested whether CLIP-170 knockdown affects autophagy that is induced by rapamycin or L-690330. The loss of CLIP-170 prevented the increase in the LC3B II/LC3B I ratio and LC3 foci formation that were caused by both autophagy inducers (Fig. 5I–K). These findings suggest that the dynactin interaction with the AP-2 adaptor complex requires autophagy initiation, which depends on the presence of CLIP-170.
p150Glued–AP-2β interaction is induced by autophagy even when mTOR activity is preserved. A Western blot analysis of endogenous actin, LC3B I, LC3B II, P-S6 (Ser235/236), and S6 levels in Rat2 fibroblasts that were treated for 3 h with 0.1% DMSO or 100 μM L-690330. Shown is a representative example from N = 3 independent experiments. B Densitometry analysis of normalized LC3B II/LCB I ratio in Rat2 cells that were treated as in A. The data are presented as mean of the normalized ratio of LC3B II to LC3B I levels ± SEM. N = 3 independent experiments. *p < 0.05 (one-tailed Mann–Whitney test). C Representative images of Rat2 cells that were treated for 3 h with 0.1% DMSO or 100 μM L-690330 with immunofluorescently labeled endogenous LC3B (green) and nuclei stained with Hoechst 33,258 (blue). Scale bar = 10 µm. D Representative images of Rat2 fibroblasts that were treated for 3 h with 0.1% DMSO or 100 μM L-690330 with p150Glued–AP-2β PLA signals (magenta), immunofluorescently labeled tubulin (green), and DAPI-stained nuclei (blue). Scale bar = 10 μm. E Quantification of the number of p150Glued–AP-2β PLA puncta in cells that were treated as in D. The data are expressed as the mean number of PLA puncta per cell, normalized to the control variant (DMSO) ± SEM. N = 5 independent experiments. n = 200 cells (DMSO), 181 cells (L-690330). **p < 0.01 (Student’s t-test). F Western blot analysis of endogenous CLIP-170, p150Glued, and AP-2β levels and co-immunoprecipitation of endogenous AP-2β with p150Glued in HEK293T cells that were transfected with control siRNA (siCtrl) or siRNA against human CLIP-170 (siCLIP-170) and treated for 3 h with 0.1% DMSO or 100 μM L-69330. Input, 10% of lysate used for immunoprecipitation. Shown is a representative example from N = 2 independent experiments. G Representative images of Rat2 fibroblasts that were transfected with siCtrl or rat siCLIP-170 and treated for 3 h with 0.1% DMSO or 100 μM L-690330 with PLA p150Glued–AP-2β signals (magenta), immunofluorescently labeled tubulin (green), and DAPI-stained nuclei (blue). Scale bar = 10 μm. H Quantification of the number of p150Glued–AP-2β PLA puncta in cells that were treated as in G. The data are expressed as the mean number of PLA puncta per cell, normalized to the control variant (siCtrl + DMSO) ± SEM. N = 4 independent experiments. n = 167 cells (siCtrl + DMSO), 163 cells (siCtrl + L-690330), 146 cells (siCLIP-170 + DMSO), 164 cells (siCLIP-170 + L-690330). **p < 0.01, ns, nonsignificant (two-way ANOVA followed by Tukey’s multiple-comparison post hoc test). I Western blot analysis of endogenous CLIP-170, P-S6 (Ser235/236), S6, tubulin, LC3B I, and LC3B II levels in Rat2 fibroblasts that were transfected with siCtrl or rat siCLIP-170 and treated for 2 h with 0.1% DMSO or 100 nM rapamycin (RAPA) or treated for 3 h with 100 μM L-690330. Shown is a representative example from N = 6 independent experiments. J Densitometry analysis of normalized LC3B II/LCB I ratio in Rat2 cells that were treated as in I. The data are presented as mean of the normalized ratio of LC3B II to LC3B I levels ± SEM. N = 6 independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, ns, nonsignificant (two-way ANOVA followed by Tukey’s multiple-comparison post hoc test). K Representative images of Rat2 cells that were transfected with siCtrl or rat siCLIP-170 and treated for 2 h with 0.1% DMSO or 100 nM rapamycin (RAPA) or treated for 3 h with 100 μM L-690330 with immunofluorescently labeled endogenous LC3B (green) and nuclei stained with Hoechst 33258 (blue). Scale bar = 10 µm
ABMA and chloroquine prevent rapamycin-induced p150 Glued –AP-2β interaction
Based on the data that were obtained, the initiation of autophagy appears to play a key role in the p150Glued–AP-2β-protein interaction, but unknown is whether undisturbed autophagic flux is also required. Therefore, we treated cells with rapamycin in the presence of chloroquine (CQ) and ABMA, two compounds that affect this process via different mechanisms [78, 79]. ABMA stimulates the formation of amphisomes, which however are unable to fuse with lysosomes to finish the autophagy [71]. Chloroquine appears to have pleiotropic effects that include direct blockade of the fusion of autophagosomes with lysosomes by preventing the recruitment of SNAP29 to the fusion site, slowing the acidification of lysosomes or disturbing endosomal flow [79, 80]. Rat2 cells were treated with rapamycin in the presence of ABMA (60 µM) or CQ (50 µM) for 2 h. Cells that were treated with DMSO, ABMA, or CQ alone served as controls. Although fluorescence analysis did not show a profound enhancement of LC3B protein cluster formation after treatment with ABMA (Fig. S10A), Western blot confirmed that it increased the LC3B II/LC3B I ratio as expected (Fig. S10B, C) [78]. Treatment with CQ also resulted in autophagic flux inhibition as described previously (Fig. S10D–F) [79]. Both ABMA and CQ blocked the increase in the p150Glued–AP-2β PLA signal that was caused by rapamycin (Fig. 6A–D). ABMA and CQ prevent the fusion of autolysosome-preceding compartments with lysosomes but may also have additional effects on the endomembrane system. Therefore, we tested whether the knockdown of SNAP29, which plays a key role in autophagosome-lysosome fusion [81], affects the rapamycin-induced p150Glued–AP-2β interaction. Rat2 cells were transfected with control siRNA or siRNA against Snap29 (siSnap29) for 72 h. The qRT-PCR analysis of RNA that were isolated from siRNA-transfected cells showed a significant decrease in Snap29 mRNA levels compared with controls (Fig. S10G). Additionally, immunofluorescent staining and Western blot analysis showed an increase in p62/SQSTM1 protein (Fig. S10H–J), which is expected to accumulate in the cell in the absence of Snap29 and with autophagosome-lysosome fusion inhibition. However, Snap29 knockdown did not prevent the enhancement of the p150Glued–AP-2β PLA signal by rapamycin (Fig. 6E, F). Thus, we concluded that both ABMA and CQ effectively prevented the rapamycin-induced p150Glued–AP-2β interaction, but SNAP29-dependent fusion unexpectedly did not appear to be required.
ABMA and chloroquine prevent rapamycin-induced p150Glued–AP-2β interaction. A Representative images of Rat2 fibroblasts that were treated for 2 h with 0.1% DMSO, 100 nM rapamycin (RAPA), 60 μM ABMA (ABMA), or 60 μM ABMA and 100 nM rapamycin (ABMA + RAPA) with p150Glued–AP-2β PLA signals (magenta), immunofluorescently labeled tubulin (green), and DAPI-stained nuclei (blue Scale bar = 10 μm. B Quantification of the number of p150Glued–AP-2β PLA puncta in cells that were treated as in A. The data are expressed as the mean number of PLA puncta per cell, normalized to the control variant (DMSO) ± SEM. N = 4 independent experiments. n = 140 cells (DMSO), 144 cells (RAPA), 148 cells (ABMA), 134 cells (ABMA + RAPA). *p < 0.05, ns nonsignificant (one-way ANOVA followed by Bonferroni multiple-comparison post hoc test). C Representative images of Rat2 fibroblasts that were treated for 2 h with 0.1% DMSO, 100 nM rapamycin (RAPA), 50 μM chloroquine (CQ), or 50 μM chloroquine and 100 nM rapamycin for 2 h (CQ + RAPA) with PLA p150Glued–AP-2β signals (magenta), immunofluorescently labeled tubulin (green), and DAPI-stained nuclei (blue). Scale bar = 10 μm. D Quantification of the number of p150Glued–AP-2β PLA puncta in cells that were treated as in C. The data are expressed as the mean number of PLA puncta per cell, normalized to the control variant (DMSO) ± SEM. N = 3 independent experiments. n = 142 cells (DMSO), 143 cells (RAPA), 135 cells (CQ), 154 cells (CQ + RAPA). **p < 0.01, ns nonsignificant (one-way ANOVA followed by Bonferroni multiple-comparison post hoc test). E Representative images of Rat2 fibroblasts that were transfected with siCtrl or rat siSnap29 for 72 h and then treated for 2 h with 0.1% DMSO or 100 nM rapamycin (RAPA), with p150Glued–AP-2β PLA signals (magenta), immunofluorescently labeled tubulin (green), and DAPI-stained nuclei (blue). Scale bar = 10 μm. F Quantification of the number of p150Glued–AP-2β PLA puncta in cells that were treated as in E. The data are expressed as the mean number of PLA puncta per cell, normalized to the control variant (siCtrl + DMSO) ± SEM. N = 4 independent experiments. n = 172 cells (siCtrl + DMSO), 161 cells (siCtrl + RAPA), 172 cells (siSnap29 + DMSO), 179 cells (siSnap + RAPA). *p < 0.05, ns nonsignificant (two-way ANOVA followed by Tukey’s multiple-comparison post hoc test). G Representative images of Rat2 fibroblasts that were treated with 0.1% DMSO for 2 h, 100 nM rapamycin (RAPA) for 2 h, 20 mM NH4Cl (NH4Cl) for 3 h, or pretreated with 20 mM NH4Cl for 1 h and treated with 100 nM rapamycin for 2 h (NH4Cl + RAPA) with p150Glued-AP-2β PLA signals (magenta), immunofluorescently labelled tubulin (green), and DAPI-stained nuclei (blue). Scale bar = 10 μm. H Quantification of the number of p150Glued–AP-2β PLA puncta in cells that were treated as in G. The data are expressed as the mean number of PLA puncta per cell, normalized to the control variant (DMSO) ± SEM. N = 3 independent experiments, n = 122 cells (DMSO), 109 cells (RAPA), 125 cells (NH4Cl), 114 cells (NH4Cl + RAPA). *p < 0.05, ns nonsignificant (one-way ANOVA followed by Bonferroni multiple-comparison post hoc test)
Because CQ can alkalize the lysosome environment [80] and because ABMA was not tested for it in Rat2 cells, we used lysotracker staining to investigate effects of rapamycin, CQ, ABMA, and their combination on lysosomal acidification in our experimental model. As a control, we treated cells for 2 h with the vATPase inhibitor Baf A1. Two hours of treatment with rapamycin significantly increased the intensity of lysotracker staining and the number of positive lysotracker structures (Fig. S11). In contrast, both ABMA and CQ alone and in the presence of rapamycin significantly decreased both parameters (Fig. S11). However, effects of CQ and ABMA were different. Chloroquine had a stronger effect that was comparable to treatment with Baf A1, while influence of ABMA was much milder (Fig. S11). Thus, we concluded that although autophagosome fusion with the endolysosomal pathway did not affect the p150Glued–AP-2β interaction, ABMA and CQ might affect it, leading to improper lysosome acidification or disruption of the endolysosomal pathway as previously shown [78, 79]. To test the hypothesis that the correct pH in the cell is necessary for the rapamycin-induced interaction of p150Glued and AP-2β, we repeated the experiment, this time treating the cells with 20 mM NH4Cl to alkalize the cells before administering rapamycin and running PLA (Fig. 6G, H). As in the previous experiments, rapamycin caused an increase in p150Glued–AP-2β PLA. At the same time, administration of NH4Cl under basal conditions had no significant effect on PLA signal. However, incubation with NH4Cl counteracted the increase in p150Glued–AP-2β PLA in response to rapamycin administration, supporting the hypothesis that appropriate acidification of the endolysosomal pathway is critical for the interaction of p150Glued and AP-2β.
Rapamycin enhances the lysosomal p150 Glued –AP-2β interaction and affects Lamp-1 mobility
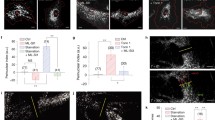
Based on the above observations, we investigated where in Rat2 cells p150Glued and AP-2β interact. We used the PLA that was adjusted for EM, revealing that the p150Glued–AP-2β PLA signal in rapamycin-treated Rat2 cells localized primarily to organelles that contained electron dense material that is characteristic for lysosomes (43% of cells with PLA-EM signal) and in double-membrane organelles, which we classified as autophagosomes (57% of cells with PLA-EM signal). In control, in DMSO-treated cells, the PLA-EM signal was also spotted but less frequently and rarely on lysosome-like structures (Fig. 7A; 14% vs. 43% of cells with PLA-EM signal; see also Fig. S12). Because the PLA procedure did not allow the preservation of a high-quality ultrastructure, we additionally analyzed in rapamycin-treated Rat2 cells the co-occurrence of the p150Glued–AP-2β PLA signal with the LC3B or lysosomal marker Lamp-1, either endogenous or overexpressed as a GFP fusion, using AiryScan high-resolution light microscopy. For LC3B, some co-occurrence was detected but not frequently (Fig. S13). For Lamp-1, we noticed an apparent, although still partial, localization of the PLA signal in proximity of lysosomes (19.86% ± 2.76% of PLA signals in proximity of Lamp1-GFP objects, N = 2 independent experiments, number of cells n = 26 [DMSO] and 22 [RAPA]) (Fig. 7B, C). Thus, the combined observations from PLA-EM and Airyscan images indicated that lysosomes are the primary localization of the p150Glued–AP-2β interaction under rapamycin treatment, raising the issue of whether rapamycin affects the mobility of Lamp-1-positive compartments.
Rapamycin induces p150Glued-AP-2β interaction on lysosomes and regulates lysosomal mobility. A Representative electron microscopy images of Rat2 fibroblasts after 2 h treatment with 0.1% DMSO or 100 nM rapamycin (RAPA) and PLA analysis of p150Glued–AP-2β with and without (negative control) primary antibodies. White arrows point to PLA signals that co-occurred with organelles that resembled lysosomes or autophagosomes. Black boxes indicate the regions shown in higher magnification. n = 30 cells per variant. N = 3 independent experiments. Scale bar = 500 nm. B Representative images of Rat2 cells treated with 100 nM rapamycin (RAPA) with p150Glued–AP-2β PLA signals (magenta) immunofluorescently labeled endogenous Lamp1 (green) and DAPI-stained nuclei (blue). Images were acquired using the AiryScan module. Scale bar = 10 μm. (Upper panel) Representative photograph of a single cell. (Lower panel) Close-up of a site with a high intensity of PLA-Lamp1 co-localization. C Representative images of Rat2 cells transfected with Lamp1-GFP (green) for 24 h and then treated with 100 nM rapamycin (RAPA) with p150Glued–AP-2β PLA signals (magenta), and DAPI-stained nuclei (blue). Images were acquired using the AiryScan module. Scale bar = 10 μm. (Top) Representative photograph of a single cell. (Bottom) Close-up of a site with a high intensity of PLA-Lamp1-GFP co-localization. D Representative images of cells that expressed Lamp1-GFP. The upper row shows the first frames from the time-lapse movies. The area inside the golden circle is considered the “center” compartment, and all movements outside this area are “peripheries.” The lower row shows trajectories (tracks) that were identified by the ImageJ “TrackMate” plugin that were longer than 6.8 mm (100 pixels). Trajectories were color-coded based on their directions, which were established using Pearson correlation coefficient (PCC) calculated by change in the distance from the cell center in time. If the distance was increasing with consecutive frames (PCC: 0.5 to 1) tracks were considered to move to the cell membrane. If distance was decreasing (PCC: − 1 to − 0.5), direction was described as moving to the center. Values in between were marked as oscillating. E Difference in speed of Lamp1-GFP vesicles' movements between rapamycin-treated (RAPA) and control (DMSO) cells. The values are mean trajectories that were identified by the ImageJ “TrackMate” plugin that were at least 6.8 µm (100 pixels) long, divided according to their initial location (center or periphery) and direction (center or cell membrane) as indicated above the graphs. The single dot represents the mean value from one measured cell. N = 4 independent experiments. n = 25 cells for both RAPA and DMSO. *p < 0.05, ns, nonsignificant (Mann–Whitney test)
We found that rapamycin significantly increased lysosome acidification (Fig. S11), and lysosome acidification was previously related to their position in the cell [82]. Therefore, we re-analyzed the data from the experiments with lysotracker staining described in the previous section, but this time in the context of the cellular distribution of the signal. For this purpose, we defined the perinuclear region (2.5 µm from the nuclear edge) and the peripheral region (2.5 or 7.5 µm from the plasma membrane). In the case of the restricted peripheral zone (2.5 µm from the PM), the intensity of the lysotracker spots was lower in the peripheral zone than in the perinuclear zone (Fig. S14; see also https://iimcb-lmcn-2024.shinyapps.io/shiny/ for more depth exploration of these data in R Shiny application for interactive data visualization). This was probably due to differences in the size of the spots at these two sites (Fig. S14). When we increased the area of the peripheral zone to 7.5 µm, the average size in the perinuclear and peripheral zones became comparable and the intensity was similar in both zones (Fig. S14). However, treatment with rapamycin increased the lysotracker intensity in both zones, regardless of the area of the peripheral zone. The number of lysotracker spots also increased in the peripheral zone after rapamycin treatment, although the spot size remained virtually unchanged (Fig. S14). From this analysis, we concluded that rapamycin treatment increases the pool of acidified organelles in the call also at the cell periphery. Therefore, we next investigated whether the rapamycin-induced p150Glued–AP-2β interaction could contribute to the mobility of lysosomes. We performed the live imaging of Rat2 cells that were transfected with Lamp-1-GFP and treated with 100 nM rapamycin for 2 h. Two distinct populations of Lamp-1-GFP objects were spotted. One aggregated near the cell center, and the other was more motile around the peripheries. When we focused on the latter one, we observed that rapamycin increased the speed of long-distance movements (> 6 µm) toward the cell center. Movement from the central Lamp-1-positive organelle pool toward the cell periphery was unaffected (Fig. 7D, E, Movies 19, 20). In the last series of experiments, we investigated whether the prevention of autophagy initiation, lysosome acidification or dynein function will result in decrease of long-distance Lamp-1-GFP movement velocity induced by rapamycin. To this end, we treated the cells with rapamycin in combination with SBI-0206965, BafA or ciliobrevin D (30 µm, 30 min before imaging and 1.5 h after addition of rapamycin). In all three cases, we did not observe any effects of the drugs on velocity of long-distance movement of Lamp-1 vesicles towards the cell center. However, all three drugs reduced the number of Lamp-1-GFP objects moving towards the cell center. In the case of BafA and ciliobrevin, the effect was statistically significant (Rapa vs. BafA: 35.41 ± 3.112 [RAPA], 23.08 ± 2.706 [BafA], p < 0.01 in Student’s t-test; Rapa vs. ciliobrevin: 35.41 ± 3.112 [RAPA], 26.44 ± 3.085 [ciliobrevin]), p < 0.05 in Mann–Whitney test; N = 3 independent experiments; n = 27 cells [RAPA], 26 cells [BafA], 25 cells [ciliobrevin]), while in the case of SBI-0206965 the difference did not reach statistical significance (30.16 ± 2.902, N = 3 independent experiments; n = 25).
Discussion
Recent work demonstrated that AP-2 is an adaptor protein for the dynein-dynactin transport of amphisomes along neuronal axons [22, 24]. To date, however, important questions about mechanistic details of the regulation of the dynactin–AP-2 interaction have not been answered. Here, we demonstrate that AP-2 and dynactin cooperate in neurons and non-neuronal cells under autophagy-permissive conditions, including, but not limited to, mTOR inactivation. Furthermore, we show that the co-occurrence of AP-2β with p150Glued does not require binding the latter to microtubule plus ends or microtubule dynamics. However, this interaction requires the presence of CLIP-170, which contributes to autophagy initiation. Finally, we show that the autophagy-induced p150Glued–AP-2β interaction likely occurs on lysosomes, possibly increasing their mobility toward autophagosomes in the perinuclear area (Fig. 8).
Postulated mechanism of autophagy initiation-induced recruitment of p150Glued and AP-2β to lysosomes. Administration of rapamycin or L-69030 initiates autophagosome formation (AP), which depends on the presence of CLIP-170. Simultaneously, autophagy inducers lead to a decrease in lysosomal pH (LY pH) and recruitment of the dynatin-AP2 complex to the lysosome (LY), presumably leading to intensification of LY retrograde transport to the perinuclear region and subsequent LY fusion with the mature AP. knd knockdown, SBI-0206965 - Ulk inhibitor
Autophagy induction and a properly functioning endolysosomal pathway are essential for the p150Glued–AP-2β interaction
The initial finding that stimulated our research was the observation that rapamycin enhanced AP-2–dynactin complex formation in neurons. This discovery raised the issue of how mTORC1 inhibition stimulates this interaction. The present findings show that AP-2β or p150Glued is not an mTORC1 substrate, instead supporting the scenario that mTORC1 inhibition promotes p150Glued–AP-2β complex formation via autophagy initiation (Fig. 4). Moreover, the pharmacological induction of autophagy that does not involve mTORC1 inhibition was sufficient to induce this interaction (Fig. 5), indicating that the direct trigger for the p150Glued–AP-2 interaction is autophagy. Furthermore, it explains why the coexistence of AP-2β and p150Glued is readily visible in axons, whereas it is barely detectable in non-neuronal cells under basal conditions. In cultured neurons, axonal autophagy is relatively high and stable at a steady-state level. Autophagosomes are continuously formed at the axonal growth cone or at presynaptic sites and likely fuse with late endosomes, forming amphisomes, and are transported toward the cell soma [24, 28, 45, 83,84,85]. Thus, there is a continual need for AP-2–dynactin complexes in axons to transport amphisomes [24]. However, if axonal autophagy is constitutive and if autophagy is sufficient to drive the p150Glued–AP-2β interaction, then how does mTORC1 inhibition potentiate it? Although some studies show that mTORC1 inhibition does not increase autophagy in neurons [41, 42], other studies reported that it is nevertheless possible [43, 44]. Such observations show that this process can still be upregulated, despite its relatively high basal level. However, we cannot exclude the possibility that the effects of rapamycin we observed are due to enhanced autophagy in the neuronal perikaryon and not in the axon, which is transferred to axonal amphisomes, although the mechanism (e.g. cytoplasmic or vesicle pH change, see further discussion) remains to be defined. In contrast to neuronal axons, autophagy in many cultured non-neuronal cells is at a low level under basal conditions, and its induction requires additional stimuli (e.g., mTORC1 inactivation). Consequently, the degree of the p150Glued–AP-2β interaction in such cells is likely to be adjusted to the current level of autophagy in the cell. In the context of requiring the initiation of autophagy for the p150Glued–AP-2β interaction, it is also an interesting observation that this interaction requires the presence of CLIP-170. However, its important canonical functions (e.g., the regulation of microtubule dynamics) seem unnecessary. In contrast, we have shown that CLIP-170 is required for proper autophagy, but unclear is how CLIP-170 regulates this process. Thus, determining precisely how CLIP-170 regulates autophagy will require further studies.
Our previous experiments showed that AP-2β interacts with p150Glued in neurons to transport amphisomes. Thus, we investigated whether unperturbed autophagosomal flux and the fusion of autophagosomes with organelles of the endolysosomal pathway are also required for this interaction in non-neuronal cells. Both inhibitors of these processes that we used (CQ and ABMA) significantly prevented the increase in the p150Glued–AP-2β PLA signal. This result may suggest that unperturbed autophagic flux is indeed crucial. Because both ABMA and CQ counteract the fusion of autophagosomes and amphisomes with lysosomes, this could suggest that this step is critical. Indeed, previous studies showed that AP-2 attaches to autolysosomes when autophagic lysosome reformation (ALR) is initiated. Autophagic lysosome reformation is a process that links autophagy, lysosomes, and AP-2 [86]. During ALR, protolysosomes emerge from autolysosomes and mature into lysosomes. Protolysosome formation requires clathrin and AP-2 [26]. Because ALR requires the merging of autophagosomes with the endolysosomal pathway, this would support the importance of this event for the p150Glued–AP-2β interaction. However, the lack of an effect of SNAP29 knockdown on the p150Glued–AP-2β interaction likely excludes this possibility. Thus, a question arises about how ABMA and CQ can block the p150Glued–AP-2β interaction. A common feature of these two compounds is their negative effect on the normal ultrastructure of the endolysosomal pathway [78, 79]. Our results clearly demonstrate that in Rat2 cells both ABMA and CQ lead to a significant decrease in lysotracker-positive organelles and a decrease in its intensity in the remaining ones. A similar effect of CQ on the pH of acidic organelles has been reported [80], although its absence has also been reported [79], suggesting that this effect may depend on the cell type. ABMA is a relatively recently described compound [78, 87], and its effect may also depend on the cell line and should always be tested experimentally. Thus, our results suggest that formation of the p150Glued–AP-2β complex requires proper function of the endocytic pathway and/or lysosome acidification. The latter possibility is further supported by the results of experiments with NH4Cl, which directly indicate that alkalinization of the cell prevents the increase in the p150Glued–AP-2β interaction upon rapamycin administration that initiates autophagy.
Autophagy-induced p150Glued-AP-2β interaction occurs on lysosomes
Our results indicate that autophagy is a critical cellular process for the interaction of AP-2β with p150Glued. Nevertheless, our results did not identify autophagosomes as the primary p150Glued–AP-2β interaction site in non-neuronal cells. An important question is where the p150Glued–AP-2β interaction occurs upon autophagy induction. As mentioned above, the AP-2–dynactin complex in neurons is involved in the transport of signaling amphisomes in axons [24]. However, in non-neuronal cells, amphisomes are considered temporary structures [32, 88]. Moreover, ABMA, which potentiates their formation [78], did not enhance the p150Glued–AP-2β interaction. Finally, our PLA-EM and PLA-AiryScan confocal microcopy findings in Rat2 cells revealed a p150Glued–AP-2β PLA signal that often co-occurred with lysosomes. Notably, the presence of lysosomal AP-2 was previously reported and not only in the context of ALR, which corroborates our observation [89]. Thus, another issue emerges about the purpose of p150Glued–AP-2β complex formation on lysosomes.
Korolchuk et al. [82] showed that the dynein-dynactin-dependent transport of lysosomes toward the cell nucleus under cell starvation conditions is crucial for their pH regulation and fusion with autophagosomes. Hence, recruitment of the p150Glued–AP-2β complex to lysosomes at the onset of autophagy may be designed to ensure the proper fusion of these two organelles at the end of the process. A similar mechanism was previously reported for another microtubular transport lysosomal adaptor, ALG2 [51]. Under amino acid starvation or mTOR inhibition conditions, ALG2 and dynein are recruited to lysosomes in a Ca2+-dependent manner for their retrograde transport. Previous studies showed that RILP protein is responsible for lysosome transport by dynein-dynactin in response to changes in cholesterol levels [90]. This suggests that non-neuronal cells use several transport systems for one organelle, depending on the cellular conditions. This raises the question about what could trigger p150Glued–AP-2β complex binding to lysosomes upon mTOR inhibition. Based on our findings that rapamycin increased and CQ and ABMA decreased lysotracker fluorescence intensity, a tempting speculation is that changes in pH could serve as one such mechanism (Fig. 8), but further research is needed to confirm this possibility.
Our results suggest that the sequence of events in non-neuronal cells provides a coherent mechanism to support the processivity of autophagy. In contrast, it is unclear how and to what end mTOR inhibition via upregulation of autophagy would support the autophagosome/amphisome trafficking in axons that we have previously described. The current model suggests that autophagosomes, which are formed constitutively in axon terminals and mature by fusion with late endosomes, are transported as amphisomes toward the cell body, lowering their pH as they approach the cell body, where they eventually fuse with lysosomes [91,92,93]. In this model, axonal transport of lysosomes, which is crucial for autophagosome maturation, depends mainly on kinesins. However, our previous data indicated that AP2-dynactin colocalizes with amphisomes in axons and not with lysosomes as in non-neuronal cells [24]. This could be related, for example, to the fact that amphisomes are very volatile structures in the latter, whereas they have an important transport function in neuronal axons. It is therefore possible that the interaction we investigated takes place in different cell types at different organelles that are critical for the correct autophagy flux. However, why autophagy would lead to an increase in the presence of motors that transport TrkB-containing vesicles towards the cell body is difficult to explain given the available and often conflicting data on axonal autophagosome transport, particularly in the context of TrkB transport [24, 28, 83, 94, 95]). Interestingly, data of Sidibe et al. and our unpublished observations suggest that retrograde transport of autophagosomes [94] or structures positive for AP2-p150Glued or TrkB (Tempes, unpublished data) is enhanced by BDNF administration. It is therefore possible that when autophagy is increased in a cell, the transport of autophagosomes/amphisomes and TrkB into the cell body is prioritized to utilize their contents more efficiently or to block the signaling of TrkB. At the same time, this could prevent signaling amphisomes from remaining at the en passant synapses [28] under conditions that are unfavorable for their intensive function. Further studies are certainly needed to understand how the mechanism we have described in detail in non-neuronal cells is transferable to the development and proper functioning of neurons.
In summary, our study provides new insights into the mechanisms that regulate formation of the p150Glued–AP-2β complex, which is essential for cargo transport along microtubules. Importantly, we showed that autophagy initiation is necessary and sufficient to trigger the formation of this complex. This finding exemplifies a basic mechanism that allows the coordination of various elements that are involved in a vital cellular process.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary materials. Raw data from all quantitatively analyzed experiments are available from the corresponding author upon reasonable request.
Abbreviations
- ABMA:
-
1-Adamantyl(5-bromo-2-methoxybenzyl)amine
- ANOVA:
-
Analysis of variance
- ALR:
-
Autophagic lysosome reformation
- Arp-1:
-
Actin-related protein 1
- AP-2:
-
Adaptor protein 2
- Baf A1:
-
Bafilomycin A1
- BSA:
-
Bovine serum albumin
- CHAPS:
-
3-([3-Cholamidopropyl]dimethylammonio)-1-propanesulfonate
- CHX:
-
Cycloheximide
- CLIP-115:
-
Cytoplasmic linker protein 115
- CLIP-170:
-
Cytoplasmic linker protein 170
- CQ:
-
Chloroquine
- DIV:
-
Day in vitro
- DMEM:
-
Dulbecco’s modified Eagle’s medium
- DMSO:
-
Dimethylsulfoxide
- EB3:
-
End binding protein 3
- EDTA:
-
Ethylenediamine tetraacetic acid
- EGFP:
-
Enhanced green fluorescent protein
- EGTA:
-
Ethylene glycol-bis(β-aminoethyl ether)-N,N,N’,N’-tetraacetic acid
- EM:
-
Electron microscopy
- FBS:
-
Fetal bovine serum
- GST:
-
Glutathione-S-transferase
- IP:
-
Immunoprecipitation
- HRP:
-
Horseradish peroxidase
- Lamp-1:
-
Lysosomal-associated membrane protein 1
- LC3:
-
Microtubule-associated protein 1A/1B-light chain 3
- mTORC1:
-
Mammalian/mechanistic target of rapamycin complex 1
- PBS:
-
Phosphate-buffered saline
- PCR:
-
Polymerase chain reaction
- PFA:
-
Paraformaldehyde
- PLA:
-
Proximity ligation assay
- S6:
-
Ribosomal protein S6
- RILP:
-
Rab-interacting lysosomal protein
- SDS-PAGE:
-
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- + TIP:
-
Plus end tracking protein
- TrkB:
-
Tropomyosin receptor kinase B
- vATPase:
-
Vacuolar (H +)-adenosine triphosphatase
References
Britt DJ, Farías GG, Guardia CM, Bonifacino JS (2016) Mechanisms of polarized organelle distribution in neurons. Front Cell Neurosci 10:88. https://doi.org/10.3389/fncel.2016.00088
Maday S, Twelvetrees AE, Moughamian AJ, Holzbaur ELF (2014) Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron 84:292–309. https://doi.org/10.1016/j.neuron.2014.10.019
Desai A, Mitchison TJ (1997) Microtubule polymerization dynamics. Annu Rev Cell Dev Biol 13:83–117. https://doi.org/10.1146/annurev.cellbio.13.1.83
Hirokawa N, Noda Y, Tanaka Y, Niwa S (2009) Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol 10:682–696. https://doi.org/10.1038/nrm2774
Schroer TA (2004) Dynactin. Annu Rev Cell Dev Biol 20:759–779. https://doi.org/10.1146/annurev.cellbio.20.012103.094623
Höök P, Vallee RB (2006) The dynein family at a glance. J Cell Sci 119:4369–4371. https://doi.org/10.1242/jcs.03176
Reck-Peterson SL, Redwine WB, Vale RD, Carter AP (2018) The cytoplasmic dynein transport machinery and its many cargoes. Nat Rev Mol Cell Biol 19:382–398. https://doi.org/10.1038/s41580-018-0004-3
Moughamian AJ, Osborn GE, Lazarus JE, Maday S, Holzbaur ELF (2013) Ordered recruitment of dynactin to the microtubule plus-end is required for efficient initiation of retrograde axonal transport. J Neurosci 33:13190–13203. https://doi.org/10.1523/JNEUROSCI.0935-13.2013
Ross JL, Wallace K, Shuman H, Goldman YE, Holzbaur ELF (2006) Processive bidirectional motion of dynein-dynactin complexes in vitro. Nat Cell Biol 8:562–570. https://doi.org/10.1038/ncb1421
Chowdhury S, Ketcham SA, Schroer TA, Lander GC (2015) Structural organization of the dynein-dynactin complex bound to microtubules. Nat Struct Mol Biol 22:345–347. https://doi.org/10.1038/nsmb.2996
Urnavicius L, Zhang K, Diamant AG, Motz C, Schlager MA, Yu M, Patel NA, Robinson CV, Carter AP (2015) The structure of the dynactin complex and its interaction with dynein. Science 347:1441–1446. https://doi.org/10.1126/science.aaa4080
Carter AP, Diamant AG, Urnavicius L (2016) How dynein and dynactin transport cargos: a structural perspective. Curr Opin Struct Biol 37:62–70. https://doi.org/10.1016/j.sbi.2015.12.003
Fu M, Holzbaur ELF (2014) Integrated regulation of motor-driven organelle transport by scaffolding proteins. Trends Cell Biol 24:564–574. https://doi.org/10.1016/j.tcb.2014.05.002
Hoogenraad CC, Akhmanova A (2016) Bicaudal D family of motor adaptors: linking dynein motility to cargo binding. Trends Cell Biol 26:327–340. https://doi.org/10.1016/j.tcb.2016.01.001
Yeh T-Y, Quintyne NJ, Scipioni BR, Eckley DM, Schroer TA (2012) Dynactin’s pointed-end complex is a cargo-targeting module. Mol Biol Cell 23:3827–3837. https://doi.org/10.1091/mbc.E12-07-0496
Vaughan KT, Tynan SH, Faulkner NE, Echeverri CJ, Vallee RB (1999) Colocalization of cytoplasmic dynein with dynactin and CLIP-170 at microtubule distal ends. J Cell Sci 112(Pt 10):1437–1447. https://doi.org/10.1242/jcs.112.10.1437
Lansbergen G, Komarova Y, Modesti M, Wyman C, Hoogenraad CC, Goodson HV, Lemaitre RP, Drechsel DN, van Munster E, Gadella TW Jr, Grosveld F, Galjart N, Borisy GG, Akhmanova A (2004) Conformational changes in CLIP-170 regulate its binding to microtubules and dynactin localization. J Cell Biol 166:1003–1014. https://doi.org/10.1083/jcb.200402082
Watson P, Stephens DJ (2006) Microtubule plus-end loading of p150(Glued) is mediated by EB1 and CLIP-170 but is not required for intracellular membrane traffic in mammalian cells. J Cell Sci 119:2758–2767. https://doi.org/10.1242/jcs.02999
Nirschl JJ, Magiera MM, Lazarus JE, Janke C, Holzbaur ELF (2016) α-Tubulin tyrosination and CLIP-170 phosphorylation regulate the initiation of dynein-driven transport in neurons. Cell Rep 14:2637–2652. https://doi.org/10.1016/j.celrep.2016.02.046
Collins BM, McCoy AJ, Kent HM, Evans PR, Owen DJ (2002) Molecular architecture and functional model of the endocytic AP2 complex. Cell 109:523–535
Traub LM (2003) Sorting it out: AP-2 and alternate clathrin adaptors in endocytic cargo selection. J Cell Biol 163:203–208. https://doi.org/10.1083/jcb.200309175
Bera S, Camblor-Perujo S, Calleja Barca E, Negrete-Hurtado A, Racho J, De Bruyckere E, Wittich C, Ellrich N, Martins S, Adjaye J, Kononenko NL (2020) AP-2 reduces amyloidogenesis by promoting BACE1 trafficking and degradation in neurons. EMBO Rep 21:47954
Imai K, Hao F, Fujita N, Tsuji Y, Oe Y, Araki Y, Hamasaki M, Noda T, Yoshimori T (2016) Atg9A trafficking through the recycling endosomes is required for autophagosome formation. J Cell Sci 129:3781–3791. https://doi.org/10.1242/jcs.196196
Kononenko NL, Claßen GA, Kuijpers M, Puchkov D, Maritzen T, Tempes A, Malik AR, Skalecka A, Bera S, Jaworski J, Haucke V (2017) Retrograde transport of TrkB-containing autophagosomes via the adaptor AP-2 mediates neuronal complexity and prevents neurodegeneration. Nat Commun 8:14819. https://doi.org/10.1038/ncomms14819
Popovic D, Dikic I (2014) TBC1D5 and the AP2 complex regulate ATG9 trafficking and initiation of autophagy. EMBO Rep 15:392–401. https://doi.org/10.1002/embr.201337995
Rong Y, Liu M, Ma L, Du W, Zhang H, Tian Y, Cao Z, Li Y, Ren H, Zhang C, Li L, Chen S, Xi J, Yu L (2012) Clathrin and phosphatidylinositol-4,5-bisphosphate regulate autophagic lysosome reformation. Nat Cell Biol 14:924–934. https://doi.org/10.1038/ncb2557
Tian Y, Chang JC, Fan EY, Flajolet M, Greengard P (2013) Adaptor complex AP2/PICALM, through interaction with LC3, targets Alzheimer’s APP-CTF for terminal degradation via autophagy. Proc Natl Acad Sci USA 110:17071–17076. https://doi.org/10.1073/pnas.1315110110
Andres-Alonso M, Ammar MR, Butnaru I, Gomes GM, Acuña Sanhueza G, Raman R, Yuanxiang P, Borgmeyer M, Lopez-Rojas J, Raza SA, Brice N, Hausrat TJ, Macharadze T, Diaz-Gonzalez S, Carlton M, Failla AV, Stork O, Schweizer M, Gundelfinger ED, Kneussel M, Spilker C, Karpova A, Kreutz MR (2019) SIPA1L2 controls trafficking and local signaling of TrkB-containing amphisomes at presynaptic terminals. Nat Commun 10:5448. https://doi.org/10.1038/s41467-019-13224-z
Dikic I, Elazar Z (2018) Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol 19:349–364. https://doi.org/10.1038/s41580-018-0003-4
Levine B, Klionsky DJ (2004) Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 6:463–477
Yin Z, Pascual C, Klionsky D (2016) Autophagy: machinery and regulation. Microb Cell 3:588–596
Zhao YG, Codogno P, Zhang H (2021) Machinery, regulation and pathophysiological implications of autophagosome maturation. Nat Rev Mol Cell Biol 22:733–750. https://doi.org/10.1038/s41580-021-00392-4
Jung CH, Ro S-H, Cao J, Otto NM, Kim D-H (2010) mTOR regulation of autophagy. FEBS Lett 584:1287–1295. https://doi.org/10.1016/j.febslet.2010.01.017
Russell RC, Yuan H-X, Guan K-L (2014) Autophagy regulation by nutrient signaling. Cell Res 24:42–57. https://doi.org/10.1038/cr.2013.166
Ganley IG, Lam DH, Wang J, Ding X, Chen S, Jiang X (2009) ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 284:12297–12305. https://doi.org/10.1074/jbc.M900573200
Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan J-L, Oshiro N, Mizushima N (2009) Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 20:1981–1991. https://doi.org/10.1091/mbc.E08-12-1248
Huang H, Ouyang Q, Zhu M, Yu H, Mei K, Liu R (2021) mTOR-mediated phosphorylation of VAMP8 and SCFD1 regulates autophagosome maturation. Nat Commun 12:6622. https://doi.org/10.1038/s41467-021-26824-5
Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH (2009) ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 20:1992–2003. https://doi.org/10.1091/mbc.E08-12-1249
Khobrekar NV, Quintremil S, Dantas TJ, Vallee RB (2020) The Dynein adaptor RILP controls neuronal autophagosome biogenesis, transport, and clearance. Dev Cell 53:141-153.e4. https://doi.org/10.1016/j.devcel.2020.03.011
Kim Y-M, Jung CH, Seo M, Kim EK, Park J-M, Bae SS, Kim D-H (2015) mTORC1 phosphorylates UVRAG to negatively regulate autophagosome and endosome maturation. Mol Cell 57:207–218. https://doi.org/10.1016/j.molcel.2014.11.013
Fox JH, Connor T, Chopra V, Dorsey K, Kama JA, Bleckmann D, Betschart C, Hoyer D, Frentzel S, Difiglia M, Paganetti P, Hersch SM (2010) The mTOR kinase inhibitor Everolimus decreases S6 kinase phosphorylation but fails to reduce mutant huntingtin levels in brain and is not neuroprotective in the R6/2 mouse model of Huntington’s disease. Mol Neurodegener 5:26. https://doi.org/10.1186/1750-1326-5-26
Tsvetkov AS, Miller J, Arrasate M, Wong JS, Pleiss MA, Finkbeiner S (2010) A small-molecule scaffold induces autophagy in primary neurons and protects against toxicity in a Huntington disease model. Proc Natl Acad Sci U S A 107:16982–16987. https://doi.org/10.1073/pnas.1004498107
Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA (2008) Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. J Neurosci 28:6926–6937. https://doi.org/10.1523/JNEUROSCI.0800-08.2008
Hernandez D, Torres CA, Setlik W, Cebrián C, Mosharov EV, Tang G, Cheng H-C, Kholodilov N, Yarygina O, Burke RE, Gershon M, Sulzer D (2012) Regulation of presynaptic neurotransmission by macroautophagy. Neuron 74:277–284. https://doi.org/10.1016/j.neuron.2012.02.020
Cason SE, Carman PJ, Van Duyne C, Goldsmith J, Dominguez R, Holzbaur ELF (2021) Sequential dynein effectors regulate axonal autophagosome motility in a maturation-dependent pathway. J Cell Biol 220:e202010179. https://doi.org/10.1083/jcb.202010179
Kimura S, Noda T, Yoshimori T (2008) Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct Funct 33:109–122. https://doi.org/10.1247/csf.08005
Stavoe AKH, Holzbaur ELF (2019) Autophagy in Neurons. Annu Rev Cell Dev Biol 35:477–500. https://doi.org/10.1146/annurev-cellbio-100818-125242
Cabukusta B, Neefjes J (2018) Mechanisms of lysosomal positioning and movement. Traffic 19:761–769. https://doi.org/10.1111/tra.12587
Pu J, Guardia CM, Keren-Kaplan T, Bonifacino JS (2016) Mechanisms and functions of lysosome positioning. J Cell Sci 129:4329–4339. https://doi.org/10.1242/jcs.196287
Bouhamdani N, Comeau D, Turcotte S (2021) A compendium of information on the lysosome. Front Cell Dev Biol 9:798262. https://doi.org/10.3389/fcell.2021.798262
Li X, Rydzewski N, Hider A, Zhang X, Yang J, Wang W, Gao Q, Cheng X, Xu H (2016) A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat Cell Biol 18:404–417. https://doi.org/10.1038/ncb3324
Willett R, Martina JA, Zewe JP, Wills R, Hammond GRV, Puertollano R (2017) TFEB regulates lysosomal positioning by modulating TMEM55B expression and JIP4 recruitment to lysosomes. Nat Commun 8:1580. https://doi.org/10.1038/s41467-017-01871-z
Fu M-M, Nirschl JJ, Holzbaur ELF (2014) LC3 binding to the scaffolding protein JIP1 regulates processive dynein-driven transport of autophagosomes. Dev Cell 29:577–590. https://doi.org/10.1016/j.devcel.2014.04.015
Malik AR, Urbanska M, Gozdz A, Swiech LJ, Nagalski A, Perycz M, Blazejczyk M, Jaworski J (2013) CYR61, a matricellular protein, is needed for dendritic arborization of hippocampal neurons. J Biol Chem. https://doi.org/10.1074/jbc.M112.411629
de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, Strouboulis J (2003) Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc Natl Acad Sci U S A 100:7480–7485. https://doi.org/10.1073/pnas.1332608100
Urbanska AS, Janusz-Kaminska A, Switon K, Hawthorne AL, Perycz M, Urbanska M, Bassell GJ, Jaworski J (2017) ZBP1 phosphorylation at serine 181 regulates its dendritic transport and the development of dendritic trees of hippocampal neurons. Sci Rep 7:1876. https://doi.org/10.1038/s41598-017-01963-2
Koscielny A, Malik AR, Liszewska E, Zmorzynska J, Tempes A, Tarkowski B, Jaworski J (2018) Adaptor complex 2 controls dendrite morphology via mTOR-dependent expression of GluA2. Mol Neurobiol 55:1590–1606. https://doi.org/10.1007/s12035-017-0436-3
Swiech L, Blazejczyk M, Urbanska M, Pietruszka P, Dortland BR, Malik AR, Wulf PS, Hoogenraad CC, Jaworski J (2011) CLIP-170 and IQGAP1 cooperatively regulate dendrite morphology. J Neurosci 31:4555–4568. https://doi.org/10.1523/JNEUROSCI.6582-10.2011
Jaworski J, Kapitein LC, Gouveia SM, Dortland BR, Wulf PS, Grigoriev I, Camera P, Spangler SA, Di Stefano P, Demmers J, Krugers H, Defilippi P, Akhmanova A, Hoogenraad CC (2009) Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron 61:85–100. https://doi.org/10.1016/j.neuron.2008.11.013
Komarova YA, Akhmanova AS, Kojima S, Galjart N, Borisy GG (2002) Cytoplasmic linker proteins promote microtubule rescue in vivo. J Cell Biol 159:589–599. https://doi.org/10.1083/jcb.200208058
Hoogenraad CC, Akhmanova A, Howell SA, Dortland BR, De Zeeuw CI, Willemsen R, Visser P, Grosveld F, Galjart N (2001) Mammalian Golgi-associated Bicaudal-D2 functions in the dynein-dynactin pathway by interacting with these complexes. EMBO J 20:4041–4054. https://doi.org/10.1093/emboj/20.15.4041
Farías GG, Guardia CM, De Pace R, Britt DJ, Bonifacino JS (2017) BORC/kinesin-1 ensemble drives polarized transport of lysosomes into the axon. Proc Natl Acad Sci U S A 114:E2955–E2964. https://doi.org/10.1073/pnas.1616363114
Chi S, Cao H, Chen J, McNiven MA (2008) Eps15 mediates vesicle trafficking from the trans-Golgi network via an interaction with the clathrin adaptor AP-1. Mol Biol Cell 19:3564–3575. https://doi.org/10.1091/mbc.e07-10-0997
Ciechomska IA, Gabrusiewicz K, Szczepankiewicz AA, Kaminska B (2013) Endoplasmic reticulum stress triggers autophagy in malignant glioma cells undergoing cyclosporine a-induced cell death. Oncogene 32:1518–1529. https://doi.org/10.1038/onc.2012.174
Hoogenraad CC, Akhmanova A, Grosveld F, De Zeeuw CI, Galjart N (2000) Functional analysis of CLIP-115 and its binding to microtubules. J Cell Sci 113(Pt 12):2285–2297
Tinevez J-Y, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, Eliceiri KW (2017) TrackMate: An open and extensible platform for single-particle tracking. Methods 115:80–90. https://doi.org/10.1016/j.ymeth.2016.09.016
Mangeol P, Prevo B, Peterman EJG (2016) KymographClear and KymographDirect: two tools for the automated quantitative analysis of molecular and cellular dynamics using kymographs. Mol Biol Cell 27:1948–1957. https://doi.org/10.1091/mbc.E15-06-0404
Robitaille AM, Christen S, Shimobayashi M, Cornu M, Fava LL, Moes S, Prescianotto-Baschong C, Sauer U, Jenoe P, Hall MN (2013) Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science 339:1320–1323. https://doi.org/10.1126/science.1228771
Schwarz JJ, Wiese H, Tölle RC, Zarei M, Dengjel J, Warscheid B, Thedieck K (2015) Functional proteomics identifies acinus L as a direct insulin- and amino acid-dependent mammalian target of rapamycin complex 1 (mTORC1) substrate. Mol Cell Proteom 14:2042–2055. https://doi.org/10.1074/mcp.M114.045807
Hong Z, Yang Y, Zhang C, Niu Y, Li K, Zhao X, Liu J-J (2009) The retromer component SNX6 interacts with dynactin p150(Glued) and mediates endosome-to-TGN transport. Cell Res 19:1334–1349. https://doi.org/10.1038/cr.2009.130
Johansson M, Rocha N, Zwart W, Jordens I, Janssen L, Kuijl C, Olkkonen VM, Neefjes J (2007) Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor betalll spectrin. J Cell Biol 176:459–471. https://doi.org/10.1083/jcb.200606077
Keyel PA, Thieman JR, Roth R, Erkan E, Everett ET, Watkins SC, Heuser JE, Traub LM (2008) The AP-2 Adaptor β2 Appendage Scaffolds Alternate Cargo Endocytosis. MBoC 19:5309–5326. https://doi.org/10.1091/mbc.e08-07-0712
Saito K, Murayama T, Hata T, Kobayashi T, Shibata K, Kazuno S, Fujimura T, Sakurai T, Toyoshima YY (2020) Conformational diversity of dynactin sidearm and domain organization of its subunit p150. MBoC 31:1218–1231. https://doi.org/10.1091/mbc.E20-01-0031
Cheong FKY, Feng L, Sarkeshik A, Yates JR, Schroer TA (2014) Dynactin integrity depends upon direct binding of dynamitin to Arp1. Mol Biol Cell 25:2171–2180. https://doi.org/10.1091/mbc.E14-03-0842
Echeverri CJ, Paschal BM, Vaughan KT, Vallee RB (1996) Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J Cell Biol 132:617–633. https://doi.org/10.1083/jcb.132.4.617
De Zeeuw CI, Hoogenraad CC, Goedknegt E, Hertzberg E, Neubauer A, Grosveld F, Galjart N (1997) CLIP-115, a novel brain-specific cytoplasmic linker protein, mediates the localization of dendritic lamellar bodies. Neuron 19:1187–1199
Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M, Cook LJ, Rubinsztein DC (2005) Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol 170:1101–1111. https://doi.org/10.1083/jcb.200504035
Wu Y, Boulogne C, Carle S, Podinovskaia M, Barth H, Spang A, Cintrat J-C, Gillet D, Barbier J (2020) Regulation of endo-lysosomal pathway and autophagic flux by broad-spectrum antipathogen inhibitor ABMA. FEBS J 287:3184–3199. https://doi.org/10.1111/febs.15201
Mauthe M, Orhon I, Rocchi C, Zhou X, Luhr M, Hijlkema KJ, Coppes RP, Engedal N, Mari M, Reggiori F (2018) Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. https://doi.org/10.1080/15548627.2018.1474314
Pasquier B (2016) Autophagy inhibitors. Cell Mol Life Sci 73:985–1001. https://doi.org/10.1007/s00018-015-2104-y
Morelli E, Ginefra P, Mastrodonato V, Beznoussenko GV, Rusten TE, Bilder D, Stenmark H, Mironov AA, Vaccari T (2014) Multiple functions of the SNARE protein Snap29 in autophagy, endocytic, and exocytic trafficking during epithelial formation in Drosophila. Autophagy 10:2251–2268. https://doi.org/10.4161/15548627.2014.981913
Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts EA, Imarisio S, Jahreiss L, Sarkar S, Futter M, Menzies FM, O’Kane CJ, Deretic V, Rubinsztein DC (2011) Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol 13:453–460. https://doi.org/10.1038/ncb2204
Cheng X-T, Zhou B, Lin M-Y, Cai Q, Sheng Z-H (2015) Axonal autophagosomes recruit dynein for retrograde transport through fusion with late endosomes. J Cell Biol 209:377–386. https://doi.org/10.1083/jcb.201412046
Maday S, Holzbaur ELF (2014) Autophagosome biogenesis in primary neurons follows an ordered and spatially regulated pathway. Dev Cell 30:71–85. https://doi.org/10.1016/j.devcel.2014.06.001
Maday S, Wallace KE, Holzbaur ELF (2012) Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol 196:407–417. https://doi.org/10.1083/jcb.201106120
Chen Y, Yu L (2017) Recent progress in autophagic lysosome reformation. Traffic 18:358–361. https://doi.org/10.1111/tra.12484
Wu Y, Pons V, Goudet A, Panigai L, Fischer A, Herweg J-A, Kali S, Davey RA, Laporte J, Bouclier C, Yousfi R, Aubenque C, Merer G, Gobbo E, Lopez R, Gillet C, Cojean S, Popoff MR, Clayette P, Le Grand R, Boulogne C, Tordo N, Lemichez E, Loiseau PM, Rudel T, Sauvaire D, Cintrat J-C, Gillet D, Barbier J (2017) ABMA, a small molecule that inhibits intracellular toxins and pathogens by interfering with late endosomal compartments. Sci Rep 7:15567. https://doi.org/10.1038/s41598-017-15466-7
Ganesan D, Cai Q (2021) Understanding amphisomes. Biochem J 478:1959–1976. https://doi.org/10.1042/BCJ20200917
Traub LM, Bannykh SI, Rodel JE, Aridor M, Balch WE, Kornfeld S (1996) AP-2-containing clathrin coats assemble on mature lysosomes. J Cell Biol 135:1801–1814. https://doi.org/10.1083/jcb.135.6.1801
Jordens I, Fernandez-Borja M, Marsman M, Dusseljee S, Janssen L, Calafat J, Janssen H, Wubbolts R, Neefjes J (2001) The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol 11:1680–1685
Ferguson SM (2019) Neuronal lysosomes. Neurosci Lett 697:1–9. https://doi.org/10.1016/j.neulet.2018.04.005
Roney JC, Cheng X-T, Sheng Z-H (2022) Neuronal endolysosomal transport and lysosomal functionality in maintaining axonostasis. J Cell Biol 221:e202111077. https://doi.org/10.1083/jcb.202111077
Kuijpers M, Azarnia Tehran D, Haucke V, Soykan T (2021) The axonal endolysosomal and autophagic systems. J Neurochem 158:589–602. https://doi.org/10.1111/jnc.15287
Sidibe DK, Kulkarni VV, Dong A, Herr JB, Vogel MC, Stempel MH, Maday S (2022) Brain-derived neurotrophic factor stimulates the retrograde pathway for axonal autophagy. J Biol Chem 298:102673. https://doi.org/10.1016/j.jbc.2022.102673
Kulkarni VV, Stempel MH, Anand A, Sidibe DK, Maday S (2022) Retrograde axonal autophagy and endocytic pathways are parallel and separate in neurons. J Neurosci 42:8524–8541. https://doi.org/10.1523/JNEUROSCI.1292-22.2022
Acknowledgements
The authors thank Dr. Anna Akhmanova, Dr. Juan Bonifacino, Dr. Hong Cao, Dr. Iwona Ciechomska, Dr. Casper Hoogenraad, Dr. Volker Haucke, Dr. Mark McNiven, Dr. Lukasz Swiech, and Dr. Agata Zieba-Wicher for reagents, Dr. Rafaella de Pace, Dr. Carlos Guardia, and Dr. Juan Bonifacino for their support with establishing the protocol for the live imaging of neurons, Dr. Iwona Cymerman for help with designing cloning strategy, Dr. Katarzyna Poleszak for help with establishing protein production protocols, and Dr. Alexander Heberle for help with analyzing existing mTOR phosphoproteome datasets. We are also grateful to Dr. Iwona Ciechomska, Dr. Kathrin Thedieck, Dr. Viktor Korolchuk, Dr. Anne Spang for comments and discussions. We also thank Alina Zielinska and Marek Sarnacki from our laboratory and Tomasz Wegierski from the Microscopy and Flow Cytometry Core Facility at IIMCB for technical assistance and support, Angelika Jocek for laboratory management logistics, and Michael Arends for proofreading the manuscript.
Funding
Research was supported by Polish National Science Centre Opus grant no. 2016/21/B/NZ3/03639 to JJ. AT was partly financed by Polish National Science Centre Preludium grant no. 2017/25/N/NZ3/01280 and Opus grant no. 2017/27/B/NZ3/01358. JJ was partly financed by the TEAM grant from the Foundation for Polish Science (POIR.04.04.00-00-5CBE/17-00). AM was partly financed within the Parent-Bridge program of the Foundation for Polish Science, co-financed by the European Union under the European Regional Development Fund (POMOST/2013-7/10) and by I.3.4 Action of the Excellence Initiative—Research University Programme at the University of Warsaw.
Author information
Authors and Affiliations
Contributions
AT, KB, AB, JW, AM, and JJ designed the experiments. AT, KB, AB, JW, MMa, OT, AL, MC-K, TB, AAS, KO, MMl, SK, EL, KM, MB, and AM performed the experiments. AT, KB, AB, JW, MMa, OT, TB, EL, MB, TR, AM, and JJ analyzed the data. AT, KB, AB, JW, AM, and JJ wrote the manuscript. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
None of the authors have any financial or non-financial competing interests.
Ethical approval
The rats that were used to obtain neurons for further experiments were sacrificed according to protocols that complied with European Community Council Directive 2010/63/EU. Rapamycin treatment and brain protein lysate isolation were performed according to a protocol that was approved by the 1st Ethical Committee in Warsaw (Poland; decision no. 843/2008 and 288/2012), which was also in compliance with European Community Council Directive 2010/63/EU.
Consent to participate
This study did not involve human subjects.
Consent to publish
This study did not involve human subjects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 (MP4 1161 KB)
Supplementary file2 (MP4 89 KB)
Supplementary file3 (MP4 1114 KB)
Supplementary file4 (MP4 90 KB)
Supplementary file5 (AVI 8422 KB)
Supplementary file6 (AVI 19031 KB)
Supplementary file7 (AVI 5872 KB)
Supplementary file8 (AVI 22011 KB)
Supplementary file9 (MP4 11453 KB)
Supplementary file10 (MP4 11402 KB)
Supplementary file11 (MP4 11232 KB)
Supplementary file12 (MP4 11338 KB)
Supplementary file13 (MP4 11477 KB)
Supplementary file14 (MP4 11460 KB)
Supplementary file15 (MP4 11434 KB)
Supplementary file16 (MP4 11179 KB)
Supplementary file17 (MP4 11415 KB)
Supplementary file18 (MP4 11348 KB)
Supplementary file19 (MP4 15825 KB)
Supplementary file20 (MP4 25860 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tempes, A., Bogusz, K., Brzozowska, A. et al. Autophagy initiation triggers p150Glued–AP-2β interaction on the lysosomes and facilitates their transport. Cell. Mol. Life Sci. 81, 218 (2024). https://doi.org/10.1007/s00018-024-05256-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-024-05256-6