Abstract
During in vitro culture, human pluripotent stem cells (hPSCs) often acquire survival advantages characterized by decreased susceptibility to mitochondrial cell death, known as “culture adaptation.” This adaptation is associated with genetic and epigenetic abnormalities, including TP53 mutations, copy number variations, trisomy, and methylation changes. Understanding the molecular mechanisms underlying this acquired survival advantage is crucial for safe hPSC-based cell therapies. Through transcriptome and methylome analysis, we discovered that the epigenetic repression of CHCHD2, a mitochondrial protein, is a common occurrence during in vitro culture using enzymatic dissociation. We confirmed this finding through genetic perturbation and reconstitution experiments in normal human embryonic stem cells (hESCs). Loss of CHCHD2 expression conferred resistance to single cell dissociation-induced cell death, a common stress encountered during in vitro culture. Importantly, we found that the downregulation of CHCHD2 significantly attenuates the activity of Rho-associated protein kinase (ROCK), which is responsible for inducing single cell death in hESCs. This suggests that hESCs may survive routine enzyme-based cell dissociation by downregulating CHCHD2 and thereby attenuating ROCK activity. These findings provide insights into the mechanisms by which hPSCs acquire survival advantages and adapt to in vitro culture conditions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Contrasting somatic cells, human pluripotent stem cells (hPSCs) have distinctive cellular and molecular properties that preserve the genomic integrity [1,2,3]. In particular, hESCs have active DNA repair mechanisms and are considerably susceptible to genotoxic stimuli [4, 5]. These characteristics cause rapid cell death induction upon DNA damage through mitochondria-mediated apoptosis, preventing progeny from inheriting mutations [1]. Mitochondria induce rapid cell death in hESCs upon DNA damage through ‘high mitochondrial priming’ [6] due to high pro-apoptotic gene expression [7, 8] and the immediate active BAX translocation from the Golgi complex upon genotoxic insults [9]. In addition, when DNA damage occurs, cytoplasmic p53 stabilizes and moves into the mitochondria, increasing mitochondrial membrane permeability to induce swift apoptosis [7, 10]. Consistently, mutation accumulation of hPSCs is lower than somatic stem cells, when cultured in vitro [11].
Genetic abnormalities, such as recurrent copy number variations (CNVs) [12, 13] and chromosomal abnormalities [14] as well as abnormal mitosis [15], frequently arise when hPSCs are maintained in vitro, compromising hPSC-based cell therapy safety [16, 17]. Notwithstanding, the clinical consequences and underlying mechanisms of culture-induced (epi)genetic abnormalities in hPSCs remain uncertain [17]. Survival trait acquisition, known as culture adaptation [14], is a common phenotype during long-term culture [18,19,20] associated with not only BCL2L1 induction [20] and recurrent CNV at 20q11.21 [18, 19, 21], but also dominant negative TP53 mutations [22], which impede the mitochondrial apoptosis. In addition, high mitochondrial priming of hESCs, through NOXA depletion, BCL2L1 induction [8] or caspase inhibition [15], augments aneuploid populations, suggesting that survival trait acquisition during in vitro culture leads to further genetic alterations as previously proposed [23]. Accordingly, mutations that randomly occur in vitro culture favor culture-induced stress survival and the mutant clone becomes dominant to win the competition [23]. Such clonal dominance is attained through genetic variations with vigorous YAP activity [24] from a high TPX2 expression [25]. Furthermore, epigenetic aberrations including DNA methylation change, loss of parental imprinting, and variable X chromosome inactivation have also been reported in the prolonged culture of hPSCs [26]. Specifically, abnormal hypermethylation has been observed in tumor suppressor genes [27]and antioxidant genes [28]. However, only limited studies have been conducted to examine the biological consequence of these epigenetically repressed genes [29].
Coiled-coil-helix-coiled-coil-helix domain containing 2 (CHCHD2), coded by the CHCHD2 gene (located on human chromosome 7p11.2), encodes a bi-organelle protein located in the mitochondria and nucleus [30]. The roles of this protein are multifaced, serving as a regulatory protein for mitochondrial energy metabolism and apoptosis, while also functioning as a transcription factor [31]. The significance of CHCHD2 extends to various diseases. It has been implicated in cancers (i.e., induction) [32, 33], Parkinson’s disease (i.e., mutations) [34] and mitochondrial encephalomyopathy (i.e., repression). However, the complete scope of CHCHD2 functions remains to be fully determined [30]. Mitochondrial CHCHD2, initially identified as a mitochondrial chaperone protein binding partner, inhibits apoptosis through BCL-xL interaction in cancer cell models [35] and regulates mitochondrial morphology in Drosophila models [36]. Interestingly, CHCHD2 expression in human embryonic stem cells (hESCs) primes neuroectodermal differentiation by sequestering SMAD4 in the mitochondria [37].
This study demonstrated that repeated hESCs enzymatic dissociation led to epigenetic repression of CHCHD2, closely associated with increased survival under diverse stresses encountered during in vitro culture. In addition, this effect was mediated through Rho A-ROCK signaling modulation. Consequently, hESCs lacking CHCHD2 expression exhibited clonal dominance during long-term in vitro culture. These results suggest that loss of CHCHD2 occurring through enzymatic dissociation is another cellular adaptation to favor cell survival under conditions of culture stress.
Materials and method
Cell culture
hESCs [WA09 (H9); WiCell Research Institute], BJ-iPSCs [38], and CHCHD2 KO/reconstitution subclones were maintained in mTeSR1 (Stem Cell Technology, #85,851) or MACS-iPSC brew medium (Milltany Biotechnology, #130-104-368) on plates coated with Matrigel (Corning, #354,277) diluted at 1:80 in hESC basal medium (DMEM/F12 supplemented with 1% non-essential amino acids, 0.1% β-mercaptoethanol, and 0.1% gentamicin, Gibco) for feeder-free conditions. Cells were incubated in 37 °C, 5% CO2 conditions. The medium was replaced every day up to passaging, and the cells were enzymatically dissociated using a dispase solution (Gibco, #17,105,041) with 10 μM of Y-27632 (Biogems, #1,293,823). 1 ~ 2 × 105 cells were seeding in 60Ø plate for transfer. 4 h mitomycin C-treated mouse embryonic fibroblast (MEF) was used as feeder cells for H9 feeder culture. Inverted microscope [Olympus, CKX-41 (Light source: 6 V/30W halogen lamp, Software: ProgRes Capture Pro)] was used to capture images for live cells.
RNA extraction, quantitative real-time PCR, and genome-wide gene expression profiling
Total RNA was extracted using Easy-blue reagent (Intron, #17,061) in accordance with the manufacturer’s instructions. PrimeScript™ RT reagent kit (Takara, #RR014A) was used to generate total RNA to cDNA following the manufacturer’s protocol. Quantitative real-time PCR (qPCR) was performed using TB green premix Taq (Takara, #RR820A) on a LightCycler 480 Instrument II (Roche) according to the manufacturer’s instructions. Primer information is shown in Table 1.
For library construction, we used the TruSeq Stranded mRNA Library Prep Kit (Illumina, San Diego, CA). Briefly, the strand-specific protocol included the following steps: (1) strand cDNA synthesis, (2) strand synthesis using dUTPs instead of dTTPs, (3) end repair, A-tailing, and adaptor ligation, and (4) PCR amplification. Each library was then diluted to 8 pM for 76 cycles of paired-read sequencing (2 × 75 bp) on an Illumina NextSeq 500 following the manufacturer’s recommended protocol. Read quality was assessed using FastQC (v) and poor-quality bases (Phred score < 20) were eliminated using TrimGalore (v0.6.6). Trimmed reads were aligned to the human reference genome (GRCh38) using the STAR aligner (v2.7.9a) with default parameters. Gene-level expression values such as transcripts per million (TPM) and read counts were calculated using RSEM (v1.3.3.) with human gene annotation (GRCh38.84). FASTQ format files, gene-level count data, and TPM of all samples are available in the Gene Expression Omnibus. Differential gene expression analysis for P1 ~ P4 data was performed using the ‘DESeq2’ package (v3.15) in R (v4.2.1). Differential gene expression analysis for CHCHD2 KO data was performed using the ‘EdgeR’ package (v3.15) in R (v4.2.1). Transcripts were considered significant if their fold change was above or below 1 (in log2 scale) and the p value corrected by FDR was below 0.05.
Chromatin immunoprecipitation (ChIP)
Cells were cross-linked with 1.21% formaldehyde for 10 min, and then quenched with Glycine for 5 min at room temperature. After fixation, chromatin samples were sonicated for 10 s 25 times to generate 300–500 bp fragments. Chromatin was immunoprecipitated with antibodies against rabbit IgG (Invitrogen, #31,235), H3K9me3 (CST, #13,969), H3K27me3 (CST, #9733), and H3K27Ac (Abcam, #ab4729). The eluted DNA was purified with phenol–chloroform. ChIP-qPCR was performed for quantification using CHCHD2 primers (F: 5ʹ-CAG GCC TGA AGT TCA TTG GAA-3ʹ, R: 5ʹ-ACT TCC GGG TTT TAA AGA TCC T-3ʹ), following the qRT-PCR protocols above.
Teratoma forming assay
For the teratoma forming assay, H9-hESCs with CHCHD2 wild type and knockout were cultured for 4 days. Cells were dissociated by accutase and counted as 1 × 106 cells for injection. The cells were injected into the right testis of 4-weeks-old BALB/c-nude mice along with Y-27632 in mTeSR1 medium. The left testis was used as the control. After 4 weeks, the first teratoma formation was observed, and after an additional 2 weeks, the mice were killed and the teratoma was extracted. A paraffin block was made using extracted teratoma, and H&E staining was performed.
Promoter methylation analysis using endonuclease digestion
For DNA methylation analysis in CHCHD2 promoter regions, genomic DNAs were extracted using genomic DNA purification kit (Promega, #A1120) according to the manufacturer’s instructions. After the extraction, 1 μg of genomic DNA was digested by methylcytosine-sensitive endonuclease McrBC (Takara, #1234A) for 4 h at 37 °C and inactivated at 65 °C for 20 min. Genomic DNAs without McrBC with the same reaction and amount were used as a control. Samples after digestion were diluted to a final volume of 400 μl, and 9 μl for each qRT-PCR was used. Primers were designed to be located in CpG island of the CHCHD2 promoter region. Sequence information of forward and reverse primers was as follows: F: 5ʹ-GTGAGTCACTCTTAAGGTTGGA-3ʹ, R: 5ʹ-AAGAGCTAAGCGACTTCTGAG-3ʹ.
Temporal trend classification of gene expression
Gene expression was normalized using Z-score or mean-centering normalization. Z-score was applied after collapsing all replicates to their average at each time point. Next, we calculated gene distances using Euclidean distance and used complete clustering methods.
Re-analysis of published transcriptomics data
The current publication utilized processed data from the following publications: Garitaonandia et al. (GSE34982). Data was downloaded directly from GEO repository. Transcript intensities after processing were log2 transformed and normalized using “Loess” (from limma package in R). After that, the R limma program was used to estimate the difference genes between groups. Transcripts were considered differentially expressed if their fold change was above or below 1 (in log2 scale) and the p value corrected by FDR was below 0.01.
Determination of copy number variations
Whole-genome genotyping was performed using the Illumina HumanOmni1-Quad Beadchip (Illumina) containing 1,140,419 genetic markers across the human genome. Samples were processed according to the specifications of the Illumina Infinium HD super assay. Briefly, each sample was whole genome amplified, fragmented, precipitated, and re-suspended in an appropriate hybridization buffer. Denatured samples were hybridized on a prepared BeadChip for a minimum of 16 h at 48 °C. Following hybridization, the bead chips were processed for the single-base extension reaction, stained, and imaged on an Illumina iScan system. Normalized bead intensity data for each sample were loaded into the GenomeStudio software package (Illumina). Ratios of signal intensity were calculated using the Log R Ratio (LRR: logged ratio of observed probe intensity to expected intensity; any deviations from zero in this metric are evidence for copy number change) and allelic intensity was determined by the B allele frequency for all samples. Values were exported using Illumina GenomeStudio. Analysis for structural variants was performed using the sliding window approach (window size 10).
Re-analysis of published methylation profile data
The current publication utilized processed data from the following publications: Garitaonandia et al., (GSE34982). Methylation profiles were downloaded as β values directly from the GEO repository. By range scaling to control samples of unmethylated, fully methylated, and half-methylated DNA, methylation levels for each probe were produced [39]. Methylation differences were calculated by subtracting the ß-values. For instance, if the difference in methylation was larger than 0.2 and the change in expression was greater than 2, an included gene of these category was deemed to be hypermethylated and silenced.
Immunoblotting and immunofluorescent assay
Immunoblotting and immunofluorescent assay were performed as described previously [40]. Antibody for CHCHD2 (#19,424-1-AP) was purchased from Proteintech. Antibodies for cleaved caspase-3 (C.Casp-3, #9664), OCT-4A (#2840), pPTEN (#9551), and pMYPT1 (#5613) were purchased from Cell Signaling Technology. Antibodies for α-tubulin (#sc-5286) and β-actin (#sc-47778) were purchased from Santa Cruz Biotechnology. Antibody for BCL-xL (#ab32370) and phosphor-BAX (#ab111391) were purchased from Abcam. Antibodies for Flag (#F1804) and active BAX (#MABC1176M) were purchased from Sigma-Aldrich. Quantification of blots was performed by Fusion software (Vilber Lourmat) in accordance with the manufacturer’s protocol. For immunofluorescent assay, secondary antibodies for mouse primary antibody conjugated to Alexa Fluor 488 (A11029) and Alexa Fluor 594 (A11032) fluorophores as well as for rabbit primary antibody conjugated to Alexa Fluor 488 (A11034) and Alexa Fluor 594 (A11037) were purchased from Invitrogen. Nucleus staining reagent 4’,6-diamidini-2-phenylindole (DAPI, #D1306) was purchased from Thermo Fisher Scientific. Fluorescence microscopy [Olympus, BX53 (Light source: 103W mercury lamp/12V 100W halogen lamp, Software: CellSense)] was used for imaging samples.
Immunoprecipitation
For immunoprecipitation, 1 mg of total proteins was incubated with 2 μg of CHCHD2, BCL-xL, or Flag antibody at 4 °C for 16 h, followed by the addition of Protein-A or G agarose beads (Santa cruz, #sc-2001 and #sc-2002) and incubated at 4 °C for an additional 4 h. The precipitates were washed with tissue lysis buffer (TLB) for 15 min each more than three times, followed by immunoblotting performed in accordance with the protocol reported previously [40] using second HRP antibody (Jackson Laboratory, #111-035-003 and #115-035-003) or Variblot (Abcam, #ab131366).
Alkaline phosphatase (AP) assay
Alkaline phosphatase (AP) staining assay was performed as per the manufacturer’s protocol (Sigma-Aldrich, #SCR004), and bright field images were captured by digital single-lens reflex camera (Nikon, D80).
Mitochondrial isolation
Mitochondrial isolations from human embryonic stem cells were performed by mitochondria isolation kit for cultured cells (ThermoFisher, #89,874) according to the manufacturer’s instructions. Mitochondria lysis for immunoblot was preserved by RIPA buffer or 2% CHAPS in TBS as described elsewhere.
Flow cytometry
For antibody staining flow cytometry, cells were washed twice with PBS and ixed with fix-permeabilization solution (BD Bioscience, #554,722). Cells were washed with permeabilization-wash solution twice (BD Bioscience, #554,723) and stained with primary antibodies in 3% BSA solution for 1 h, followed by 1 h of incubation with fluorescent-conjugated secondary antibodies.
Clonogenic assay
For the clonogenic assay, human embryonic stem cells were detached as single cell by accutase in 37 °C CO2 incubator for 3 min. Single cell-dissociated cells were seeded 5 × 104 cells for each wall into Matrigel-coated six-wall plate and cultured 5 ~ 7 days. After culture, 4% paraformaldehyde (PFA) solution was used for 10 min for fixing, and 0.1% of crystal violet solution was used for 1 h for staining the cells. After the crystal violet staining, the dye was destained using distilled water (DIW) twice and dried overnight to remove residual humidity. Colony images were captured by an optical camera (Nikon #NKR-D80(B)) and analyzed by Image J software.
Cell death analysis
Cell death was analyzed by flow cytometry as described previously [25]. For Annexin V/7-AAD staining, cells at 24 h after treatment of each flavonoid were washed twice with PBS and stained with FITC-conjugated Annexin V antibody (BD Bioscience, # 556,419) and 7-AAD (BD Bioscience, #559,925) for an additional 45–60 min at room temperature in the dark. Cells stained with Annexin V/7-AAD were analyzed by FACS Calibur or FACS Lyric (BD Bioscience). For the bright field images captured, light channel of optical microscope (Olympus, CKX-41) or JuLI-stage (NanoEntek, Korea) was used in accordance with the manufacture’s protocol. JuLI-STAT was used for analysis of the data from JuLI-stage (NanoEntek, Korea). The activity of caspase-3 was analyzed by colorimetric active caspase-3 assay kit (Sigma-Aldrich, #CASP3C) in accordance with the manufacturer's protocol.
Construction of CHCHD2 sgRNA and knockout hESCs
To construct the CHCHD2 knockout (KO) hESCs, two sgRNAs targeting for intron 2 and 3 to eliminate Exon 3 of CHCHD2 were predicted by RGEN Tool (http://www.rgenome.net/cas-designer/). Two sgRNAs were cloned into pX330A and pRGEN-U6-GFP vector and transfected into H9-hESCs by electroporation with the CRISPR/Cas9 vector. Cells were double selected by GFP and puromycin and single cell cloning was performed for equal genetic background.
Construction of doxycycline-inducible CHCHD2 vector
To construct the doxycycline-inducible CHCHD2 and rtTA piggybac plasmid, Flag-tagged (in C-terminal) human CHCHD2 in pCL-Neo backbone vector, kindly provided by Dr. Lawrence I. Grossman, was cloned into piggybac plasmid pB-TET. Two piggybac plasmids were transfected into normal or long-term cultured H9 and CHA3 hESCs using electroporation (NEPA) and selected using a 200 μg/mL of G418 (Sigma-Aldrich, #A1720) 3 days later. After the G418 selection, Flag-tagged CHCHD2 expressing a minimal dosage of doxycycline (Sigma-Aldrich, #D9891) was selected using colony picking, and single cell cloning was performed.
Statistical analysis
The graphical and quantification data were presented as mean ± S.D. Statistical significance among the three groups and between groups was determined using one-way or two-way analysis of variance (ANOVA) following Tukey post-test and Student’s t test, respectively. Statistical analysis was performed with GraphPad Prism 8 software (https://www.graphpad.com/scientific-software/prism/). Significance was assumed for p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), p < 0.0001 (****).
Results
Identifying CHCHD2 as a potential hESCs repetitive culture biomarker
To identify potential long-term cultured [or late passage (LP)] hESC biomarkers, we capitalized on four passage-dependent H9 hESC variants (P1: < 50, P2: 100 s, P3: 200 s, and P4: 300 s passages), in which survival trait (or survival advantage) [20] and abnormal mitosis [15] were apparent after 200 passages (P3 and P4) (Fig. 1A). In addition, we performed RNA sequencing on these four hESC sublines to explore hESCs’ long-term culture effect on its gene expression. We observed a clear difference in overall gene expression (Fig. 1B) and distinct clustering (Fig. 1C) between early (EP cluster: P1 and P2) and late passages (LP cluster: P3 and P4). We indicated the differentially expressed genes (DEGs) via volcano plot, revealing the mostly distinct CHCHD2 repression from upregulated and downregulated genes in LP-hESCs (Fig. 1D). To search for the ‘potential biomarker’ to reflect the culture adaptation, we utilized transcriptome datasets from one large-scale study (GSE34982) previously reported [41] to monitor potential effects of diverse culture conditions [passaging methods (mechanical vs. enzymatic) and culture matrices (extracellular matrix (ECM) vs. mouse feeder) for different passaging numbers (early vs. late)] (Fig. S1A). We further compared significantly altered gene expression under repetitive culture conditions [mechanical (Me) or enzymatic (En) dissociation], obtained from the dataset of GSE34982 (Fig. 1E) with transcriptome data from the in-house model (H9 hESCs) for early (E) and late (L) passage (Fig. 1F). Compared to altered LP-hESCs genes with transcriptome data from different culture conditions (Fig. 1F), CHCHD2 loss, distinctly observed in LP-hESCs (Fig. 1D), was a common repetitive enzymatic dissociation event (Fig. 1G).
Identify CHCHD2 as a potential gene marker of LP-hESCs. A Schematic images of long-term cultured hESCs model (P1: 40 s, P2: 100 s, P3: 200 s, P4: over 300 passages). B Heatmap of RNA sequencing for passage-dependent hESCs (EP early passage, LP late passage). C PCA analysis of RNA sequencing for passage-dependent hESCs. D Volcano plot of DEGs for passage-dependent hESCs. E PCA analysis of passage-dependent hESCs with enzymatic or mechanically cultured hESCs was displayed. F Diagram of downregulated intersection genes between passage and culture method. G List of downregulated genes for early passage and different culture methods (Me mechanical, En enzymatic) in hESCs. H Scatter plots of RNA expression and DNA methylation differences in high passage compared to low passage samples in data from GSE34982 (Hypo: low methylation, Hypr: high methylation, Up: high expression, Down: low expression) (left). The RNA expression and DNA methylation data of the probes in the Hyper-Down dataset showed opposite pattern, as shown in the heatmaps. (right). I Differentially methylated region (DMR) analysis at CHCHD2 promoter region of different passages with cultured enzymatically in data from GSE34982. Beta value, DMR, CpG islands, DNase clusters, and reference gene are shown below each plot
The methylome data, previously reported to show significant epigenetic aberrations in long-term culture conditions [41], were further analyzed in detail. As described earlier [41], there were distinct alterations in methylation patterns at CpG sites were observed in hESCs that had been cultured for an extended period (Fig. S1B). Where a larger number of differentially methylated CpG (DMC) sites were observed in mechanically passaged hESCs over time in culture (Fig. S1B), a different methylation profile was observed between mechanical and enzymatic passaging methods (Fig. S1C). Notably, CHCHD2, hypermethylated in enzymatic passaging (Fig. S1D), was identified as one of the hypermethylated genes that exhibited strong repression, as shown in Fig. 1H. In more detail, differentially methylated region (DMR) analysis clearly revealed that hypermethylation occurred at the CHCHD2 promoter region in long-term enzymatic passaging (Fig. 1I).
CHCHD2 depletion during prolonged hESC culture
Notably, enzymatic dissociation has been used as hESCs culture methodology since the Rho-associated protein kinase (ROCK) chemical inhibitor (Y-27632) was identified to block single cell-induced cell death [42]. Before this report, P1 hESCs were maily maintained by mechanical dissociation, so that they were much less exposed to enzymatic dissociation with Y-27632 (Fig. 2A). As predicted in silico in Fig. 1, CHCHD2 expression was drastically repressed in P3 and P4 hESCs where BCL2L1, a known marker for ‘culture adaptation’ or ‘survival advantage’ [20], was highly upregulated (Fig. 2B). Drastic CHCHD2 protein repression in P3 and P4 hESCs was made observable through immunofluorescence (Fig. 2C) and flow cytometry (Fig. 2D). In P2 hESC, representing a part of the total 100s passages was carried out, enzymatic dissociation revealed a significant decrease in CHCHD2 expression compared to P1 hESCs (Fig. 2E and F). The normal copy of 7p11.2, where CHCHD2 is located, failed to account for the distinct repression of CHCHD2 (Fig. 2G).
Depletion of CHCHD2 in long-term cultured hESCs. A Schematic images of the LP-hESCs model cultured with ROCK inhibitor (Y-27632). B mRNA expression of BCL2L1 in passage-dependent cultured hESCs (n = 2). C Immunofluorescent assay for CHCHD2 in passage-dependent cultured hESCs (scale bar = 25 μm). D Flow cytometry (left) and quantification graph (right) for CHCHD2 with fluorescent conjugated staining in low and high passage hESCs (P1: 40 s, P3: 200 s, P4: over 300 passages), and IgG for negative control of CHCHD2 primary antibody (n = 2). E Flow cytometry (left) and quantification graph (right) for CHCHD2 with fluorescent conjugated staining in passage-dependent cultured hESCs (P1: 40 s, P2: 100 s, P4: over 300 passages), and IgG for negative control of CHCHD2 primary antibody (n = 2). F Immunoblot assay for CHCHD2 protein expression in passage-dependent cultured hESCs, and α-tubulin for equal protein loading control. G Log R ratio (LRR) plot (i.e., a normalized measure of the total signal intensity for two alleles of the SNP) of P1 and P4 hESCs for chromosome 7 indicating the relative abundance of the genomic DNA around the SNP, which is expected to correlate with the copy number. H qRT-PCR-based methylation level analysis in the CHCHD2 promoter region using the methylation-sensitive restriction enzyme McrBC. Samples are shown as a relative value to the CHCHD2 exon (n = 3). I Chip-qPCR analysis of methylation (left) and acetylation (right) of two passage-dependent (early: P1, late: P4) hESCs in the CHCHD2 promoter region (n = 2)
As similar as strong epigenetic repression of CHCHD2 (Fig. 1H) and their promoter region (Fig. 1I) by prolonged enzymatic culture of hESCs from the previous dataset, hypermethylation at the CHCHD2 promoter was evident in P4 hESCs, as determined by the reactivity to McrBC, the methyl-specific DNA nuclease (Fig. 2H). In addition, tri-methylation of histone 3 lysine 9 (H3K9me3), typical histone modifications associated with gene repression [43], was increased, while acetylation of histone 3 lysine 27 (H3K27Ac) was markedly diminished in CHCHD2 promoter region in P4 hESCs (Fig. 2 I). CHCHD2 repression through repetitive in vitro culture (with enzymatic dissociation) was also apparent in another hESCs line (CHA3-hESCs) [44] (Fig. S2A and B) and iPSCs (BJ-iPSCs) with different passage numbers (EP: 67 CHA3-hESC passages, 35 BJ-iPSC passages; LP: 328 CHA3-hESC passages, 165 BJ-iPSCs passages] (Fig. S2C and D).
CHCHD2 knockout hESC establishment and initial characterization
Next, we established a CHCHD2 knockout (KO) model in P1 hESCs (i.e., normal) using the previously described CRISPR/Cas9 technique [45] to monitor biological consequences from CHCHD2 repression during hESCs enzymatic dissociation culture. Two sgRNA targeting sequences at introns 2 and 3 were designed to excise CHCHD2’s exon 3 (Fig. S3A). Simple genotyping analysis (Fig. S3B) and Sanger sequencing demonstrated the single clone establishment, lacking CHCHD2’s exon 3 in P1 hESCs at two alleles (hereafter KO hESCs) (Fig. S3C). Unlike the KO approach using CRISPR/Cas9 with one sgRNA to induce frame-shift after indel (insertion and deletion) formation, exon three deletion for CHCHD2 KO allowed us to verify complete CHCHD2 KO through simple RT-PCR analysis as it is similar to P4 hESCs’ level (Fig. 3A).
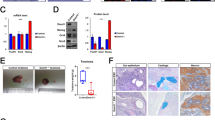
Establishment of CHCHD2 knockout hESCs with CRISPR/Cas9 (A, B). A mRNA expression of CHCHD2 in normal early passage (P1), long-term cultured (P4), CHCHD2 WT, and KO hESCs, B mRNA expression of POU5F1 for pluripotent gene marker (n = 2). C Immunofluorescent assay for CHCHD2 expression, DAPI for nucleus staining (scale bar = 50 μm). D Immunoblot assay for CHCHD2 protein expression, α-tubulin for equal protein loading control. E Alkaline phosphatase activity assay in WT and CHCHD2 KO-hESCs. F mRNA expression during in vitro spontaneous differentiation of WT and CHCHD2 KO-hESCs, POU5F1, and SOX2 for pluripotent gene marker (n = 2). G Teratoma formation for confirmed three-germ layer differentiation in WT and CHCHD2 KO-hESCs (scale bar = 50 μm)
Of note, the POU5F1 level in KO hESCs was comparable to the parent P1 hESCs (Fig. 3B). Immunostaining and immunoblotting analysis confirmed that complete mitochondrial CHCHD2 protein loss in KO hESCs was similar to that of P4 hESCs (Fig. 3C and D). The initial KO hESCs pluripotency characterization was determined through key pluripotent mRNA level markers (data not shown) and the comparable colony level with alkaline phosphatase activity (Fig. 3E). One study has demonstrated that CHCHD2 expression primes neuroectodermal differentiation by sequestering SMAD4 at mitochondria through direct protein binding [37]. Consistently, clear ‘TGFβ receptor signaling’ enrichment in the WikiPathway (Fig. S3D) and ID1 induction, a common TGFβ downstream gene, in KO hESCs revealed functional CHCHD2 KO was achieved (Fig. S3E). Accordingly, PAX6, a typical neuroectodermal determinant [46], was markedly attenuated in KO hESCs during spontaneous differentiation, while comparable level of mesoderm, endoderm, and pluripotency markers in KO hESCs (Fig. 3F). These data imply that CHCHD2 loss solely impairs neuroectodermal differentiation, as previously described [37]. However, KO hESCs-derived teratoma also developed a neural rosette structure similar to WT hESCs, and no distinct alteration was observed in the three germ layers (Fig. 3G), which was consistent with the normal development of CHCHD2 KO mice [47]. Additionally, we noted that hallmark ‘Apoptosis’ was associated with KO hESCs (Fig. S3F). In particular, there is a report that mitochondrial CHCHD2 interacts with Bcl-xL to inhibit mitochondrial apoptosis in a cancer cell line model [35].
Survival trait acquisition from CHCHD2 loss
According to the study that CHCHD2 inhibits mitochondrial apoptosis [35], we examined whether KO hESCs were more susceptible to mitochondrial apoptosis than WT. To trigger hESC cell death, we utilized YM155, a small molecule, inducing the selective cell death of undifferentiated hPSCs [7] via selective SLC35F2 cellular import, a solute carrier protein, highly expressed in hPSCs [45, 48]. Unexpectedly, KO hESCs were rather less sensitive to YM155 than WT (Fig. 4A and Movie S1) despite their comparable SLC35F2 expression levels (Fig. 4B). KO’s unexpected YM155 resistance was confirmed through flow cytometry analysis (Fig. 4C), caspase activity assay (Fig. 4D), and a YM155 dose-dependent challenge (Fig. 4E). KO hESCs were also more resistant to genotoxic agents such as etoposide (Eto, Fig. 4F and G) and doxorubicin to the parent control (Doxo, Fig. 4H and I). The resistance of KO hESCs to doxorubicin was highlighted by apoptosis level in a dose-dependent manner (Fig. 4J). Notably, resistance to these genotoxic agents was observed in culture-adapted hESCs (P3 and P4 hESCs), where BCL2L1 was highly induced [20]. These data imply that CHCHD2 loss in hESCs through repetitive enzymatic dissociation culture (Figs. 1 and 2) favors survival.
Acquire resistance of genotoxic stress in CHCHD2 KO-hESCs. A Time-dependent optical microscopic image upon 10 nM of YM155 in WT and CHCHD2 KO-hESCs (Scale bar = 500 μm). B mRNA expression of SLC35F2 in WT and CHCHD2 KO-hESCs (n = 2). C Flow cytometry analysis (left) and bar graph for quantification (right) with 10 nM of YM155 in WT and CHCHD2 KO-hESCs (n = 3). D Graphical presentation of caspase-3 activity at 24 h after treatment of 10 nM of YM155 in WT and CHCHD2 KO-hESCs (n = 3). E Flow cytometry for Annexin V/7-AAD analysis (left) and quantification graph (right) of WT and CHCHD2 KO-hESCs in YM155-dependent manner (n = 2). F Immunoblot assay with 50 nM of etoposide (Eto) in WT and CHCHD2 KO-hESCs, and α-tubulin for equal protein loading control. (G) Flow cytometry for Annexin V/7-AAD analysis (left) and quantification graph (right) of WT and CHCHD2 KO-hESCs in 100 nM of etoposide (n = 2). H Immunoblot assay with 50 nM of doxorubicin (Doxo) in WT and CHCHD2 KO-hESCs, and α-tubulin for equal protein loading control. (I) Quantification graph for trypan blue assay of WT and CHCHD2 KO-hESCs in 50 nM of doxorubicin (n = 2). J Immunoblot assay of WT and CHCHD2 KO-hESCs in Doxo-dependent manner, and α-tubulin for equal protein loading control
CHCHD2 loss rescues hESCs from dissociation-induced cell death
Culture-adapted hESCs survive diverse stresses derived from culture conditions (e.g., dissociation-induced apoptosis from enzymatic dissociation) [49] by inducting BCL2L1, a common factor for ‘culture adaptations’ [18] and TP53 mutations [22]. Thus, aberrant clones eventually become dominant through ‘winning’ the competition. As CHCHD2 repression was the most evident in hESCs late passage through the ‘enzymatic dissociation’ culture method (Fig. 1G), we surmised that CHCHD2 repression is an adapted response from ‘enzymatic dissociation’ and would favor survival upon dissociation-induced apoptosis.
Despite comparable growth rate of KO compared to WT (Fig. 5A and Movie S2), KO hESCs formed more colonies after enzymatic dissociation without Y-27632 supplementation (to mimic a dissociation-induced apoptosis condition) (Fig. 5B). The survival difference between WT and KO hESCs disappeared with Y-27632 treatment (Fig. 5C). Of note, culture-adapted hESCs with distinct CNV at 20q11.21 (P4 hESCs) retained high BCL2L1 expressions [20] and YAP activity [25], showing a more distinct survival phenotype than CHCHD2 loss under the same conditions (Fig. 5C). However, KO hESCs expressed a comparable level of BCL2L1 unlike that of P4 hESCs (Fig. 5D, S4A and B). Thus, it is intriguing that KO hESCs independently survived from single cell dissociation-induced apoptosis in BCL2L1.
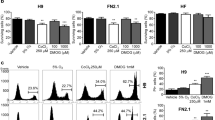
Acquire survival trait under single cell dissociation-induced cell death by wane of CHCHD2. A Growth curve of WT and CHCHD2 KO-hESCs for 72 h (n ≥ 3). B Image of clonogenic assay (left) and quantification bar graph (right) of single cell-dissociated WT, CHCHD2 KO-hESCs without ROCK inhibitor (Y-27632) (n = 3). C Image of clonogenic assay (left) and quantification bar graph (right) of single cell-dissociated WT and CHCHD2 KO hESCs with or without Y-27632, and late passage (P4) hESCs for positive single cell-dissociated stress resistance control (n = 4). D mRNA expression of CHCHD2 and BCL2L1 in WT, CHCHD2 KO, and late passage (P4) hESCs (n = 2). E mRNA expression of CHCHD2 in H9 variants with different culture methods. F Image of clonogenic assay (left) and quantification graph (right) of single cell-dissociated H9 variants with different culture methods. G and H Log R ratio (LRR) plot of H9 sublines for chromosome 20 (G) and chromosome 7 (H) indicating the relative abundance of the genomic DNA around the SNP, which is expected to correlate with the copy number. I mRNA expression level of CHCHD2 in WT and CNV (in 20q11.21 loci) occurred hiPSCs (n ≥ 4)
Since ROCK inhibitor (Y-27632) rescues hPSCs’ specific dissociation-induced apoptosis [42], which has been a considerable hurdle for routine cell culture, Y-27632 was widely used to improve cell viability during hPSC culture [50]. Thus, other than prompt metabolic change by transient exposure [51] and change in actin filament by prolonged exposure [52], we surmise that repetitive exposure of Y-27632 would affect CHCHD expression. To this end, we collected three additional H9 hESCs from two independent institutes in Korea, which have been maintained under different culture protocols (Fig. S4C). CHCHD2 expression levels from P1 hESCs experiencing 15 passages with enzymatic dissociation (55 passages, hereafter P1) from the original stock (40 passage), hESCs from Soonchunhyang university (hereafter SCH), and hESCs from Korea Research Institue of Bioscience and Biotechnology (KRIBB, hereafter KR1) maintained in different culture conditions (Fig. S4C) were examined and compared to H9 hESCs expanded from the original WiCell stock (WiCell, passage number 28) by one passage under our culture condition. Consistently, KR1 hESCs maintained in the feeder without Y-27632 supplementation (Fig. S4C) kept CHCHD2 levels similar to WiCell hESCs (Fig. 5E). In parallel, KR1 hESCs and WiCell expressed a more distinct sensitivity to dissociation-induced apoptosis, unlike other hESCs (Fig. 5F).
One study elucidated CHCHD2 downregulation as a 20q11.21 gain marker [53]. However, the set of H9 hESCs with different expression level of CHCHD2 (Fig. 5E) were all normal copy numbers in 20q11.21 (Fig. 5G) as well as 7p11.2 (Figs. 2G and 5H) where CHCHD2 gene is located. Additionally, there was no noticeable correlation of CHCHD2 expression to 20q11.21 gain in nine iPSCs (5 iPSCs with normal copy number: WT; 4 iPSCs with 20q11.21 gain: CNV) maintained without Y-27632 supplement (under 35 passages) (Figs. 5I and S4D).
CHCHD2 expression for cell death susceptibility
Next, we confirmed CHCHD2’s role in cell death susceptibility by producing doxycycline (Dox) inducible CHCHD2 in KO hESCs (KO-iC2) (Fig. 6A). POU5F1 expression levels were not significantly altered regardless of CHCHD2 expression (Fig. S5A). Simple Dox treatment markedly increased CHCHD2 levels with no distinct change of OCT4 protein levels (Fig. 6B). Expressed mitochondrial CHCHD2 (with FLAG-tag) was further validated through immunofluorescence with FLAG antibody (Fig. 6C). As predicted, CHCHD2 reconstitution in KO-hESCs re-sensitized KO hESCs to Doxo- (Fig. 6D) and YM155-induced cell death (Fig. 6E and S5B) without significant BCL2L1 and SLC35F2 alteration, respectively (Fig. S5C). Survived colonies after single cell dissociation in KO-iC2 were markedly reduced by CHCHD2 reconstitution with Dox (Fig. 6F).
Reverted survival trait by reconstitution of CHCHD2 in hESCs. A Schematic images for construction of doxycycline-inducible CHCHD2 reconstitution (iC2) cell model in CHCHD2 KO-hESCs. B Immunoblot assay of WT, CHCHD2 KO, and iC2 hESCs with 0.25 μg/mL of Dox, and α-tubulin for equal protein loading control. C Immunofluorescent assay for BCL-xL and CHCHD2-Flag in WT and iC2 hESCs with 0.1 μg/mL Dox, DAPI for nucleus staining (Scale bar = 25 μm). D Immunoblot assay of WT, KO, and iC2 hESCs with 40 nM of Doxo, and α-tubulin for equal protein loading control. E Flow cytometry for Annexin V/7-AAD analysis (left) and quantification bar graph (right) with 10 nM of YM155 in WT, CHCHD2 KO, and iC2 hESCs (n = 2). F Optical microscopic images of clonogenic assay (left) and quantification bar graph (right) of single cell-dissociated early passage (P1) and CHCHD2 reconstitution hESCs with or without 0.25 μg/mL Dox without ROCK inhibitor (n = 5)
CHCHD2 expression for ROCK activity
We were surprised that CHCHD2 loss favored survival under diverse culture stresses in hESCs (Figs. 4 and 5) because CHCHD2 was previously determined to inhibit apoptosis through direct BCL-xL interaction in cancer cell lines [35]. First, we examined whether CHCHD2 interacts with BCL-xL in hESCs. BCL-xL (with FLAG tag) was ectopically expressed in WT and KO hESCs, and co-immunoprecipitation with CHCHD2 was carried out to emphasize this interaction. Unexpectedly, neither ectopically (Exo) expressed nor endogenous (Endo) BCL-xL was pulled down alongside CHCHD2 immunoprecipitation (Fig. 7A). Additionally, CHCHD2 co-immunoprecipitation repetition in KO-iC2 after Dox treatment also failed to draw BCL-xL out (Fig. 7B). Thus, we surmised that the lack of CHCHD2 and BCL-xL interaction in hESCs account for the contradictory CHCHD2 effect on apoptosis in hESCs.
CHCHD2 regulates ROCK activity. A Immunoprecipitation assay of WT and CHCHD2 KO-hESCs with exogeneous expression of Bcl-xL-Flag, heavy chain for equal primary antibody control and α-tubulin for equal protein loading control. B Immunoprecipitation assay of iC2 hESCs with exogeneous expression of Bcl-xL-Flag. C Immunoblot assay of WT and CHCHD2 KO-hESCs with incubation after single cell dissociation, and β-actin for equal protein loading control. D Immunoblot assay of CHCHD2 KO and iC2 hESCs with incubation after single cell dissociation, and β-actin for equal protein loading control. E Kinase activity assay for ROCK2 kinase in single cell dissociated WT and CHCHD2 KO hESCs with 0.25 μg/mL of Dox (n = 4). F Kinase activity assay for ROCK2 kinase in single cell dissociated WT, CHCHD2 KO, and iC2 hESCs with 0.25 μg/mL of Dox (n = 4)
Repetitive enzymatic dissociation was likely to repress CHCHD2 (Fig. 1) and favor survival from dissociation-induced apoptosis. Thus, we surmised that CHCHD2 expression affected ROCK activity, a primary cell death determinant, of which attenuation occurs in culture adapted hPSCs [54]. To this end, phosphorylated myosin phosphatase target subunit 1 (MYPT1), a direct ROCK substrate, [55] levels were monitored after single cell dissociation. MYPT1 phosphorylated 10 min after single cell dissociation was markedly diminished in KO hESCs, suggesting that CHCHD2 expression contributes to ROCK activation (Fig. 7C). Consistently, ROCK-regulated PTEN phosphorylation levels [56] notably increased through CHCHD2 reconstitution in KO-iC2 hESCs after single cell dissociation (Fig. 7D). Additionally, phosphorylation of cofilin (pCofilin), occurring in ROCK activity-dependent manner through LIM-kinase activation [57], was also noticeably decreased in KO hESCs upon single cell dissociation (Fig. S6A). The level of pCofilin upon dissociation was recovered by reconstitution of CHCHD2 expression (Fig. S6B). A biochemical assay quantified ROCK activity after single cell dissociation to validate these results. ROCK activity drastically increased immediately after cell dissociation and remained elevated for 180 min (Fig. S6C). Maximum ROCK activity (30 min after dissociation) was markedly attenuated in KO hESCs (Fig. 7E) and was regained after CHCHD2 reconstitution in KO-iC2 hESCs (Fig. 7F). Unlike cancer cells, where ROCK activity governs actin organization to promote cell migration, the level of CHCHD2 expression gave only marginal effect on migration capacity (Fig. S6D and E). These data conclusively imply that CHCHD2 epigenetic repression during in vitro culture (through enzymatic dissociation in particular) is another cellular adaptive event to endow ‘survival traits,’ through repression of ROCK activity upon dissociation, leading to cellular dominancy like TP53 mutations [22], BCL2L1 induction [18], or YAP activation [25].
Discussion
Tissue regeneration through hPSCs-derived cell therapy has drawn substantial attention since the first human autologous stem cell therapy’s promising clinical outcome [58] and recent FDA approval for Parkinson’s disease hESC-based phase I trial (run by ‘BlueRock Therapeutics’). However, hPSC-based cell therapy safety has been a consistent societal concern due to the uncertainty of cellular and genomic profile alteration during in vitro culture [17]. ‘Survival trait’ acquisition during in vitro culture (or culture adaptation [14]) due to BCL2L1 induction [18, 20, 53], TP53 mutation [22], or YAP activation [25] leads to abnormal clonal dominance and exacerbates genetic aberrations by escaping abnormal mitosis [8]. We confirmed that epigenetic repression of CHCHD2, occurred specifically through repetitive ‘enzymatic dissociation’ culture by in silico analysis of methylome data [41] (Fig. 1H and I), McrBC reactivity and ChIP assay with an in-house model (Fig. 2H and I). This cellular event is associated with the acquisition of ‘survival trait’ through ROCK activation interference.
A previous study substantiated that CHCHD2 loss, as a 20q11.21 gain marker, affects neuroectodermal differentiation [53]. Similarly, we observed that neuroectodermal lineage, determined by PAX6 expression during spontaneous differentiation, was impaired in CHCHD2 KO (Fig. 3F). P3 and P4 hESCs with apparent 20q11.21 gain completely repressed CHCHD2 (Fig. 2) [25]. However, CHCHD2 expression level in early passaged iPSCs varied regardless of 20q11.21 gain (Fig. 5I). Instead, CHCHD2 levels in the relatively early passage of four different hESCs (collected from independent Korean institutes through different culture methods) (Figs. 5E and S4C) were consistent with transcriptome analyses from early and late hPSCs maintained with different culture conditions [41]. Accordingly, we suggest that CHCHD2 repression would be a ‘repetitive enzymatic dissociation’ marker rather than gain of 20q11.21. Considering the complete CHCHD2 repression, possibly due to closed chromatin accessibility of CHCHD2 promoter region (Fig. 2H and I) in P4 hESCs with a 20q11.21 gain and the previous study [53], we could not exclude the possibility that 20q11.21 gain in hPSCs could have resulted from the repetitive culture with ‘enzymatic dissociation’. This possibility requires additional follow-up studies.
Unlike previous cancer cell line studies demonstrating that mitochondrial CHCHD2 interacts with BCL-xL to inhibit apoptosis [35], CHCHD2 KO in hESCs inhibited mitochondrial cell death induced by genotoxic insults (Fig. 4), which was re-sensitized by CHCHD2 reconstitution (Fig. 6). Despite multiple attempts, CHCHD2 in BCL-xL interactions could not be reproduced in hESCs (Fig. 7A and B), implying that CHCHD2’s role in apoptosis is distinct in hESCs. We also tested whether CHCHD2 reconstitution in P4 hESCs with clear 20q11.21, consequent BCL2L1 induction, and YAP activation would sensitize cell death induced by ‘single cell dissociation’ or genotoxic stress. As high BCL-xL expression, an anti-apoptotic protein, desensitizes ‘mitochondrial apoptosis’ priming, it endows ‘the strong selective advantage’ [18] in P4 hESCs; CHCHD2 expression failed to overcome the survival trait (data not shown). These data imply that high BCL-xL expression or YAP activity is major determinant for ‘survival advantage’ in hESCs.
Given the lack of an early marker for ‘survival trait acquisition’ or ‘culture adaptation’ cellular events other than BCL2L1 induction or CNV at 20q11.21, CHCHD2 expression level may be used as a ‘repetitive culture with enzymatic dissociation’ and ‘survival trait’ from ‘dissociation induced cell death’ indicator useful in routine hPSCs assessments.
Data availability
Source data are available from the Cellular and Molecular Life Sciences online or corresponding authors upon request.
Abbreviations
- hPSCs:
-
Human pluripotent stem cells
- hESCs:
-
Human embryonic stem cells
- iPSCs:
-
Induced pluripotent stem cells
- CHCHD2:
-
Coiled-coil-helix-coiled-coil-helix domain containing 2
- CNV:
-
Copy number variation
- EP:
-
Early passage
- LP:
-
Late passage
- Me:
-
Mechanical
- En:
-
Enzymatic
- DMC:
-
Differentially methylated CpG
- DMRs:
-
Differentially methylated regions
- ROCK:
-
Rho-associated protein kinase
- WT:
-
Wild type
- KO:
-
Knockout
- RT-PCR:
-
Real-time polymerase chain reaction
- Dox:
-
Doxycycline
- Doxo:
-
Doxorubicin
- Eto:
-
Etoposide
- Endo:
-
Endogenous
- Exo:
-
Ectopically
- ChIP:
-
Chromatine immunoprecipitation
References
Weissbein U, Benvenisty N, Ben-David U (2014) Quality control: genome maintenance in pluripotent stem cells. J Cell Biol 204(2):153–163
Maynard S, Swistikowa A, Lee J, Liu Y, Liu S, Da Cruz A, Rao M, de Souza-Pinto N, Zeng X, Bohr V (2008) Human embryonic stem cells have enhanced repair of multiple forms of DNA damage. Stem Cells 26:2266–2274
Park JC, Jang HK, Kim J, Han JH, Jung Y, Kim K, Bae S, Cha HJ (2022) High expression of uracil DNA glycosylase determines C to T substitution in human pluripotent stem cells. Mol Ther Nucleic Acids 27:175–183
Smith AJ, Nelson NG, Oommen S, Hartjes KA, Folmes CD, Terzic A, Nelson TJ (2012) Apoptotic susceptibility to DNA damage of pluripotent stem cells facilitates pharmacologic purging of teratoma risk. Stem Cells Transl Med 1(10):709–718
Liu JC, Lerou PH, Lahav G (2014) Stem cells: balancing resistance and sensitivity to DNA damage. Trends Cell Biol 24(5):268–274
Liu JC, Guan X, Ryan JA, Rivera AG, Mock C, Agrawal V, Letai A, Lerou PH, Lahav G (2013) High mitochondrial priming sensitizes hESCs to DNA-damage-induced apoptosis. Cell Stem Cell 13(4):483–491
Lee MO, Moon SH, Jeong HC, Yi JY, Lee TH, Shim SH, Rhee YH, Lee SH, Oh SJ, Lee MY et al (2013) Inhibition of pluripotent stem cell-derived teratoma formation by small molecules. Proc Natl Acad Sci U S A 110(35):E3281-3290
Zhang J, Hirst AJ, Duan F, Qiu H, Huang R, Ji Y, Bai L, Zhang F, Robinson D, Jones M et al (2019) Anti-apoptotic mutations desensitize human pluripotent stem cells to mitotic stress and enable aneuploid cell survival. Stem Cell Rep 12(3):557–571
Dumitru R, Gama V, Fagan BM, Bower JJ, Swahari V, Pevny LH, Deshmukh M (2012) Human embryonic stem cells have constitutively active Bax at the Golgi and are primed to undergo rapid apoptosis. Mol Cell 46:573–583
TeSlaa T, Setoguchi K, Teitell MA (2016) Mitochondria in human pluripotent stem cell apoptosis. Semin Cell Dev Biol 52:76–83
Kuijk E, Jager M, van der Roest B, Locati MD, Van Hoeck A, Korzelius J, Janssen R, Besselink N, Boymans S, van Boxtel R et al (2020) The mutational impact of culturing human pluripotent and adult stem cells. Nat Commun 11(1):2493
Draper JS, Smith K, Gokhale P, Moore HD, Maltby E, Johnson J, Meisner L, Zwaka TP, Thomson JA, Andrews PW (2004) Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat Biotechnol 22(1):53–54
Lefort N, Feyeux M, Bas C, Feraud O, Bennaceur-Griscelli A, Tachdjian G, Peschanski M, Perrier AL (2008) Human embryonic stem cells reveal recurrent genomic instability at 20q11.21. Nat Biotechnol 26(12):1364–1366
Baker DE, Harrison NJ, Maltby E, Smith K, Moore HD, Shaw PJ, Heath PR, Holden H, Andrews PW (2007) Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol 25(2):207–215
Jeong HC, Go YH, Shin JG, Kim YJ, Cho MG, Gwon D, Cheong HS, Lee H, Lee JH, Jang CY et al (2023) TPX2 amplification-driven aberrant mitosis in culture adapted human embryonic stem cells with gain of 20q11.21. Stem Cell Rev Rep 19:1466–1481
Assou S, Bouckenheimer J, De Vos J (2018) Concise review: assessing the genome integrity of human induced pluripotent stem cells: what quality control metrics? Stem Cells 36(6):814–821
Andrews PW, Ben-David U, Benvenisty N, Coffey P, Eggan K, Knowles BB, Nagy A, Pera M, Reubinoff B, Rugg-Gunn PJ et al (2017) Assessing the safety of human pluripotent stem cells and their derivatives for clinical applications. Stem Cell Rep 9(1):1–4
Avery S, Hirst AJ, Baker D, Lim CY, Alagaratnam S, Skotheim RI, Lothe RA, Pera MF, Colman A, Robson P et al (2013) BCL-XL mediates the strong selective advantage of a 20q11.21 amplification commonly found in human embryonic stem cell cultures. Stem Cell Rep 1(5):379–386
Nguyen HT, Geens M, Mertzanidou A, Jacobs K, Heirman C, Breckpot K, Spits C (2014) Gain of 20q11.21 in human embryonic stem cells improves cell survival by increased expression of Bcl-xL. Mol Hum Reprod 20(2):168–177
Cho SJ, Kim KT, Jeong HC, Park JC, Kwon OS, Song YH, Shin JG, Kang S, Kim W, Shin HD et al (2018) Selective elimination of culture-adapted human embryonic stem cells with BH3 mimetics. Stem Cell Rep 11(5):1244–1256
International Stem Cell I, Amps K, Andrews PW, Anyfantis G, Armstrong L, Avery S, Baharvand H, Baker J, Baker D, Munoz MB et al (2011) Screening ethnically diverse human embryonic stem cells identifies a chromosome 20 minimal amplicon conferring growth advantage. Nat Biotechnol 29(12):1132–1144
Merkle FT, Ghosh S, Kamitaki N, Mitchell J, Avior Y, Mello C, Kashin S, Mekhoubad S, Ilic D, Charlton M et al (2017) Human pluripotent stem cells recurrently acquire and expand dominant negative P53 mutations. Nature 545(7653):229–233
Halliwell J, Barbaric I, Andrews PW (2020) Acquired genetic changes in human pluripotent stem cells: origins and consequences. Nat Rev Mol Cell Biol 21(12):715–728
Price CJ, Stavish D, Gokhale PJ, Stevenson BA, Sargeant S, Lacey J, Rodriguez TA, Barbaric I (2021) Genetically variant human pluripotent stem cells selectively eliminate wild-type counterparts through YAP-mediated cell competition. Dev Cell 56(17):2455-2470 e2410
Kim YJ, Go YH, Jeong HC, Kwon EJ, Kim SM, Cheong HS, Kim W, Shin HD, Lee H, Cha HJ (2023) TPX2 prompts mitotic survival via the induction of BCL2L1 through YAP1 protein stabilization in human embryonic stem cells. Exp Mol Med 55:32–42
Bar S, Benvenisty N (2019) Epigenetic aberrations in human pluripotent stem cells. EMBO J 38(12):e101033
Calvanese V, Horrillo A, Hmadcha A, Suarez-Alvarez B, Fernandez AF, Lara E, Casado S, Menendez P, Bueno C, Garcia-Castro J et al (2008) Cancer genes hypermethylated in human embryonic stem cells. PLoS One 3(9):e3294
Konki M, Pasumarthy K, Malonzo M, Sainio A, Valensisi C, Soderstrom M, Emani MR, Stubb A, Narva E, Ghimire B et al (2016) Epigenetic silencing of the key antioxidant enzyme catalase in karyotypically abnormal human pluripotent stem cells. Sci Rep 6:22190
Weissbein U, Plotnik O, Vershkov D, Benvenisty N (2017) Culture-induced recurrent epigenetic aberrations in human pluripotent stem cells. PLoS Genet 13(8):e1006979
Modjtahedi N, Tokatlidis K, Dessen P, Kroemer G (2016) Mitochondrial proteins containing coiled-coil-helix-coiled-coil-helix (CHCH) domains in health and disease. Trends Biochem Sci 41(3):245–260
Kee TR, Espinoza Gonzalez P, Wehinger JL, Bukhari MZ, Ermekbaeva A, Sista A, Kotsiviras P, Liu T, Kang DE, Woo JA (2021) Mitochondrial CHCHD2: disease-associated mutations, physiological functions, and current animal models. Front Aging Neurosci 13:660843
Chehade H, Purandare N, Fox A, Adzibolosu N, Jayee S, Singh A, Tedja R, Gogoi R, Aras S, Grossman LI et al (2023) MNRR1 is a driver of ovarian cancer progression. Transl Oncol 29:101623
Karapanagioti A, Nasiri-Ansari N, Moustogiannis A, Trigas GC, Zografos G, Aggeli C, Kyriakopoulos G, Choreftaki T, Philippou A, Kaltsas G et al (2023) What is the role of CHCHD2 in adrenal tumourigenesis? Endocrine 81(2):357–367
Funayama M, Ohe K, Amo T, Furuya N, Yamaguchi J, Saiki S, Li Y, Ogaki K, Ando M, Yoshino H et al (2015) CHCHD2 mutations in autosomal dominant late-onset Parkinson’s disease: a genome-wide linkage and sequencing study. Lancet Neurol 14(3):274–282
Liu Y, Clegg HV, Leslie PL, Di J, Tollini LA, He Y, Kim TH, Jin A, Graves LM, Zheng J et al (2015) CHCHD2 inhibits apoptosis by interacting with Bcl-x L to regulate Bax activation. Cell Death Differ 22(6):1035–1046
Liu W, Duan X, Xu L, Shang W, Zhao J, Wang L, Li JC, Chen CH, Liu JP, Tong C (2020) Chchd2 regulates mitochondrial morphology by modulating the levels of Opa1. Cell Death Differ 27(6):2014–2029
Zhu L, Gomez-Duran A, Saretzki G, Jin S, Tilgner K, Melguizo-Sanchis D, Anyfantis G, Al-Aama J, Vallier L, Chinnery P et al (2016) The mitochondrial protein CHCHD2 primes the differentiation potential of human induced pluripotent stem cells to neuroectodermal lineages. J Cell Biol 215(2):187–202
Bang JS, Choi NY, Lee M, Ko K, Lee HJ, Park YS, Jeong D, Chung H-M, Ko K (2018) Optimization of episomal reprogramming for generation of human induced pluripotent stem cells from fibroblasts. Anim Cells Syst 22(2):132–139
Nazor KL, Altun G, Lynch C, Tran H, Harness JV, Slavin I, Garitaonandia I, Muller FJ, Wang YC, Boscolo FS et al (2012) Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell 10(5):620–634
Kwon OS, Lee H, Kong HJ, Kwon EJ, Park JE, Lee W, Kang S, Kim M, Kim W, Cha HJ (2020) Connectivity map-based drug repositioning of bortezomib to reverse the metastatic effect of GALNT14 in lung cancer. Oncogene 39(23):4567–4580
Garitaonandia I, Amir H, Boscolo FS, Wambua GK, Schultheisz HL, Sabatini K, Morey R, Waltz S, Wang YC, Tran H et al (2015) Increased risk of genetic and epigenetic instability in human embryonic stem cells associated with specific culture conditions. PLoS One 10(2):e0118307
Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, Takahashi JB, Nishikawa S, Nishikawa S, Muguruma K et al (2007) A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol 25(6):681–686
Kimura H (2013) Histone modifications for human epigenome analysis. J Hum Genet 58(7):439–445
Cho SW, Moon SH, Lee SH, Kang SW, Kim J, Lim JM, Kim HS, Kim BS, Chung HM (2007) Improvement of postnatal neovascularization by human embryonic stem cell derived endothelial-like cell transplantation in a mouse model of hindlimb ischemia. Circulation 116(21):2409–2419
Kim K-T, Park J-C, Jang H-K, Lee H, Park S, Kim J, Kwon O-S, Go Y-H, Jin Y, Kim W (2020) Safe scarless cassette-free selection of genome-edited human pluripotent stem cells using temporary drug resistance. Biomaterials 262:120295
Zhang X, Huang CT, Chen J, Pankratz MT, Xi J, Li J, Yang Y, Lavaute TM, Li XJ, Ayala M et al (2010) Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell 7(1):90–100
Sato S, Noda S, Torii S, Amo T, Ikeda A, Funayama M, Yamaguchi J, Fukuda T, Kondo H, Tada N et al (2021) Homeostatic p62 levels and inclusion body formation in CHCHD2 knockout mice. Hum Mol Genet 30(6):443–453
Go YH, Lim C, Jeong HC, Kwon OS, Chung S, Lee H, Kim W, Suh YG, Son WS, Lee MO et al (2019) Structure-activity relationship analysis of YM155 for inducing selective cell death of human pluripotent stem cells. Front Chem 7:298
Ohgushi M, Matsumura M, Eiraku M, Murakami K, Aramaki T, Nishiyama A, Muguruma K, Nakano T, Suga H, Ueno M et al (2010) Molecular pathway and cell state responsible for dissociation-induced apoptosis in human pluripotent stem cells. Cell Stem Cell 7(2):225–239
Emre N, Vidal JG, Elia J, O’Connor ED, Paramban RI, Hefferan MP, Navarro R, Goldberg DS, Varki NM, Marsala M et al (2010) The ROCK inhibitor Y-27632 improves recovery of human embryonic stem cells after fluorescence-activated cell sorting with multiple cell surface markers. PLoS One 5(8):e12148
Vernardis SI, Terzoudis K, Panoskaltsis N, Mantalaris A (2017) Human embryonic and induced pluripotent stem cells maintain phenotype but alter their metabolism after exposure to ROCK inhibitor. Sci Rep 7:42138
Gao L, Nath SC, Jiao X, Zhou R, Nishikawa S, Krawetz R, Li X, Rancourt DE (2019) Post-Passage rock inhibition induces cytoskeletal aberrations and apoptosis in Human embryonic stem cells. Stem cell research 41:101641
Markouli C, De Deckersberg EC, Regin M, Nguyen HT, Zambelli F, Keller A, Dziedzicka D, De Kock J, Tilleman L, Van Nieuwerburgh F et al (2019) Gain of 20q11.21 in human pluripotent stem cells impairs TGF-beta-dependent neuroectodermal commitment. Stem Cell Rep 13(1):163–176
Weissbein U, Peretz M, Plotnik O, Yanuka O, Sagi I, Golan-Lev T, Benvenisty N (2019) Genome-wide screen for culture adaptation and tumorigenicity-related genes in human pluripotent stem cells. iScience 11:398–408
Kawano Y, Fukata Y, Oshiro N, Amano M, Nakamura T, Ito M, Matsumura F, Inagaki M, Kaibuchi K (1999) Phosphorylation of myosin-binding subunit (MBS) of myosin phosphatase by Rho-kinase in vivo. J Cell Biol 147(5):1023–1038
Li Z, Dong X, Wang Z, Liu W, Deng N, Ding Y, Tang L, Hla T, Zeng R, Li L et al (2005) Regulation of PTEN by Rho small GTPases. Nat Cell Biol 7(4):399–404
Maekawa M, Ishizaki T, Boku S, Watanabe N, Fujita A, Iwamatsu A, Obinata T, Ohashi K, Mizuno K, Narumiya S (1999) Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science 285(5429):895–898
Schweitzer JS, Song B, Herrington TM, Park TY, Lee N, Ko S, Jeon J, Cha Y, Kim K, Li Q et al (2020) Personalized iPSC-derived dopamine progenitor cells for Parkinson’s disease. N Engl J Med 382(20):1926–1932
Acknowledgements
The human CHCHD2 expression vector (pCL-Neo-CHCHD2-Flag) was generously provided by Dr. Lawrence I. Grossman. Additionally, we would like to extend our gratitude to Drs. Man Ryul Lee and Ok-Seon Kwon for kindly providing the H9 hESCs subline for our research.
Funding
This research was supported by the National Research Foundation of Korea (NRF) no. NRF-2023M3A9E4009624 and a grant from the Korean Fund for Regenerative Medicine funded by Ministry of Science and ICT, and Ministry of Health and Welfare no. RS-2022-00070316.
Author information
Authors and Affiliations
Contributions
HJ-C and YH-G: conceived the overall study design and led the experiments. YH-G and J-K: conducted the experiments and critical discussion of the results. EJ-K: performed in silico analysis. YJ-K established cell lines. DK and YZ-S performed the revision experiments. MO-L characterized H9 sublines. D.G and JH-K provided key CNV analysis data. LHK and HDS: conducted CNV analysis of hESCs. All authors contributed to manuscript writing and revising and endorsed the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
This study using hESCs lines was approved by the Public Institutional Bioethics Committe designated by the Ministry of Health and Welfare (Seoul, Republic of Korea; IRB no. P01-201409-ES-01).
Consent for publication
In this manuscript, the data of any individual patient was not included.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file2 (MP4 9320 KB)
Supplementary file3 (MP4 8966 KB)
Supplementary file4 (MP4 7340 KB)
Supplementary file5 (MP4 11321 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, J., Kwon, EJ., Kim, YJ. et al. Epigenetic repression of CHCHD2 enhances survival from single cell dissociation through attenuated Rho A kinase activity. Cell. Mol. Life Sci. 81, 38 (2024). https://doi.org/10.1007/s00018-023-05060-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-023-05060-8