Abstract
The Notch pathway is an evolutionarily conserved signaling system that is intricately regulated at multiple levels and it influences different aspects of development. In an effort to identify novel components involved in Notch signaling and its regulation, we carried out protein interaction screens which identified non-muscle myosin II Zipper (Zip) as an interacting partner of Notch. Physical interaction between Notch and Zip was further validated by co-immunoprecipitation studies. Immunocytochemical analyses revealed that Notch and Zip co-localize within same cytoplasmic compartment. Different alleles of zip also showed strong genetic interactions with Notch pathway components. Downregulation of Zip resulted in wing phenotypes that were reminiscent of Notch loss-of-function phenotypes and a perturbed expression of Notch downstream targets, Cut and Deadpan. Further, synergistic interaction between Notch and Zip resulted in highly ectopic expression of these Notch targets. Activated Notch-induced tumorous phenotype of larval tissues was enhanced by over-expression of Zip. Notch-Zip synergy resulted in the activation of JNK pathway that consequently lead to MMP activation and proliferation. Taken together, our results suggest that Zip may play an important role in regulation of Notch signaling.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Notch signaling is an evolutionarily conserved pathway that plays a fundamental role in various developmental events like cell fate determination, proliferation, apoptosis, and stem cell maintenance [1,2,3,4]. Notch is synthesized as a 300 kDa precursor protein that is subjected to its first cleavage in the trans-Golgi network by furin-like convertases (S1 cleavage), resulting in a 180 kDa N-terminal extracellular domain NECD (Notch extracellular domain) and a 120 kDa C-terminal transmembrane intracellular domain, N™ [5]. This heterodimer of Notch receptor is targeted to the cell membrane where it interacts with ligands of the DSL family (Drosophila Delta and Serrate (Jagged in mammals) and C. elegans LAG-2). Binding of ligands expressed on an adjacent cell to NECD leads to second proteolytic cleavage (S2) by ADAM family of metalloproteases in the extracellular portion of the NTM [6]. Ligand endocytosis is thought to generate mechanical force to induce a conformational change in the bound Notch receptor. This conformational change promotes the second cleavage of the receptor by metalloprotease [7, 8]. The second cleavage creates a membrane-tethered intermediate form of Notch, the NEXT (Notch extracellular truncation). This is followed by an intramembranous cleavage (S3) by γ-secretase complex (Presenilin, Nicastrin, PEN-2, and APH-1) and results in the release of Notch intracellular domain (NICD) from the membrane [9, 10]. The NICD then translocates to the nucleus with the aid of Importin α3 [11], where it transduces Notch signals by regulating the transcription of downstream target genes. In nucleus, NICD associates with the DNA binding protein CSL (mammalian CBF1/Drosophila Su(H)/C. elegans Lag-1) and facilitates the displacement of transcriptional co-repressors. The NICD–CSL complex then recruits Mastermind and other transcriptional coactivators leading to activation of Notch target genes such as the Enhancer of Split [E(spl)] complex genes in Drosophila [12,13,14,15]. These bHLH transcription factors, in turn, repress achaete–scute complex (As-C) proneural genes.
The same pathway can be deployed in numerous cellular contexts to play varied and critical roles for the development of an organism. The versatility of this pathway to influence different aspects of development comes from its multiple levels of regulation. We carried out different protein interaction screens to identify novel components involved in Notch signaling and its regulation. Independent protein interaction screens identified Zipper (Zip), also known as non-muscle myosin II (NM II), as an interacting partner of Notch. Non-muscle myosin II (NM II) is a hexameric actin-binding protein that consists of two heavy chains, two essential light chains and two regulatory light chains. It has contractile properties and is regulated by the phosphorylation of its light and heavy chains. In Drosophila, heavy chain of NM II is encoded by the zipper, and regulatory light chain is encoded by spaghetti squash [16, 17]. Zip plays a major role in regulating cell adhesion, cell migration, and determination of the polarity of migrating cells [18]. It has also been reported that Zip plays a crucial role in axon patterning, head involution and dorsal closure during embryonic development [19]. In the context of Notch signaling, it has already been shown that mechanical force has considerable implication in the activation of Notch receptor [20,21,22]. Recent studies have demonstrated that non-muscle myosin II-dependent tension across the Notch–Delta complex contributes to Notch activation [23]. Actomyosin contractility in cells having extensive lateral contacts with other cells has been shown to promote Notch activation and it was postulated that endocytosis and myosin-dependent pulling may both contribute to force-dependent Notch activation in these cells [23]. Further, presence of non-muscle myosin II at the nuclear periphery and its co-localization with the linker of nucleoskeleton and cytoskeleton (LINC) protein Nesprin2 and apical actin caps suggests that it plays role in transmission of cytoplasmic signals to the nucleus. Earlier it was reported that NM IIs can act as a mechano-transducer at the perinuclear area supporting the notion of actomyosin-mediated gene regulation through LINC [23].
In the present study, we have identified Zip as a Notch interactor using two independent protein–protein interaction screens and characterized the functional significance of this interaction. Using genetic and molecular studies, we have shown that Zip positively regulates Notch. Our co-immunoprecipitation experiments reconfirmed their physical interaction. Zip also genetically interacted with the components of Notch pathway. Further, loss-of-function of zip resulted in wing phenotypes identical to Notch loss-of-function wing phenotypes and compromised signaling, validating the integration of Zip with Notch signaling. Notch loss-of-function wing phenotype was rescued by over-expressing zip in the background. We have also shown the synergistic interaction between Notch and Zip. Additionally, increased levels of Notch targets, Cut, and Dpn upon zip co-expression with Notch clearly indicated the positive regulation of Notch by Zip.
Materials and methods
Yeast two-hybrid
A 393 bp Drosophila Notch cDNA (accession number M11664) fragment which encodes amino acids 1765–1895 containing NLS was amplified by polymerase chain reaction (PCR) and cloned in frame with the sequence encoding the LexA DNA-binding domain of bait vector. This construct was used as bait to screen oligo(dT)-primed D. melanogaster 0–24 h embryo cDNA libraries cloned in pGAD prey vectors containing GAL4 activation domains. A yeast two-hybrid screen was carried out as described previously [24]. Sequencing was performed for all three positive pGAD plasmids from His + colonies.
Protein extraction, immunoprecipitation, immunoblotting, and mass spectrometry
Protein lysate was made using 1X RIPA lysis buffer (50 mM Tris–HCl pH 7.4, 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA). The lysate containing 3 mg of protein was incubated with 5 μl antibody (anti-Notch antibody (C17.9C6), Developmental Studies Hybridoma Bank). 30 μl of A/G agarose beads was added to the lysate and it was rotated for overnight at 4 °C. After washing three times with 1X RIPA buffer, the samples were denatured and run at 12% SDS-PAGE gel to separate the peptides. This was followed by overnight transfer of proteins from gel onto a PVDF membrane. After blocking with 4% skimmed milk in TBST, the membrane was incubated with primary antibody followed by AP-conjugated secondary antibody. Chromogenic detection of signal was performed using Sigma FAST NBT/BCIP. 5.0 μl of anti-Notch antibody (C17.9C6) (Developmental Studies Hybridoma Bank) was used for immunoprecipitation. The primary antibodies used for western blotting were rabbit anti-Zip (1:1500) (kindly provided by Prof. Daniel P. Kiehart, Department of Biology, Duke University, Durham, NC) and rabbit anti-GFP (1:2000, Invitrogen).
For mass spectrometry, the separated protein samples in gel were stained with Coomassie Brilliant Blue. The sample preparation for mass spectrometry analysis was done as per the protocol of Bruker Daltonics adapted from Shevchenko et al., 1996 [25]. At first, the gel was excised into pieces with the help of scalpel and treated with washing solution (1:1 ratio of acetonitrile and 100 mM NH4HCO3) for 30 min. This step was accompanied with occasional vortexing at 300–350 RPM in order to destain the gel pieces. The gel pieces were then dehydrated by treating it with 100% acetonitrile for 3 min, followed by air drying. Dehydration was followed by reduction reaction where the gel pieces were treated with 10 mM dithiothreitol (DTT) dissolved in 100 mM NH4HCO3 for 30 min at 50 °C. Alkylation reaction was performed after this by treating the sample with 50 mM iodoacetamide (IAA) in100mM NH4HCO3 for 30 min in the dark at room temperature. The alkylated gel pieces were washed with a washing solution (1:1 ratio of acetonitrile and 100 mM NH4HCO3), and were dehydrated using 100% acetonitrile and subsequently air-dried. Overnight digestion was set up with 20 μl of 25 μg/ml proteomics grade trypsin solution (Promega Gold, USA) at 37 °C in a water bath. 0.1% trifluoroacetic acid (TFA) in 50% acetonitrile was used to extract the digested peptides from the gel pieces by vortexing for 10 min. This extraction step was repeated twice. The extracted peptides were concentrated using Speed-Vac and subjected to mass spectrometric analysis using AUTOFLEX speed MALDI-TOF/TOF instrument (Bruker Daltonics, Bremen, Germany).
Proteomic analysis
Identification of the interacting partners of Zip was done by co-immunoprecipitation of the interacting proteins from GFP-Zip over-expression protein lysate with anti-GFP antibody followed by in-solution trypsin digestion and analysis by high-resolution mass spectrometry. The peptides identified in the HRMS screening were sorted on the basis of the sequence score (> 40). STRING database (https://string-db.org/) was used to generate the protein interactome based on identification of Zip interacting partners. Functional enrichment of the biological processes in the identified network represented as log 10 having p values with Benjamini–Hochberg correction has been shown in the bar graph. High-Resolution Accurate Mass Spectrometry System (Orbitrap Eclipse Tribrid Mass Spectrometer) from Thermo Fischer Scientific was used for the MS screening of Zip interacting proteins.
Drosophila genetics
All fly stocks were maintained on standard cornmeal/yeast/molasses/agar medium at 25 °C as per standard procedures. Oregon-R flies were used as wild-type controls. UAS-GFP-zip, UAS-GFP-Zip DN (UAS-Myo II-Neck-Rod), UAS-Myo II-Rod, and UAS-Myo II-Rod (delta Nterm58) [26] were obtained as a gift from Prof. Daniel P. Kiehart (Department of Biology, Duke University, Durham, NC). UAS-Notch-FL [27], UAS-Notch-ICD, UAS-Notch-DN [28], and Notch pathway components were kindly provided by Prof. S. Artavanis-Tsakonas (Department of Cell Biology, Harvard Medical School, Boston, MA).
-
cn1 bw1 sp1 zip1/CyO (BDSC 4199),
-
P{FRT(whs)}G13 zip2/CyO (BDSC 8739),
-
P{lacZ.w+}276, y1 sc* v1 sev21; P{TRiP.GL00623}attP40 (BDSC 37480), y1 sc* v1 sev21; P{TRiP.HMS01618}attP2 (BDSC 65947),
-
SqhAX3-GFP (BDSC 57144), UAS-Bsk-DN (BDSC 6409), vg-GAL4 (BDSC 8222), GMR-GAL4 (BDSC 8121), ptc-GAL4 (BDSC 2017), en-GAL4 (BDSC 30564), ap-GAL4 (BDSC 56807), dpp-GAL4 (BDSC 1553), and C96-GAL4 (BDSC 43343) stocks were obtained from Bloomington stock center. All crosses were performed at 25 °C. The combination lines vg-GAL4/UAS-GFP-zip and UAS-Notch-DN/C96-GAL4 were made with the help of appropriate genetic crosses.
Wing mounting
For the preparation of wings from adult flies, F1 progeny flies were taken and their wings were separated with the help of needle and scalpel. The wings were then transferred on a glass slide and washed with isopropyl alcohol. This was followed by mounting the wings immediately in 70% glycerol. Wing images were taken using Brightfield Nikon Eclipse Ni microscope.
Immunocytochemistry and confocal microscopy
Drosophila third instar larvae were dissected in chilled 1X PBS and fixed in 4% Paraformaldehyde for 20 min. The tissues were then washed with washing solution (0.1% BSA in Tri-PBS) for 4 times, 15 min each. This was followed by blocking the tissues using blocking solution (PBST with 8% of serum) for 30 min to an hour. The tissues were then incubated with primary antibody for overnight at 4 °C. On the next day, the tissues were washed again for 4 times, 20 min each followed by blocking for 30 min to 1 h at room temperature. This was followed by incubating the tissues with fluorophore-conjugated secondary antibody for 90 min at room temperature. Tissues were then washed again for 4 times, 20 min each followed by a wash in 1X PBS. DAPI was added in the dark for 20–30 min. The tissues were washed again with washing solution for 4 times and once with 1X PBS for 15 min each. The discs were finally dissected in chilled 1X PBS and incubated in DABCO for overnight. Next day, the tissues were mounted and observed under Carl Zeiss LSM 780 laser scanning confocal microscope. The images were processed using Adobe Photoshop 7. The primary antibodies used in this study are as follows: mouse anti-Notch (C17.9C6; 1:300), mouse anti-Notch (C458.2H; 1:100), mouse anti-Cut (2B10; 1:100), mouse anti-MMP1 (3A6B4; 1:100), mouse anti-Delta (1:100), Guinea Pig anti-Rab5 (1:1000, a generous gift from Prof. Akira Nakamura, Institute of Molecular Embryology and Genetics, Kumamoto, Japan), mouse anti-Armadillo (N2 7A1; 1:100) (all from Developmental Studies Hybridoma Bank), rabbit anti-Cleaved Caspase 3 (1:50), rabbit phospho-JNK (1:100), rabbit anti-phospho myosin regulatory light chain 2 (1:50) (Cell Signaling Technology), rabbit anti-PH3 (1:50) (Merck), rabbit anti-Dpn (1:100) (kindly provided by Prof. Yuh N Jan, Howard Hughes Medical Institute, University of California San Francisco, California), rabbit anti-Zip (1:200). Phalloidin stain was used to detect actin. Alexa488-, and Alexa555-conjugated secondary antibodies (1:200, Molecular Probes) were used to detect the primary antibodies.
Statistical analysis
Intensity profiling in the Drosophila imaginal discs and quantification of the area covered by tumorigenic discs representing the size of the discs was done using Image J. 5–10 imaginal discs were used for the quantification purpose in each case. Integrated density/area of the domain indicated the intensity of the staining in confocal images. Image J was also used to quantify the number of PH3 positive cells in the wing imaginal discs where all the cells were counted using analyze particles feature in the software. The RGB plot profile feature of Image J was also used to denote the intensity of Notch targets in Zip compromised condition compared to the internal control. The line icon was used to measure the area of the pouch region of the wing discs with Cut and Dpn expression. RGB plot profile feature from plugins section was then used to plot intensity graphs. The error bars in the graphs denoted the standard error of the mean value from the replicated experiments. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test and unpaired t test was used to determine the extent of significance among different genotypes. p value < 0.05 was accepted as statistically significant. GraphPad Prism 5 was used for plotting graphs.
Results
Zip is an interacting partner of Notch
In an effort to identify novel components involved in Notch signaling and its regulation, we carried out two independent protein interaction screens, one based on the identification of cellular protein complexes using immunoprecipitation followed by mass spectrometry and other based on yeast two-hybrid system. Both the screens identified Zip as an interacting partner of Notch. For immunoprecipitation, tissue lysate was prepared from hyperplastic wing imaginal discs over-expressing Notch-ICD under vg-GAL4 driver and from wild type wing discs as well that served as the control. Anti-Notch antibody was used to precipitate Notch along with its interactors from both the lysates. The protein bands exclusively present in the lane with over-expressed Notch lysate and absent in the control lane were excised from the SDS-PAGE and were processed for mass spectrometry. One of them was identified as Zip, a 205 kDa protein as a novel interactor of Notch (Fig. S1). Zip was also recovered in a standard yeast two-hybrid screen in which approximately 6 × 106 cDNAs from a random primed Drosophila 0–24 h embryonic library were screened with a cDNA corresponding to amino acids 1765–1895 of Drosophila Notch fused in frame to the LexA DNA binding domain used as a bait [24]. In the same screen, multiple positive clones of Su(H), a well-established binding partner of Notch-ICD, were also identified, which validated our approach. Sequence analysis of three identified positive clones of Zipper (Zip) revealed that the carboxy-terminal part of Zip (amino acids 1692–1972) binds to Notch-ICD. In addition, we have also carried out a large-scale proteomic analysis of Zip based on co-immunoprecipitation of Zip along with its interacting proteins followed by mass spectrometry (MS). Notch was identified as one of the many interacting partners in our MS analysis. Along with Notch, many other cytoskeletal components and endocytic machinery proteins were also identified in the MS analysis (Fig. S7).
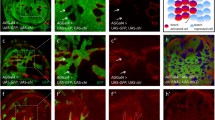
Physical interaction of Notch with Zip was further validated by co-immunoprecipitation studies. Protein lysate was prepared from adult head tissues co-expressing full-length Notch (Notch-FL) and GFP-Zip driven by GMR-GAL4. Anti-Notch antibody was used to immunoprecipitate Notch along with its interacting partners in a complex which were fractionated on SDS-PAGE. Co-immunoprecipitated GFP-conjugated Zip protein was detected on a western blot using anti-GFP antibody (Fig. 1A). In addition, we also used anti-Zip antibody that detected both endogenous and over-expressed GFP-Zip that appeared as doublet band on the western blot (Fig. 1A). In the same set of experiments, we also demonstrated the physical interaction of over-expressed Zip with endogenous Notch protein. As in case of over-expressed Notch, Zip was also detected as a physically associating partner of the endogenous Notch with anti-GFP as well as anti-Zip antibody (Fig. 1A). To rule out the presence of Zip in our western blot due to non-specific binding with AG beads, we also performed immunoprecipitation experiment without adding primary antibody. No expression of Zip could be seen in the negative control compared to the presence of a doublet band of endogenous and over-expressed GFP-tagged Zip in the experimental lanes (Fig. S1C). Conversely, we used anti-GFP antibody to immunoprecipitate GFP-tagged Zip along with its binding partners and Notch FL was found to be co-immunoprecipitated with Zip which was detected using anti-Notch antibody on the western blot (Fig. 1A). In the same set-up, we also performed co-immunoprecipitation studies using protein lysate from only Notch FL over-expressed condition. Here, anti-GFP antibody could not immunoprecipitate endogenous Zip (absence of over-expressed GFP-Zip). Hence, we did not observe presence of Notch upon immunoblotting with anti-Notch antibody (Fig. 1A). These results confirmed that Notch is indeed physically associated with Zip.
Physical interaction between Notch and Zip. A Zip was identified as an interacting partner of Notch. Co-immunoprecipitation was carried out with head tissue lysates over-expressing GFP-Zip and Notch FL. M indicates the marker lane, (+) symbol indicates the presence and (−) shows the absence of the specified reagent. Notch-FL and endogenous Notch immunoprecipitated endogenous and GFP-tagged-Zip that was detected by anti-GFP and anti-Zip antibody on the western blot. Star marks indicate the bands of GFP-tagged Zip and endogenous Zip. No GFP-Zip protein bands were observed in the negative control (Fig. S1C). In the other direction, anti-GFP immunoprecipitated Notch-FL that was detected by anti-Notch antibody on the western blot. Lower blots show the presence of the specified protein bands in the experimental and the control lysates. B–D Zip colocalizes with Notch in the over-expressed condition in the cytoplasmic compartment. Zip and Notch-FL were co-expressed under vg-GAL4. GFP-tagged Zip forms cytoplasmic aggregates upon over-expression and colocalizes with Notch-FL puncta on the cytoplasmic membrane. Anti-Notch antibody was used for the detection of over-expressed full-length Notch. Panel D is the merged image of B, C. The insat image in the panel D represents enlarged view of the image showing extent of colocalization between Notch and Zip. E–G Zip colocalizes with Notch in wild type wing discs. Merged image of panel G shows that Zip colocalizes with Notch on the cytoplasmic membrane. The antibody used for detection of wild type Zip in panel E is anti-Zip and for Notch detection in panel F is anti-Notch. The insat image in the panel G represents enlarged view of the images showing extent of colocalization between these two proteins. Scale bar: 20 μm
To corroborate our physical interaction experiments of Notch and Zip, we performed colocalization experiments through immunostaining using anti-Notch and anti-Zip antibodies. We checked their localization in GFP-tagged Zip and Notch-FL co-expressed larval wing imaginal discs. We observed that Zip colocalized with Notch in over-expressed condition on the cell membrane and in the cytoplasm (Fig. 1B–D, Fig. S1G–G″). As in case of colocalization of Notch and Zip in over-expressed condition, endogenous Zip and Notch also colocalized in the same cytoplasmic compartment in the wild-type wing imaginal disc (Fig. 1E–G). In the over-expressed condition, GFP-tagged Zip was found to form large cytoplasmic aggregates [29,30,31], and these aggregates colocalized with Notch-FL (Fig. 1B, D) indicating that the two proteins colocalize and perhaps functionally regulate developmental processes together.
zip genetically interacts with Notch pathway components
To address the functional implications of physical interaction between Zip and Notch, we studied the genetic interactions between zip mutants and mutants of Notch pathway components in trans-heterozygous combinations. We used two independent amorphic alleles of zip, zip1, and zip2. We checked their genetic interaction with the Notch mutants, the amorphic allele of Notch, N54l9, and the hypomorphic allele of Notch, Nnd3. A trans-heterozygous combination of zip alleles with N54l9 and Nnd3 resulted in an increased number of flies with wing-nicking phenotype (Fig. 2B–C″). It was observed that 28% of N54l9 mutant flies exhibited wing-nicking phenotype which was increased to 60% and 52% (42/80) when these flies were combined heterozygously with zip1 and zip2 alleles, respectively (Fig. 2B‴). Similarly, 28% of Nnd3 mutant flies exhibited wing-nicking phenotype which was increased to 37% and 35% when these flies were combined heterozygously with zip1 and zip2 alleles respectively (Fig. 2C‴). This indicated a further reduction of Notch signaling upon lowering the dose of zip in the same background thus validating a functional relevance between the two genes. The wing vein thickening phenotype of Delta (Dl5f) was also enhanced by reducing the dose of zip (Fig. 2D–D″). The null allele of deltex (dx), a cytoplasmic modulator of Notch resulted in mild wing-vein thickening. However, we noticed an enhancement in wing vein thickening phenotype in dx hemizygous condition with zip alleles. Since vein thickening is a Notch loss-of-function phenotype, the enhancement of it with zip alleles in the background indicates further decrease of Notch signaling upon lowering the dose of Zip (Fig. 2E–E″). The functional interaction predicted to be caused by the colocalization of Notch and Zip in the same cytoplasmic compartment (as shown in Fig. 1B–G) is evident from genetic interaction between zip and Notch mutant alleles (Fig. 2B–C‴). Additionally, we also checked the genetic interaction with C96-GAL4-driven dominant-negative form of C-terminal Mastermind truncation (MamH), that display a fully penetrant wing-nicking phenotype [32, 33]. Reducing the dose of zip in this background elicited enhanced wing notching combined with reduced marginal bristles. It was observed that the approximate number of bristles exhibited by C96-Mam H flies was 40 on the entire posterior margin which was reduced to 19 and 21 when the flies were heterozygously combined with zip1 and zip2 alleles (Fig. 2F‴). This modulation in the wing phenotype could be attributed to a compromised dose of Zip in the cytoplasm that ultimately affects the downstream Notch signaling processes, resulting in an enhanced wing-nicking and loss of marginal bristles in the background of C96-GAL4-driven dominant-negative Mastermind (Fig. 2F–F″). The genetic interaction between zip and Notch pathway components demonstrated that effects caused by decreased Notch signaling were further enhanced when zip mutants were brought in trans-heterozygous combination indicating that Zip positively regulates Notch signaling and its loss results in the reduction of Notch signaling (Fig. 2).
Genetic interaction of zip with Notch pathway components. Representative wings from different Notch pathway component mutants are shown in first column and in trans-heterozygous condition with zip mutants zip1 and zip2, are shown in second and third column respectively. Wing from wild type is shown in A and from zip alleles are shown in A′–A″. B-C″ Wings from heterozygote N54l9 (B) and hemizygous Nnd−3 (C) shows nicking phenotype which were further increased in number in trans-heterozygous combination with loss-of-function alleles of zip, zip1 (B′, C′), and zip2 (B″, C″). D–E″ Heterozygous wings of DL5f (D) and hemizygous dx (E) showed vein thickening phenotype which was enhanced in trans-heterozygous condition with zip mutants (D′, D″ and E′, E″) respectively. (F–F″) Representative wings of loss-of-function allele of Mastermind driven by C96-GAL4 showed serrated wing margin phenotype, which was further enhanced by combining zip alleles trans-heterozygously. (B‴, C‴ and F‴) Graphs representing the percentage of wings showing nicking phenotype (B‴ and C‴) and approximate number of bristles on the posterior margin of the wing (F‴) in trans-heterozygous combination of zip mutants with the components of Notch signaling. (B‴ and C‴) All experiments were performed in triplicates (n). For consistency a total of 80 flies/vial was observed (B‴) No. of wings with nicking phenotype N54l9/ + : n1 = 22/80, n2 = 20/80 and n3 = 24/80; N54l9/zip1: n1 = 48/80, n2 = 42/80 and n 3 = 52/80; N54l9/zip2: 42/80, n2 = 40/80 and n3 = 46/80. (C‴) No. of wings with nicking phenotype Nnd3: n1 = 22/80, n2 = 22/80 and n3 = 24/80; Nnd3/zip1: n1 = 30/80, n2 = 27/80 and n3 = 32/80; Nnd3/zip2: 28/80, n2 = 20/80 and n3 = 36/80. F‴ The genotype of the flies mentioned on the X-axis of the graph are as follows: C96-GAL4/UAS-Mam H, C96-GAL4/UAS-Mam H + zip1, C96-GAL4/UAS-Mam H + zip2. Unpaired t-test was performed to determine p-value (**p < 0.01, ***p > 0.001). Scale bar: 3 cm
The loss-of-function of zip renders wing phenotypes similar to Notch mutant phenotypes and perturbs the expression pattern of Notch targets, Cut and Deadpan
The integration of Zip with Notch signaling was further verified by loss-of-function studies in which Zip was downregulated using UAS-zip-RNAi and UAS-GFP-zip DN under various wing-specific GAL4 driver lines. vg-GAL4-driven UAS-zip-RNAi displayed slightly bent, crumpled wing phenotype with serration on the lower margin of the wing blade. Some of these wings also exhibited irregular marginal bristles with thickened veins and extra vein material. Similarly, abrogating Zip using UAS-GFP-zip DN in the ventral domain of the wings using the same vg-GAL4 driver resulted in wing-nicking phenotype (Fig. S2A–A′). Down-regulating Zip using UAS-zip-RNAi in the dorsal region of the wing with ap-GAL4 yielded outward-directed erect wings with a crumpling phenotype. These wings also harbored several other phenotypes including irregular marginal bristles, extra vein material, and extra cross-veins. Similarly, eliminating Zip using UAS-GFP-zip DN in the dorsal region of the wing resulted in pupal lethality (Fig. S2B–B″). en-GAL4-driven UAS-zip-RNAi resulted in 100% pupal lethality. However, blocking Zip using Zip-DN in this region with same Gal4 driver resulted in wings harboring various phenotypes similar to Notch loss-of-function that included wing-nicking, mispatterning of wing hair, disruption of vein pattern and the cross-veins, and wing blister (Fig. S2C–C″). C96-GAL4-driven UAS-zip-RNAi resulted in slightly bent wings with fully penetrant serration on the lower margin of the wing blade. These wings also exhibited mild crumpling with ectopic bristles on the anterior portion of the wings. Reducing the activity of Zip using UAS-GFP-zip DN in the wing margin resulted in wing-nicking phenotype (Fig. S2D–D″). Reducing the dose of zip on the anterior/posterior boundary using ptc-GAL4 resulted in pupal lethality with very small number of flies eclosing that harbored wing-nicking on the anterior/posterior region of the wing blade (Fig. S2E–E″). Down-regulating zip at the anterior/posterior boundary using dpp-GAL4 displayed disrupted wing bristles and mild wing-nicking at A/P boundary (Fig. S2F–F″). These phenotypes shown by downregulation of zip using different GAL4 lines are reminiscent to Notch loss-of-function phenotypes, indicating that lowering the dose of Zip results in further reduction of the Notch signaling (Fig. S2G).
The modulation of the Notch signaling caused by down-regulation of Zip was further validated in the wing imaginal discs by examining the expression level of Notch downstream targets, Cut, and Dpn. Cut encodes a homeodomain transcription factor that bears a significant structural and functional similarity with several vertebrate proteins. Notch is required for the activation of Cut in a cell-autonomous manner [34]. Similarly, activated Notch directs the expression of Dpn by binding to the Notch-responsive enhancer present in the regulatory region of Dpn [35]. ptc-GAL4 and en-GAL4-driven UAS-GFP-zip DN in third instar larvae resulted in the perturbation of the expression of Cut and Dpn. RGB Plot profiles display the decreased fluorescence intensity of Cut and Dpn in the regions with compromised Zip compared to the internal control (Fig. 3A1–D1). The expression of Cut and Dpn has been observed to be normal in the wing imaginal discs of wild type flies (Fig. S3A, B). These observations confirmed that Zip not only positively regulates Notch signaling but also play a vital role in the modulation of Notch downstream targets (Fig. 3). Perturbation of Notch targets, Cut, and Dpn upon down-regulating Zip at A/P boundary (using ptc-GAL4) and in posterior domain (using en-GAL4) was observed in all the discs that were examined (total number of wing discs examined = 30). Image J was used for the intensity profiling where integrated density/area of the domain was used for quantification purpose. A total number of 5 discs were used for quantification in each case which was subjected to unpaired t-test to determine the significance of our findings. Mean intensity for internal control of Cut was 29, whereas that of en-GAL4 > UAS-GFP-zip DN was 20.6. Mean intensity for internal control of Dpn was 33.4, whereas that of en-GAL4 > UAS-GFP-zip DN was 28.2. In case of ptc GAL4-driven UAS-GFP-zip DN, mean intensity of internal control for Cut expression was 31.2, whereas that of Zip DN domain was 15.2. For Dpn, mean intensity for internal control was 38, whereas UAS-GFP-zip DN was 22.
A–D Reducing the dose of Zip results in lowered Notch signaling. Representative wing discs display the expression pattern of Cut and Dpn upon eliminating Zip on the A/P boundary and in the posterior region using ptc-GAL4 and en-GAL4 respectively. Abrogating Zip in the patched and engrailed domain using UAS-GFP-zip DN results in the perturbed expression of Cut (A, C) and Dpn (B, D) in these regions. A′–D′ Merged images showing the expression pattern of Cut and Dpn along with the patched and engrailed domain marked by GFP-tagged Zip DN. A1–D1 RGB plot profiles indicating the intensity of Cut and Dpn in A, B, C and D, respectively. E–J′ Loss of Cut and Dpn expression due to Zip DN on the A/P boundary using dpp GAL4 (E and H respectively) is rescued by over-expressing Notch ICD in the background (G and J respectively). Over-expressing UAS-Notch-ICD with dpp-GAL4 resulted in ectopic expression of Cut and Dpn throughout A/P boundary (F and I respectively). (E′–G′ and H′–J′) Merged images showing the expression of Cut and Dpn in dpp domain with GFP-tagged UAS-zip-DN (E′ and H′), and GFP-tagged UAS-zip-DN + UAS-Notch ICD (G′ and J′). (F′ and I′) Merged images showing the expression of Cut and Dpn along AP boundary with over-expressed Notch ICD. Absence of GFP indicates the absence of GFP-zip DN. (D″–D‴) Graphs representing the intensity profiling of Cut and Dpn upon abrogation of Zip at the A/P boundary (D″) and in the posterior region (D‴) of the wing imaginal discs. Unpaired t-test was performed to determine the p-value (*p < 0.05, ***p < 0.001). D″″ Western blot showing the expression of Cut, Dpn and internal control Tubulin (Tub). Scale bar: 20 μm
To verify whether the diminished expression of Cut and Dpn downstream to Zip-DN is mediated by reduced activity of Notch signaling, we supplied Notch-ICD in the Zip compromised background. Over-expression of UAS-GFP-zip DN using dpp-GAL4 alone leads to Cut and Dpn reduction in the D/V boundary straddling the A/P domain of dpp-GAL4 (Fig. 3E, H), whereas both Cut and Dpn were found to be ectopically increased upon over-expressing Notch ICD in the Dpp domain (Fig. 3F, I). When UAS-Notch ICD was over-expressed in the background with dominant-negative zip, the Notch targets were found to be rescued thus indicating that loss of Cut and Dpn resulting due to perturbed Zip is Notch-mediated (Fig. 3G, J), thus highlighting the role of Zip in modulating the activity of Notch signaling. To validate the loss of Notch signaling targets in Zip DN background, we prepared protein lysates from wing discs of en-GAL4 > Oregon R and en-GAL4 > UAS-GFP-zip DN and checked for the expression of Cut and Dpn using Western blotting. Both Notch downstream targets were found to be reduced in Zip DN condition compared to the control condition (Fig. 3D″″).
The loss-of-function of zip leads to cell surface accumulation of Notch receptor and endocytic component Rab5
Since down-regulation of Zip leads to reduced expression of Notch targets, we wanted to check the status of Notch receptor in Zip compromised background. Down-regulation of Zip in the posterior domain of the wing disc in UAS-GFP-zip DN using en-GAL4 driver, resulted in altered Notch localization. The wild type membranous Notch appeared to be accumulated in Zip compromised domain. These studies indicated that loss of Zip might result in perturbation of Notch receptor processing subsequently leading to down-regulated Notch signaling (Fig. S4A–A″). An accumulation of Notch in Zip compromised domain of the wing discs was observed in 100% of the wing discs that were examined (total number of discs examined = 20). A quantification of the membranous Notch expression was used as a readout for accumulated Notch at the cell surface. About 10 discs were used for the quantification purpose using Image J. Mean intensity of the wild type expression of Notch in the internal control was 22, whereas that in the posterior domain with compromised Zip was 31.5. To verify the accumulation of Notch at the cell surface, we also checked the colocalization of accumulated Notch with membrane marker actin. Actin associates with the plasma membrane and provide mechanical support, determine cell shape, etc. [36]. Here, accumulated Notch in the posterior region with compromised Zip using UAS-zip RNAi with en-GAL4 was observed to colocalize with Phalloidin marked actin (Fig. 4A–B″). Western blotting also showed a higher band intensity of Notch FL in the Zip compromised condition compared to the wild type that served as the control (Fig. 4A‴).
Loss of Zip leads to accumulation of Notch receptor and Rab5. A–B′″ Zip downregulation with UAS-zip RNAi in the posterior region of the wing disc using en-GAL4 resulted in an accumulated expression of the Notch receptor (A′) and actin (A) on the cell membrane compared to its endogenous expression in the anterior domain (A). Panel A″ represents a merged image of A and A′ where Notch was observed to colocalize with Phal. Panel B–B′ shows the expression of Phal, Notch at a higher resolution. Panel B″ represents merged image of B and B′. (A′″) Western blotting showing the expression of Notch in control and Zip compromised condition. C–C″ Down-regulation of Zip using UAS-zip RNAi with en-GAL4 resulted in accumulated Notch (C) and Rab5 (C′) in the posterior region of the wing disc. Panel C″ is the merged image of C and C′. Scale bar: 20 µm. B‴ Graph representing the accumulation of Notch and Rab 5 upon downregulating Zip in the posterior region. Error bar denotes the error between the mean values of the specified genotypes
Accumulation of the Notch receptor at the cell surface has been correlated with impaired endocytosis of the receptor in the signal receiving cell leading to compromised signaling [37]. Myosin II is implicated in the endocytic process where via force generation, it assists in pulling the clathrin-coated pit along with the receptor toward inside of the cell leading to invagination of the membrane thus initiating the process of early endosome formation [38]. Rab5 belongs to the family of GTPases that regulate trafficking into and between the early endosomes [39]. Hence, to understand the mechanism behind accumulation of Notch receptor in the signal receiving cell upon abrogating Zip, we checked the status of early endosomal marker Rab5 in Zip compromised condition. Down-regulating Zip using UAS-zip RNAi with en-GAL4 revealed an accumulated pattern of Notch at cell surface in the posterior domain of the wing imaginal disc. Similar to Notch accumulation, Rab5 also appeared to be accumulated in the engrailed domain compared to the anterior domain with endogenous Zip. Accumulated Notch was observed to partially colocalize with Rab5 (Fig. 4C–C″). This suggested that perturbation of Zip compromised the formation of early endosome as evident from the accumulated expression pattern of Rab5. Loss of Zip led to loss of pulling force required for the formation of endocytic vesicles indispensable for receptor internalization, leading to accumulated Notch receptor at the cell surface. Our findings suggested that Zip is necessary for Notch receptor internalization in the signal receiving cell leading to activation of the signaling pathway. The accumulation of Notch along with Rab5 was observed in all the wing discs that were examined (total number of wing discs examined = 15). Among them, 5 discs were used for the quantification of the intensity.
To rule out the involvement of the regulatory light chain in the functional role of Zipper in Notch regulation, we also examined the status of Notch in regulatory light chain compromised background. The gene spaghetti squash (sqh) encodes the myosin regulatory light chain in Drosophila. Notch appeared to be unaltered in the wing imaginal discs obtained from null mutant of spaghetti squash (Fig. S4C) indicating that the absence of myosin heavy chain (encoded by zip) is mainly responsible for the accumulation of Notch receptor at the cell surface (Fig. S4).
Over-expression of zip rescues Notch loss-of-function phenotype and this rescue is facilitated by motor domain of Zip
Through epistatic interaction studies, here we wanted to check whether over-expressing Zip in the compromised Notch background can rescue Notch loss-of-function phenotypes. At this end, we over-expressed Zip in larval wing imaginal discs of UAS-GFP-zip individuals and reduced the expression of Notch in the same tissue by dominant-negative Notch using C96-GAL4 driver. Notch, upon being downregulated under C96-GAL4, yielded a highly serrated wing phenotype which was significantly rescued by over-expressing zip in the same background (Fig. 5A–C). This rescue in the wing serration upon upregulating Zip in Notch DN background using C96 GAL4 has been shown via a bar graph (Fig. 5G) where massive nicking of the wing margin has been referred to as “severe serration” (e.g., Fig. 5B) and a lesser nicking of the wing margin has been referred to as “mild serration” (e.g., Fig. 5C). Here, the experiment was performed in two batches (Batch1 and Batch2) where a total number of 80 wings were examined for each case. In batch 1, C96 GAL4-driven UAS-GFP-zip yielded all the wings with no serration, UAS-Notch DN yielded 66 wings with severe serration and 14 wings with mild serration, UAS-GFP-zip + UAS-Notch DN yielded 66 mildly serrated wings (rescued) and 14 severely serrated wings. In batch 2, C96 GAL4-driven UAS-GFP-zip yielded wild type wings with no serration, UAS-Notch DN displayed 67 wings with severe serration and 13 wings with mild serration, UAS-GFP-zip + UAS-Notch DN yielded 60 mildly serrated wings (rescued) and 20 severely serrated wings.
Over-expression of zip rescues Notch loss-of-function phenotype. A–C C96-GAL4-driven expression of UAS-Notch-DN leads to severe wing-nicking phenotype (B) that is rescued upon expression of zip in the same background (C). Over-expression of zip alone results in wild-type wing (A). Scale bar: 3 cm. G Graph representing the number of wings showing rescue in wing serration on co-expressing zip with Notch-DN using C96-GAL4. The genotype of the flies mentioned on the X-axis of the graph are as follows: C96-GAL4/UAS-GFP-zip, C96-GAL4/UAS-Notch-DN, and C96-GAL4/UAS-GFP-zip + UAS-Notch-DN. D–F′ Representative wing discs showing the expression pattern of Cut. Downregulating Notch in C96-GAL4 region resulted in loss of expression of Cut on the DV boundary (E) which was mildly rescued upon over-expressing zip in the same background (F). Over-expression of zip alone resulted in the expression of Cut similar to wild type (D). D′, F′ Merged images showing the expression pattern of Cut under C96-GAL4-driven GFP-zip and GFP-zip with Notch DN, respectively. Similarly, E′ represents the merged image showing the expression pattern of Cut under C96-GAL4-driven UAS-Notch DN. Absence of GFP in this image denotes the absence of GFP-zip. Scale bar: 20 μm. H–H‴ Zip interacts with Notch via motor domain. H Co-expression of full-length UAS-zip with UAS-Notch-DN using C96-GAL4 lead to rescued wing serration caused by Notch loss of function alone. However, this rescue failed to occur when UAS-Notch-DN was co-expressed with UAS-Myo II Neck Rod having truncated motor domain (H′), UAS-Myo II-Rod (H″) and UAS-Myo II-Rod (delta Nterm58) (H‴) having truncated head and neck domain respectively. Scale bar: 3 cm. I A depiction of full-length UAS-GFP-zip and domain truncation stocks. UAS-GFP-zip has all the domains including head domain, neck domain harboring binding sites for essential and regulatory light chains, and rod domain. UAS-Myo II Neck Rod has truncated head domain. UAS-Myo II Rod has truncated head and neck domains. UAS-Myo II-Rod (delta Nterm58) has truncated head, neck and deletion of 58 amino acids of Rod domain. J Graphical representation of the percentage of wings showing mild and severe wing serration upon co-expression of UAS-Notch-DN with full-length zip and its truncation domains. The genotype of the flies mentioned on the X-axis of the graph is as follows: C96-GAL4/UAS-Notch-DN + UAS-GFP-zip, C96-GAL4/UAS-Notch-DN + UAS-Myo II Neck Rod, C96-GAL4/UAS-Notch-DN + UAS-Myo II Rod, C96-GAL4/UAS-Notch-DN + UAS-Myo II-Rod (delta Nterm58)
This was further validated in wing discs where we wanted to assess the expression of the Notch signaling target Cut upon over-expressing zip in the lowered Notch background. Over-expression of zip alone using C96-GAL4 resulted into the expression of Cut almost like wild type (Fig. 5D). Downregulating Notch signaling using dominant-negative form of Notch under C96-GAL4 resulted in complete loss of Cut staining in the wing discs (Fig. 5E). However, the loss of Cut expression was mildly rescued (Fig. 5F) upon over-expressing zip in the same background indicating an important role of Zip in Notch signaling (Fig. 5D–F).
Subsequently, we wanted to investigate the functional domain of Zip that is responsible for the rescue of the serrated wing phenotype caused by Notch loss-of-function. Several studies suggest that not all the domains of non-muscle myosin II are required for various biological processes [40]. It has been observed that some processes require the contractility based function of non-muscle myosin II while other processes take place normally even if the motor domain is perturbed by amino acid replacements [41,42,43]. Hence, we wanted to characterize the functional domain involved in Notch and Zip interaction that would subsequently lead to a significant rescue of the nicked wing phenotype of Notch dominant-negative individuals. In Drosophila, separate genes encode each subunit of non-muscle myosin II: zip encodes the heavy chain (zip/MyoII), spaghetti squash encodes the regulatory light chain (sqh/RLC) and mlc-c encodes the essential light chain (mlc-c/ELC) [16, 17, 40, 44]. Zip i.e., each individual heavy chain consists of (1) a globular N-terminal motor or head domain (~ 800 amino acids) that contains the ATP and actin-binding sites; (2) a neck domain (~ 50 amino acids) composed of two IQ motifs that bind one ELC and one RLC; (3) a coiled coil or rod domain (~ 1100 amino acids) composed of heptad repeats; and (4) a short C-terminal segment, termed the tailpiece of ~ 34–47 amino acids in length [45]. Truncation alleles with truncated domains of non-muscle myosin II was used for the study of the interaction between Zip domain and Notch. UAS-Myo II-Neck-Rod truncation allele lacks only the motor domain, UAS-Myo II-Rod encompasses the entire rod domain, thus lacking the motor domain and the neck region and UAS-Myo II-Rod (delta Nterm58) lacks only the first 58 amino acids of the rod domain along with motor and neck domains (Fig. 5I) [40]. It was observed that no rescue of the wing-nicking phenotype could occur when UAS-Notch-DN was co-expressed with the truncated allele, UAS-Myo II-Neck-Rod, UAS-Myo II-Rod and UAS-Myo II-Rod (delta Nterm58) (Fig. 5H′–H‴). Our findings indicated that the truncation of motor domain fails to rescue the nicked wing phenotype caused by Notch loss-of-function suggesting that motor domain is indispensable for the function of Zip in regulation of Notch.
zip synergises with Notch
It is apparent from genetic interaction experiments that zip modulates Notch signaling activity. Hence, we wanted to explore the integrative effect of Zip in Notch signaling. To investigate this, we over-expressed Notch-FL and zip together using vg-GAL4 driver. Over-expression of zip alone resulted in wild-type wing phenotype and over-expressed Notch-FL alone resulted in multiple wing phenotypes, such as wing crumpling, wing blisters, ectopic outgrowth, vein disorganization, and ectopic marginal bristles. Over-expression of both zip and Notch-FL together resulted in enhancement of only Notch-FL over-expression induced wing phenotypes (Fig. S5A–C). Massive wing disorganization, such as wing duplication, large wing blisters, severe vein disorganization, etc., was observed upon co-expression of Notch-FL and zip (Fig. S5C).
In order to check the synergistic effect of Notch-FL and Zip on the wing phenotype using other GAL4 driver line, we co-expressed UAS-Notch-FL and UAS-GFP-zip in the posterior wing compartment using en-GAL4 driver. Zip yielded almost wild type wings when over-expressed using en-GAL4. Notch-FL over-expression in the posterior domain resulted in multiple wing phenotypes including fifth vein shortening, extra vein material, wing crumpling and blisters. When Zip was co-expressed with Notch-FL, it culminated in 100% pupal lethality, thus confirming a significant synergistic effect of Notch and Zip (Fig. 6A–C).
A–I′ Zip synergises with Notch-FL to lead to pupal lethality and increase in the ectopic expression of Cut and Wg on D/V boundary. A–C Zip synergy with Notch lead to pupal lethality. A Over-expression of zip by en-GAL4 resulted in wild type wings whereas Notch-FL over-expression in the posterior domain resulted in disruption of wing morphology with shortening and thickening of fourth and fifth vein, presence of extra vein material, wing crumpling, and loss of cross veins (B). Flies failed to emerge from the pupal case when UAS-Notch–FL was co-expressed with UAS-zip resulting in 100% pupal lethality (C). Scale bar: 3 cm. D–I′ Representative wing discs show the expression patterns of Cut and Dpn. Ectopic expression of Cut (H) and Dpn (I) gets enhanced upon co-expression of zip with Notch (FL) under en-GAL4 in contrast to over-expressed zip that shows wild-type expression of Cut and Dpn (D, E) and over-expressed Notch-FL that shows mild ectopic expression of Cut and Dpn (F, G). D′, H′ are the merged images showing expression pattern of Cut under en-GAL4-driven expression of GFP-Zip and GFP-zip with Notch-FL, respectively. Similarly, E′, I′ are the merged images showing the expression pattern of Dpn using GFP-zip and GFP-zip with Notch-FL, respectively, in the posterior region. F′, G′ panel represent the expression pattern of Cut and Dpn respectively under Notch (FL) driven by en-GAL4. Absence of GFP in this image represents absence of GFP-zip. J, K Graphs representing the intensity profiling of Cut and Dpn in en-GAL4 > UAS-zip, en-GAL4 > UAS-Notch FL and en-GAL4 > UAS-zip + UAS-Notch FL. Scale bar: 20 μm
To further validate the synergistic interaction between Notch and Zip, we analyzed the levels of Notch targets, Cut and Dpn. Over-expressing GFP-tagged zip alone using en-GAL4 driver did not result in any significant increase in the expression of Cut and Dpn. Over-expression of only Notch-FL in the posterior compartment of wing disc resulted in increased Cut and Dpn expression. However, co-expression of zip and Notch-FL under en-GAL4 resulted in an elevated level of Cut and Dpn in the posterior domain of wing imaginal discs (Fig. 6D–I′). Image J was used for the intensity profiling. A total of 23 discs were examined and all of them showed consistent results. 6 discs were used for the quantification. Mean intensity for Cut expression in en-GAL4-driven > UAS-GFP-zip was 19.4, for UAS-Notch FL was 21.2 and for UAS-GFP-zip + UAS-Notch FL was 26. Mean intensity for Dpn expression in en-GAL4-driven > UAS-GFP-zip was 34, for UAS-Notch FL was 44.5 and for UAS-GFP-zip + UAS-Notch FL was 52.17.
At this point, it was interesting to explore the synergistic interaction between Zip and activated Notch. Hence, we explored the co-operative effect of zip with processed Notch (Notch-ICD). To analyze this, we co-expressed both Notch-ICD and zip using C96-GAL4 driver. C96-GAL4-driven zip alone did not show any phenotype, whereas C96-GAL4-driven Notch-ICD resulted in wing margin defects with irregular marginal bristles resulting in mild crumpling of the wing. When Notch-ICD and zip were co-expressed together with C96-GAL4 driver, it resulted in increased irregular marginal bristles and severely crumpled wings (Fig. S5D–F).
In order to determine that GAL4 is not a limiting factor in our experimental set-up and there is a similar level of expression of transgenes downstream to single copy of UAS (UAS-GFP zip/UAS-Notch FL/UAS-Notch ICD) in control case and double copy of UAS (UAS-GFP zip + UAS-Notch FL) in experimental condition, we performed immunoblotting to check the expression level of Zip and Notch in protein lysates from UAS-GFP-Zip, UAS-Notch FL, UAS-Notch FL + UAS-GFP Zip and UAS-Notch ICD + UAS-GFP-Zip driven with GMR-GAL4. A similar level of Zip and Notch expression was observed in the transgenes with single UAS and double UAS copy (Fig. S1D′–F′). The graphs depicting a similar band intensity suggest that irrespective of single or double copy of UAS, the expression level of transgenes is almost same in all the cases and that GAL4 is not a limiting factor here (Fig. S1D″–F″). Similarly, we also analyzed the GFP-Zip intensity between en-GAL > UAS-GFP-zip and en-GAL > UAS-GFP-zip + UAS-NotchFL condition (Fig. 6D′, H′; E′, I′) to rule out the potential GAL4 dilution effect. The mean intensities of GFP-Zip between respective genotypes were observed to be 32.5 in Zip alone condition and 37.5 in Zip co-expression with Notch FL. The difference in the intensities between both the genotypes was calculated to be non-significant (p > 0.05), thus indicating that there has been no case of GAL4 dilution effect in our studies (Fig. S1F″).
Interactivity of Zip and activated Notch results into hyperplasia, high mitotic activity index (MAI) and compromised epithelial integrity
The integration of other genes with Notch signaling is known to modulate its downstream activity [46,47,48]. To further explore and dissect the downstream changes mediated by Zip upon integration with Notch signaling, we co-expressed zip with Notch-ICD in the ventral domain of the wing disc using vg-GAL4. We have already shown the role of Zip in modulating Notch signaling outcome by interacting with the Notch-FL. Here we wanted to explore the effect of Zip over-expression in activated Notch-mediated hyperproliferation. Over-expression of zip alone does not cause any significant changes in wing disc morphology and over-expression of Notch-ICD results in hyperplastic wing disc with deformed morphology. When zip and Notch-ICD both were co-expressed, the severity of tumorous phenotype was much more increased in terms of size and morphology (Fig. 7A–B″). Here, 3 discs of each genotype were analyzed to determine the size of the tumorous wing discs. To determine the proliferation activity in these discs, mitotically active cells were checked using Phosphohistone H3 (PH3) staining. An increase in the number of mitotically active cells was apparent when zip was co-expressed with Notch-ICD (Fig. 7C–C″). At this end, 3 discs from each genotype were analyzed to determine the number of PH3 positive cells.
A–D″ Co-expression of Zip and Notch ICD results into hyperplasia, high mitotic activity index (MAI) and compromised epithelial integrity. A–B″ Representative wing discs showing the wild type wing morphology when UAS-GFP-zip was over-expressed with vg-GAL4 (A, B). Wing morphology was observed to be hyperplastic when UAS-Notch-ICD was over-expressed with vg-GAL4 (A′, B′). Co-expression of UAS-Notch-ICD along with UAS-GFP-zip using vg-GAL4 resulted in highly deformed severely hyperplastic tumorigenic wing discs with increase in size (A″, B″). Scale bar in the panels A, A′ and A″ represents the size of the discs with respective genotypes. C–C″ Representative wing discs showing mitotic activity index similar to wild type upon over-expressing Zip in vestigial domain (C). An increased mitotic activity index was observed upon over-expressing Notch ICD by vg-GAL4 (C). However, PH3 puncta representing MAI was drastically increased upon co-expressing Notch ICD with Zip in vestigial compartment (C″). D–D″ Armadillo staining showing the status of adherent junctions when zip alone, Notch-ICD alone and Notch ICD along with zip was co-expressed using vg-GAL4. Epithelial integrity was observed to be almost wild type when zip and Notch ICD were over-expressed alone. However, highly disrupted tissue integrity was found upon co-expression of UAS-Notch-ICD and UAS-zip using vg-GAL4 (D″). B‴ Graphical illustration of the size of the imaginal disc in vg-GAL4 > UAS-zip, vg-GAL4 > UAS-Notch ICD and vg-GAL4 > UAS-zip + UAS-Notch ICD imaginal discs. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test was conducted to calculate the p values (*p < 0.05, ***p < 0.001). C‴ Graph representing the approximate number of PH3 positive cells per unit area in vg-GAL4 > UAS-zip, vg-GAL4 > UAS-Notch ICD and vg-GAL4 > UAS-zip + UAS-Notch ICD imaginal discs. Scale bar: 20 μm
Further, we examined the cell–cell junction integrity to measure the severity of tumorous-phenotype. We investigated the status of Armadillo (Arm), β-catenin homologue in fly, which is an important component of the cytoskeleton and is required to maintain the integrity of the tissue. Cell–cell junction integrity is maintained when Zip or Notch-ICD are expressed individually. However, it was observed that zip upon co-expression with Notch-ICD in the vestigial domain significantly deregulated the expression of junctional molecule as evident by the disrupted and diffused expression pattern of Arm (Fig. 7D–D″). The compromised integrity of the junctional molecule in the hyperplastic discs indicates the combinatorial effect of Zip and Notch in Notch-mediated tumorigenesis.
To check whether Notch has any role in the regulation of Zip activity, we examined the phosphorylation status of myosin regulatory light chain in the wing imaginal discs of Notch over-expressed (Notch-induced hyperplastic) condition in ventral domain using vg-GAL4 (vg-GAL4 > UAS-Notch ICD) and down-regulated condition at D/V boundary using C96-GAL4 (C96-GAL4 > UAS-Notch DN). Anti-phospho-myosin regulatory light chain 2 antibody was used for studying the phosphorylation status of the light chain. Oregon R served as the control. No alteration in the phosphorylation status of myosin regulatory light chain could be observed in Notch perturbed condition compared to the control. This indicated that Notch does not modulate Zip activity (Fig. S6A–C″).
We also examined the status of Notch ligand Delta in Notch hyperplastic condition and observed that Delta was unaltered. However, perturbation in the expression pattern of Delta occurred mainly due to the disrupted tumor morphology of the wing disc from Notch over-expressed condition compared to the control (Fig. S6D–E″).
Zip induces JNK-mediated invasiveness in tumors caused by activated Notch
To determine whether the Notch-induced tumorous phenotypes become more aggravated in presence of over-expressed Zip, we analyzed the expression of Matrix metalloproteinase 1 (MMP1) that disintegrates the ECM in Notch and Zip co-expressing wing discs.
A prerequisite for invasiveness or metastasis to occur is the penetration of the surrounding extracellular matrix (ECM) stroma and epithelial basement membranes by tumor cells [49]. The basement membrane and ECM components are degraded by enzymes, matrix metalloproteinases (MMPs) [50, 51]. Various human tumors display increased MMP expression in correlation with metastasis [52,53,54]. Hence, we studied the expression levels of MMP1 in the tumorous discs induced by over-expression of Notch alone or Notch and Zip together. No significant expression of MMP1 was observed in the case of wing discs over-expressing only activated Notch (also seen earlier by [47, 55]. Similarly, Zip over-expressed non-tumorous disc also showed no expression of MMP1. However, a high level of MMP1 was detected when zip was co-expressed with Notch-ICD (Fig. 8A–C).
A–P Zip induces JNK mediated invasiveness in tumors caused by Notch. A–C Representative wing discs showing the status of MMP1 in zip over-expressed condition with vg-GAL4 where the expression level of MMP1 was observed to be almost wild type (A). Notch ICD when over-expressed in ventral domain results in no MMP1 expression (B). However, mmp1 expression was observed to be upregulated when UAS-Notch ICD was co-expressed with UAS-GFP-zip using vg-GAL4 (C). A′–C′ Merged images showing the expression status of MMP1 under vg-GAL4-driven UAS-GFP-zip, UAS-Notch-ICD and UAS-GFP-zip along with UAS-Notch-ICD respectively. E–G Wing discs showing the expression level of Cleaved Caspase 3 upon co-expression of Notch ICD along with Zip and Notch ICD and Zip individually. A mild and no expression of Cl Casp3 was observed upon over-expression of Notch ICD and zip using vg-GAL4 respectively (E-F). However, this expression was increased when the two proteins were co-expressed in the proliferated vestigial domain (G). E′–G′ Merged images showing the expression pattern of Cleaved Caspase 3 using vg-GAL4-driven UAS-GFP-zip, UAS-Notch-ICD and UAS-GFP-zip + UAS-Notch-ICD. I–K Representative wing discs showing the status of activated JNK with phospho-JNK staining. I, J Mild expression of pJNK puncta was observed upon over-expression of UAS-GFP-zip and UAS-Notch-ICD with vg-GAL4. However, an increase in the expression level and puncta size of pJNK was observed upon co-expression of Notch-ICD with zip. I′–K′ Merged images showing the expression status of pJNK with vg-GAL4-driven UAS-GFP-zip, UAS-Notch-ICD alone and UAS-GFP-zip together with UAS-Notch-ICD. D, H, L Wing discs showing the suppressed expression status of MMP1 (D), Cleaved Caspase 3 (H) and pJNK (L) in vestigial domain upon blocking JNK using UAS-Bsk-DN in the background with co-expressed UAS-Notch-ICD and UAS-GFP-zip. D′, H′, L′ represents the merged images showing the expression status of MMP1, Cleaved Caspase 3 and pJNK with co-expressed UAS-Notch-ICD, UAS-GFP-zip and UAS-Bsk-DN using vg-GAL4. (M-P) Graphical representation of MMP1, Cleaved Caspase 3, and pJNK intensity in vg-GAL4-driven UAS-GFP-zip, UAS-Notch-ICD, UAS-GFP-zip + UAS-Notch-ICD and UAS-GFP-zip + UAS-Notch-ICD + UAS-Bsk-DN. (M and N) One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test was performed to calculate p values (ns > 0.05, **p < 0.01, ***p < 0.001). Unpaired t test was performed to calculate p values (***p < 0.001). Scale bar: 20 μm
As such caspase activation is a hallmark of cell death, however recent studies also suggest the role of these proteases in non-apoptotic functions [56, 57]. It has been reported that caspases have a functional role during tumor invasion and metastasis other than their cell death-related function [58]. These non-apoptotic roles have indicated that caspases can get activated without initiating apoptotic cascade leading to the cleavage of specific cellular substrates. One of the non-apoptotic roles of caspases being recently investigated includes controlling cell cycle and driving proliferation leading to cell survival [59, 60]. It has been demonstrated that apical caspase, cleaved caspase-3 can lead to proliferation of the cells via the activation of the JNK pathway which subsequently drives the expression of MMP1 [48, 61]. To examine the role of caspases in Notch and Zip co-expressed tumor, we examined the levels of cleaved caspase-3 in the vestigial domain of the wing discs over-expressing Notch and Zip. A high level of caspase expression was observed in the proliferated vestigial domain of the Notch-Zip co-expressed discs in comparison to the wing disc expressing Notch or Zip alone. This showed that the activation of caspases is linked to the proliferation resulting due to the collaboration of Notch and Zip (Fig. 8E–G). An elevated expression level of caspases leads to the activation of JNK pathway that consequently lead to MMP activation and proliferation [54, 57, 58, 61, 62]. Therefore, we also wanted to explore the phosphorylation status of active JNK (Bsk) in the tumors where Notch and Zip were co-expressed. At this end, we performed immunostaining of the wing imaginal discs with a phospho-JNK-specific antibody recognizing the active form of JNK. In the case of over-expression of Notch and zip alone, there was no activation of JNK. However, JNK was found to be hyper-phosphorylated in the vestigial domain when zip was co-expressed with Notch-ICD, indicating the involvement of JNK in inducing invasive tumor (Fig. 8I–K).
To further validate that JNK is the key player in causing aggressive tumor due to Notch and Zip synergy, we blocked the JNK signaling in the same background using a dominant-negative form of Basket (UAS-Bsk DN). By abrogating JNK signaling in the background co-expressing Notch-ICD and zip, we wanted to see if we can rescue the hyperproliferative activity by comparing the size, and metastatic behavior by MMP1 expression levels in these tumors. Immunostaining experiments were performed to analyze the level of phosphorylated JNK, MMP1, and Cleaved caspase3 in this background. As expected, the JNK signaling was significantly reduced upon expression of Bsk-DN (Fig. 8L). Further, the level of MMP1 and Cleaved caspase3 was also significantly reduced to near wild type levels in Notch-Zip co-expressed discs when JNK was blocked (Fig. 8D, H). These results confirmed the role of JNK in inducing hyper-proliferation and invasiveness in the tumorigenic discs resulting due to Notch and Zip synergy.
Graphs were plotted to show the intensity of the expression of MMP1, Cleaved Caspase3 and p-JNK. A total of 3 discs were used for the quantification purpose for every genotype. Mean intensity for MMP1 for different genotypes was observed as follows: vg-GAL4-driven GFP-zip: 7.25, Notch ICD: 11.29, GFP-zip + Notch ICD: 29.356 and GFP-zip + Notch ICD + Bsk DN: 6.023. Mean intensity for Cl Casp3 for different genotypes was observed as follows: vg-GAL4-driven GFP-zip: 6.334, Notch ICD: 9.77, GFP-zip + Notch ICD: 15.711 and GFP-zip + Notch ICD + Bsk DN: 5.83. Mean intensity for p-JNK for different genotypes was observed as follows: vg-GAL4-driven GFP-zip: 9.32, Notch ICD: 13.25, GFP-zip + Notch ICD: 17.47 and GFP-zip + Notch ICD + Bsk DN: 9.07.
Discussion
Here we present evidence that non-muscle myosin II (NM II) Zip physically and genetically interacts with Notch. Down-regulation of Notch signaling targets either using RNAi or through dominant-negative over-expression line clearly showed that Zip positively regulates Notch signaling. Synergistic interactions between Notch and Zip also indicated that Notch is positively regulated by Zip. Upregulation of downstream targets of the Notch pathway, Cut, and Dpn, was detected when Zip and Notch were co-expressed. Here we showed that Zip integrates with processed Notch to cause invasive tumor of the wing discs. Increased activity of JNK signaling downstream to Notch/Zip collaboration was evident by hyperphosphorylated Basket (orthologue of JNK) in the hyperplastic discs. Thus, we postulate that Notch/Zip synergy converges on JNK signaling that ultimately regulates invasive behavior of the tumorous discs.
Earlier it was proposed that DSL ligand binding to heterodimeric Notch receptor in the plasma membrane induces proteolytic cleavage (S2) via a disintegrin and metalloprotease (ADAM), which facilitates γ-secretase proteolysis (S3) within the transmembrane region to release the Notch intracellular domain (NICD) [6, 63]. Later on, it was shown that in canonical ligand-dependent Notch pathway, ligand binding and subsequent endocytosis of the NECD-bound ligand into the ligand-expressing signal-sending cell generates pulling-force on the extracellular fragment of Notch, which causes some conformational changes in the Notch receptor and consequently allows S2 cleavage to occur. Thus, NECD dissociation is not a consequence of S2 site cleavage, but rather that dissociation occurs first, and subsequently allows for S2 site cleavage by ADAMs [7, 64, 65]. Interestingly NM II plays critical role in clathrin-mediated endocytosis which is important for receptor-mediated signaling. It has been shown that clathrin-mediated endocytosis is actomyosin-dependent [38, 66]. Endocytosis is required for ligand-dependent Notch activation in both signal-sending and receiving cells. Recently, it was shown that NM II activity is required for robust Notch signaling and NM II is important for signaling in both signal-sending and receiving cells [23]. NM II contractility plays important role in Notch signaling and NM II-mediated mechanotransduction via which forces generated within the actomyosin cytoskeleton influence signaling. Actomyosin contractility in cells having extensive lateral contacts with other cells has been shown to promote Notch activation and it was postulated that endocytosis and myosin-dependent pulling may both contribute to force-dependent Notch activation in these cells [23]. Here we showed that membrane accumulation of Notch and early endosomal marker Rab5 occurred in Zip loss-of-function condition which clearly indicates that NM II Zip plays important role in endocytosis of Notch. Further, large-scale proteomic analysis based on co-immunoprecipitation of GFP-tagged Zip interacting proteins using anti-GFP antibody identified several proteins of endocytic machinery as interacting partners of Zip along with Notch. Some of these proteins were Clathrin heavy chain, Dynamin, Dynein heavy chain, Kinesin heavy chain, AP-2 complex subunit alpha, Ras-related protein Rab 3, etc. Identification of these components as interacting partners of Zip suggested that Zip may have a major role to play in the endocytosis of Notch receptor in the signal receiving cell. Biological processes, such as cytoskeletal organization, establishment of vesicle localisation, receptor-mediated endocytosis, myosin filament assembly, and receptor internalization, was observed to be highly enriched in the proteomic network thus reinforcing role of Zip in Notch receptor endocytosis.
Further, presence of non-muscle myosin IIs at the nuclear periphery and its colocalization with the linker of nucleoskeleton and cytoskeleton (LINC) protein Nesprin2 and apical actin caps suggests that it plays role in transmission of cytoplasmic signals to the nucleus [67, 68]. Recently it has been suggested that nuclear envelope proteins are involved in regulation of gene expression by signal transmission from the cytoskeleton to the nucleus [69]. Here we have shown the synergistic interaction of Zip with FL-Notch as Notch downstream targets were ectopically expressed when these two proteins were co-expressed in larval discs. We have also presented the combinatorial effect of Zip and activated Notch in Notch-mediated tumorigenesis. Recently it has been shown that NMIIs colocalize at the perinuclear area and modulate the expression of genes associated with cancer progression and it was postulated that NMIIs may act as a mechanotransducer during tumor progression [67]. We have also identified cyto- and nucleo-skeleton components that aid in nuclear transmission of signals from the cytoplasm to nucleus as interacting partners of Zip in our MS analysis. These components included Nesprin-1 and Nesprin-2, nuclear anchorage protein, Lamin, nuclear pore complex proteins, etc. The underlying mechanism is unknown at this point, but it is possible that similarly, Zip may enhance the Notch signaling activity through its mechanotransduction function.
There is an emerging realization that mutations in Notch gene and aberrant Notch signaling are crucial factors in tumor initiation and progression. It has been shown that depending on the cellular context, Notch behaves as an oncogene or tumor suppressor gene, and a large number of cancers are associated with abnormal Notch functioning. Interestingly, several studies reported that NM II also promotes different types of cancer progression as well as it can also act as a tumor suppressor suggesting its context-dependent function [70]. It has been reported that NMIIs lie downstream of the Wnt signaling pathway and can regulate the local invasion of amoeboid melanoma cells, and its metastatic potential [71,72,73]. In breast cancer cells, invasiveness was shown to be increased via phosphorylation and switching from NM II C population to NM II B. In cancer stem cells (CSC), NM II B has shown to facilitate squeezing of CSC’s nucleus to pass through tight spaces leading to dissemination [74, 75]. Upregulation of NM IIs in transforming, proliferating cells in the early stages of cancer progression, and knockdown of NM II in these cells leading to altered expression of cancer pathway-related genes further hints at the involvement of NM II in cellular reprogramming [67, 76] Further, several biological processes depend on the proper cellular responses to mechanical forces. Zip–Notch synergy opens up new avenues to study force-dependent Notch activation in different cellular contexts. Further investigation of mechano-transducer Zip-induced Notch signaling activity in different disease conditions including tumorigenesis will advance the development of novel Notch-targeted therapeutics.
References
Andersson ER, Sandberg R, Lendahl UJD (2011) Notch signaling: simplicity in design, versatility in function. Development 138(17):3593–3612
Artavanis-Tsakonas S, Rand MD, Lake RJ (1999) Notch signaling: cell fate control and signal integration in development. Science 284(5415):770–776
Fortini ME (2009) Notch signaling: the core pathway and its posttranslational regulation. Dev Cell 16(5):633–647
Guruharsha K, Kankel MW, Artavanis-Tsakonas S (2012) The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet 13(9):654–666
Blaumueller CM et al (1997) Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell 90(2):281–291
Brou C et al (2000) A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell 5(2):207–216
Nichols JT et al (2007) DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J Cell Biol 176(4):445–458
Yamamoto S, Charng W-L, Bellen HJ (2010) Endocytosis and intracellular trafficking of Notch and its ligands. Curr Top Dev Biol 92:165–200
De Strooper B et al (1999) A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature 398(6727):518–522
Struhl G, Greenwald I (1999) Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature 398(6727):522–525
Sachan N et al (2013) The Drosophila importin-α3 is required for nuclear import of Notch in vivo and it displays synergistic effects with Notch receptor on cell proliferation. PLoS ONE 8(7):e68247
Bray S, Bernard F (2010) Notch targets and their regulation. Curr Top Dev Biol 92:253–275
Fortini ME, Artavanis-Tsakonas S (1994) The suppressor of hairless protein participates in notch receptor signaling. Cell 79(2):273–282
Kopan R, Ilagan MXGJC (2009) The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137(2):216–233
Wu L et al (2000) MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet 26(4):484–489
Karess RE et al (1991) The regulatory light chain of nonmuscle myosin is encoded by spaghetti-squash, a gene required for cytokinesis in Drosophila. Cell 65(7):1177–1189
Kiehart D et al (1989) Identification of the gene for fly non-muscle myosin heavy chain: Drosophila myosin heavy chains are encoded by a gene family. EMBO J 8(3):913–922
Vicente-Manzanares M et al (2009) Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol 10(11):778–790
Young P et al (1993) Morphogenesis in Drosophila requires nonmuscle myosin heavy chain function. Genes Dev 7(1):29–41
Gordon WR et al (2015) Mechanical allostery: evidence for a force requirement in the proteolytic activation of Notch. Dev Cell 33(6):729–736
Meloty-Kapella L et al (2012) Notch ligand endocytosis generates mechanical pulling force dependent on dynamin, epsins, and actin. Dev Cell 22(6):1299–1312
Wang X, Ha T (2013) Defining single molecular forces required to activate integrin and notch signaling. Science 340(6135):991–994
Hunter GL et al (2019) A role for actomyosin contractility in Notch signaling. BMC Biol 17(1):1–15
Mukherjee A et al (2005) Regulation of Notch signalling by non-visual β-arrestin. Nat Cell Biol 7(12):1191–1201
Shevchenko A et al (1996) Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc Natl Acad Sci 93(25):14440–14445
Franke JD, Montague RA, Kiehart DP (2010) Nonmuscle myosin II is required for cell proliferation, cell sheet adhesion and wing hair morphology during wing morphogenesis. Dev Biol 345(2):117–132
Lawrence N et al (2000) Structural requirements for notch signalling with delta and serrate during the development and patterning of the wing disc of Drosophila. Development 127(14):3185–3195
Go MJ, Eastman DS, Artavanis-Tsakonas S (1998) Cell proliferation control by Notch signaling in Drosophila development. Development 125(11):2031–2040
Edwards KA, Kiehart DPJD (1996) Drosophila nonmuscle myosin II has multiple essential roles in imaginal disc and egg chamber morphogenesis. Development 122(5):1499–1511
Franke JD et al (2006) Native nonmuscle myosin II stability and light chain binding in Drosophila melanogaster. Cell Motil Cytoskelet 63(10):604–622
Wheatley S, Kulkarni S, Karess R (1995) Drosophila nonmuscle myosin II is required for rapid cytoplasmic transport during oogenesis and for axial nuclear migration in early embryos. Development 121(6):1937–1946
Helms W et al (1999) Engineered truncations in the Drosophila mastermind protein disrupt Notch pathway function. Dev Biol 215(2):358–374
Kankel MW et al (2007) Investigating the genetic circuitry of mastermind in Drosophila, a notch signal effector. Genetics 177(4):2493–2505
Micchelli CA, Rulifson EJ, Blair SS (1997) The function and regulation of cut expression on the wing margin of Drosophila: Notch, Wingless and a dominant negative role for Delta and Serrate. Development 124(8):1485–1495
San-Juán BP, Baonza A (2011) The bHLH factor deadpan is a direct target of Notch signaling and regulates neuroblast self-renewal in Drosophila. Dev Biol 352(1):70–82
Ray S (2014) The cell: a molecular approach. Yale J Biol Med 87(4):603–604
Vaccari T et al (2008) Endosomal entry regulates Notch receptor activation in Drosophila melanogaster. J Cell Biol 180(4):755–762
Chandrasekar I et al (2014) Nonmuscle myosin II is a critical regulator of clathrin-mediated endocytosis. Traffic 15(4):418–432
Christoforidis S et al (1999) The Rab5 effector EEA1 is a core component of endosome docking. Nature 397(6720):621–625
Franke JD, Montague RA, Kiehart DPJDB (2010) Nonmuscle myosin II is required for cell proliferation, cell sheet adhesion and wing hair morphology during wing morphogenesis. Develop Biol 345(2):117–132
Guha M, Zhou M, Wang Y-L (2005) Cortical actin turnover during cytokinesis requires myosin II. Curr Biol 15(8):732–736
Royou A et al (2004) Reassessing the role and dynamics of nonmuscle myosin II during furrow formation in early Drosophila embryos. Mol Biol Cell 15(2):838–850
Xu XS et al (2001) During multicellular migration, myosin ii serves a structural role independent of its motor function. Dev Biol 232(1):255–264
Edwards KA, Chang X-J, Kiehart DP (1995) Essential light chain of Drosophila nonmuscle myosin II. J Muscle Res Cell Motil 16(5):491–498
Dulyaninova NG, Bresnick AR (2013) The heavy chain has its day: regulation of myosin-II assembly. BioArchitecture 3(4):77–85
Artavanis-Tsakonas S, Muskavitch MAJCTIDB (2010) Notch: the past, the present, and the future. Curr Top Develop Biol 92:1–29
Ho DM, Pallavi S, Artavanis-Tsakonas S (2015) The Notch-mediated hyperplasia circuitry in Drosophila reveals a Src-JNK signaling axis. Elife 4:e05996
Pallavi S et al (2012) Notch and Mef2 synergize to promote proliferation and metastasis through JNK signal activation in Drosophila. EMBO J 31(13):2895–2907
Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100(1):57–70
McCawley LJ, Matrisian LM (2000) Matrix metalloproteinases: multifunctional contributors to tumor progression. Mol Med Today 6(4):149–156
Vu TH, Werb Z (2000) Matrix metalloproteinases: effectors of development and normal physiology. Genes Dev 14(17):2123–2133
Nelson AR et al (2000) Matrix metalloproteinases: biologic activity and clinical implications. J Clin Oncol 18(5):1135–1135
Stetler-Stevenson WG, Aznavoorian S, Liotta LA (1993) Tumor cell interactions with the extracellular matrix during invasion and metastasis. Annu Rev Cell Biol 9(1):541–573
Uhlirova M, Bohmann D (2006) JNK-and Fos-regulated Mmp1 expression cooperates with Ras to induce invasive tumors in Drosophila. EMBO J 25(22):5294–5304
Paul MS et al (2018) Notch signals modulate lgl mediated tumorigenesis by the activation of JNK signaling. BMC Res Notes 11(1):1–9
Mollereau B et al (2013) Compensatory proliferation and apoptosis-induced proliferation: a need for clarification. Cell Death Differ 20(1):181–181
Perez E, Lindblad JL, Bergmann A (2017) Tumor-promoting function of apoptotic caspases by an amplification loop involving ROS, macrophages and JNK in Drosophila. Elife 6:e26747
Rudrapatna VA, Bangi E, Cagan RL (2013) Caspase signalling in the absence of apoptosis drives Jnk-dependent invasion. EMBO Rep 14(2):172–177
Huh JR et al (2004) Multiple apoptotic caspase cascades are required in nonapoptotic roles for Drosophila spermatid individualization. PLoS Biol 2(1):e15
Lamkanfi M et al (2007) Caspases in cell survival, proliferation and differentiation. Cell Death Differ 14(1):44–55
Ryoo HD, Gorenc T, Steller H (2004) Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev Cell 7(4):491–501
Pinal N, Calleja M, Morata G (2019) Pro-apoptotic and pro-proliferation functions of the JNK pathway of Drosophila: roles in cell competition, tumorigenesis and regeneration. Open Biol 9(3):180256
Mumm JS et al (2000) A ligand-induced extracellular cleavage regulates γ-secretase-like proteolytic activation of Notch1. Mol Cell 5(2):197–206
Langridge PD, Struhl G (2017) Epsin-dependent ligand endocytosis activates Notch by force. Cell 171(6):1383–139. e62
Parks AL et al (2000) Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development 127(7):1373–1385
Mayya C et al (2022) The roles of dynein and myosin VI motor proteins in endocytosis. J Cell Sci 135(6):jcs259387
Halder D et al (2019) Nonmuscle myosin IIA and IIB differentially modulate migration and alter gene expression in primary mouse tumorigenic cells. Mol Biol Cell 30(12):1463–1476
Keeling MC et al (2017) Actomyosin and vimentin cytoskeletal networks regulate nuclear shape, mechanics and chromatin organization. Sci Rep 7(1):1–14
Kirby TJ, Lammerding J (2018) Emerging views of the nucleus as a cellular mechanosensor. Nat Cell Biol 20(4):373–381
Pecci A et al (2018) MYH9: structure, functions and role of non-muscle myosin IIA in human disease. Gene 664:152–167
Ishihara S et al (2013) Substrate stiffness regulates temporary NF-κB activation via actomyosin contractions. Exp Cell Res 319(19):2916–2927
Wei SC et al (2015) Matrix stiffness drives epithelial-mesenchymal transition and tumour metastasis through a TWIST1–G3BP2 mechanotransduction pathway. Nat Cell Biol 17(5):678–688
Rodriguez-Hernandez I et al (2020) WNT11–FZD7–DAAM1 signalling supports tumour initiating abilities and melanoma amoeboid invasion. Nat Commun 11(1):5315
Beach JR et al (2011) Myosin II isoform switching mediates invasiveness after TGF-β-induced epithelial-mesenchymal transition. Proc Natl Acad Sci USA 108(44):17991–17996
Thomas D et al (2016) Increased cancer stem cell invasion is mediated by myosin IIB and nuclear translocation. Oncotarget 7(30):47586–47592
Saha S et al (2011) Increased expression of nonmuscle myosin IIs is associated with 3MC-induced mouse tumor. Febs J 278(21):4025–4034
Acknowledgements
We acknowledge the Bloomington Drosophila Stock Centre, Indiana University, USA and Developmental Studies Hybridoma Bank, University of Iowa, USA for stocks and reagents. We are thankful to Spyros Artavanis-Tsakonas (Department of Cell Biology, Harvard Medical School, USA), Prof. Daniel P. Kiehart (Department of Biology, Duke University, USA), Prof. Yuh N. Jan (Howard Hughes Medical Institute, San Francisco, California, USA), Prof. Akira Nakamura (Institute of Molecular Embryology and Genetics, Kumamoto, Japan) for kindly providing us with specified reagents. We acknowledge the confocal and proteomic facilities of the Interdisciplinary School of Life Sciences, Banaras Hindu University, India and High-Resolution Mass Spectrometry Lab, SATHI-BHU, Central Discovery Centre, Banaras Hindu University, India.
Funding
This work was supported by grants from the Department of Science and Technology (DST), Government of India (CRG/2021/006975), and the Institute of Eminence Scheme, Banaras Hindu University, India. Fellowship support to D.V. and A.S. was provided by the Council of Scientific and Industrial Research (CSIR), Government of India.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Data availability
The datasets supporting the conclusions of the article are included within the article. Data available upon request.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Verma, D., Singh, A., Singh, J. et al. Regulation of Notch signaling by non-muscle myosin II Zipper in Drosophila. Cell. Mol. Life Sci. 81, 195 (2024). https://doi.org/10.1007/s00018-024-05142-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-024-05142-1