Abstract
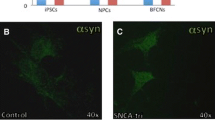
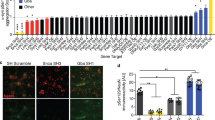
Parkinson’s disease (PD) is one of the most common neuro-degenerative diseases characterized by α-synuclein accumulation and degeneration of dopaminergic neurons. Employing genome-wide sequencing, we identified a polymorphic USP8 allele (USP8D442G) significantly enriched in Chinese PD patients. To test the involvement of this polymorphism in PD pathogenesis, we derived dopaminergic neurons (DAn) from human-induced pluripotent stem cells (hiPSCs) reprogrammed from fibroblasts of PD patients harboring USP8D442G allele and their healthy siblings. In addition, we knock-in D442G polymorphic site into the endogenous USP8 gene of human embryonic stem cells (hESCs) and derived DAn from these knock-in hESCs to explore their cellular phenotypes and molecular mechanism. We found that expression of USP8D442G in DAn induces the accumulation and abnormal subcellular localization of α-Synuclein (α-Syn). Mechanistically, we demonstrate that D442G polymorphism enhances the interaction between α-Syn and USP8 and thus increases the K63-specific deubiquitination and stability of α-Syn . We discover a pathogenic polymorphism for PD that represent a promising therapeutic and diagnostic target for PD.






Similar content being viewed by others
Availability of data and materials
The published article includes all datasets generated or analyzed during this study. The whole-genome sequencing data can be accessed from Genome Sequence Archive (https://ngdc.cncb.ac.cn/gsa, accession number: HRA002315). All datasets supporting this study are available from the lead contact, Dr. Yang Xu (xuyang2020@zju.edu.cn) upon request.
Abbreviations
- α-Syn:
-
Alpha synuclein
- USP8:
-
Ubiquitin specific protease-8
- WT:
-
Wild type
- MT:
-
USP8 D442G mutation
- PD:
-
Parkinson’s disease
- IP:
-
Immunoprecipitation
- IB:
-
Immunoblotting
- WB:
-
Western blot
- DUBs:
-
Deubiquitinases
- IF:
-
Immunofluorescence
- TH:
-
Tyrosine hydroxylase
- TUJ1:
-
Tubulin beta 3 class III
- iPSC:
-
Induced pluripotent stem cells
- NSC:
-
Neural stem cells
- hESC:
-
Human embryonic stem cell
- DAn:
-
Midbrain dopaminergic neuron cell
- EV:
-
Empty vector
References
Kalia LV, Lang AE (2015) Parkinson’s disease. Lancet 386:896–912. https://doi.org/10.1016/S0140-6736(14)61393-3
Kikuchi T, Morizane A, Doi D, Magotani H, Onoe H, Hayashi T, Mizuma H, Takara S, Takahashi R, Inoue H, Morita S, Yamamoto M, Okita K, Nakagawa M, Parmar M, Takahashi J (2017) Human iPS cell-derived dopaminergic neurons function in a primate Parkinson’s disease model. Nature 548:592–596. https://doi.org/10.1038/nature23664
Lonskaya I, Hebron ML, Desforges NM, Schachter JB, Moussa CE (2014) Nilotinib-induced autophagic changes increase endogenous parkin level and ubiquitination, leading to amyloid clearance. J Mol Med (Berl) 92:373–386. https://doi.org/10.1007/s00109-013-1112-3
Zheng N, Shabek N (2017) Ubiquitin ligases: structure, function, and regulation. Annu Rev Biochem 86:129–157. https://doi.org/10.1146/annurev-biochem-060815-014922
Yau RG, Doerner K, Castellanos ER, Haakonsen DL, Werner A, Wang N, Yang XW, Martinez-Martin N, Matsumoto ML, Dixit VM, Rape M (2017) Assembly and function of heterotypic ubiquitin chains in cell-cycle and protein quality control. Cell 171(918–933):e20. https://doi.org/10.1016/j.cell.2017.09.040
Schmidt MF, Gan ZY, Komander D, Dewson G (2021) Ubiquitin signalling in neurodegeneration: mechanisms and therapeutic opportunities. Cell Death Differ 28:570–590. https://doi.org/10.1038/s41418-020-00706-7
Lee JH, Shin SK, Jiang Y, Choi WH, Hong C, Kim DE, Lee MJ (2015) Facilitated Tau degradation by USP14 aptamers via enhanced proteasome activity. Sci Rep 5:10757. https://doi.org/10.1038/srep10757
Wang P, Joberty G, Buist A, Vanoosthuyse A, Stancu IC, Vasconcelos B, Pierrot N, Faelth-Savitski M, Kienlen-Campard P, Octave JN, Bantscheff M, Drewes G, Moechars D, Dewachter I (2017) Tau interactome mapping based identification of Otub1 as Tau deubiquitinase involved in accumulation of pathological Tau forms in vitro and in vivo. Acta Neuropathol 133:731–749. https://doi.org/10.1007/s00401-016-1663-9
Liu X, Hebron M, Shi W, Lonskaya I, Moussa CE (2019) Ubiquitin specific protease-13 independently regulates parkin ubiquitination and alpha-synuclein clearance in alpha-synucleinopathies. Hum Mol Genet 28:548–560. https://doi.org/10.1093/hmg/ddy365
Alexopoulou Z, Lang J, Perrett RM, Elschami M, Hurry ME, Kim HT, Mazaraki D, Szabo A, Kessler BM, Goldberg AL, Ansorge O, Fulga TA, Tofaris GK (2016) Deubiquitinase Usp8 regulates alpha-synuclein clearance and modifies its toxicity in Lewy body disease. Proc Natl Acad Sci U S A 113:E4688–E4697. https://doi.org/10.1073/pnas.1523597113
Du XY, Xie XX, Liu RT (2020) The role of alpha-synuclein oligomers in Parkinson’s disease. Int J Mol Sci. https://doi.org/10.3390/ijms21228645
Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, Hong H, Nakagawa M, Tanabe K, Tezuka K, Shibata T, Kunisada T, Takahashi M, Takahashi J, Saji H, Yamanaka S (2011) A more efficient method to generate integration-free human iPS cells. Nat Methods 8:409–412. https://doi.org/10.1038/nmeth.1591
Li W, Sun W, Zhang Y, Wei W, Ambasudhan R, Xia P, Talantova M, Lin T, Kim J, Wang X, Kim WR, Lipton SA, Zhang K, Ding S (2011) Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc Natl Acad Sci U S A 108:8299–8304. https://doi.org/10.1073/pnas.1014041108
Woodard CM, Campos BA, Kuo SH, Nirenberg MJ, Nestor MW, Zimmer M, Mosharov EV, Sulzer D, Zhou H, Paull D, Clark L, Schadt EE, Sardi SP, Rubin L, Eggan K, Brock M, Lipnick S, Rao M, Chang S, Li A, Noggle SA (2014) iPSC-derived dopamine neurons reveal differences between monozygotic twins discordant for Parkinson’s disease. Cell Rep 9:1173–1182. https://doi.org/10.1016/j.celrep.2014.10.023
Kriks S, Shim JW, Piao J, Ganat YM, Wakeman DR, Xie Z, Carrillo-Reid L, Auyeung G, Antonacci C, Buch A, Yang L, Beal MF, Surmeier DJ, Kordower JH, Tabar V, Studer L (2011) Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature 480:547–551. https://doi.org/10.1038/nature10648
Lees AJ, Hardy J, Revesz T (2009) Parkinson’s disease. Lancet 373:2055–2066. https://doi.org/10.1016/S0140-6736(09)60492-X
Telenti A, Pierce LC, Biggs WH, di Iulio J, Wong EH, Fabani MM, Kirkness EF, Moustafa A, Shah N, Xie C, Brewerton SC, Bulsara N, Garner C, Metzker G, Sandoval E, Perkins BA, Och FJ, Turpaz Y, Venter JC (2016) Deep sequencing of 10,000 human genomes. Proc Natl Acad Sci U S A 113:11901–11906. https://doi.org/10.1073/pnas.1613365113
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG, Exome Aggregation C (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536:285–291. https://doi.org/10.1038/nature19057
Lan T, Lin H, Zhu W, Laurent T, Yang M, Liu X, Wang J, Wang J, Yang H, Xu X, Guo X (2017) Deep whole-genome sequencing of 90 Han Chinese genomes. Gigascience 6:1–7. https://doi.org/10.1093/gigascience/gix067
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28:235–242. https://doi.org/10.1093/nar/28.1.235
Yachdav G, Kloppmann E, Kajan L, Hecht M, Goldberg T, Hamp T, Honigschmid P, Schafferhans A, Roos M, Bernhofer M, Richter L, Ashkenazy H, Punta M, Schlessinger A, Bromberg Y, Schneider R, Vriend G, Sander C, Ben-Tal N, Rost B (2014) PredictProtein—an open resource for online prediction of protein structural and functional features. Nucleic Acids Res 42:W337–W343. https://doi.org/10.1093/nar/gku366
Zhang S, Liu YQ, Jia C, Lim YJ, Feng G, Xu E, Long H, Kimura Y, Tao Y, Zhao C, Wang C, Liu Z, Hu JJ, Ma MR, Liu Z, Jiang L, Li D, Wang R, Dawson VL, Dawson TM, Li YM, Mao X, Liu C (2021) Mechanistic basis for receptor-mediated pathological alpha-synuclein fibril cell-to-cell transmission in Parkinson’s disease. Proc Natl Acad Sci U S A. https://doi.org/10.1073/pnas.2011196118
Vinueza-Gavilanes R, Inigo-Marco I, Larrea L, Lasa M, Carte B, Santamaria E, Fernandez-Irigoyen J, Bugallo R, Aragon T, Aldabe R, Arrasate M (2020) N-terminal acetylation mutants affect alpha-synuclein stability, protein levels and neuronal toxicity. Neurobiol Dis 137:104781. https://doi.org/10.1016/j.nbd.2020.104781
Savyon M, Engelender S (2020) SUMOylation in alpha-synuclein homeostasis and pathology. Front Aging Neurosci 12:167. https://doi.org/10.3389/fnagi.2020.00167
Ryan P, Xu M, Davey AK, Danon JJ, Mellick GD, Kassiou M, Rudrawar S (2019) O-GlcNAc modification protects against protein misfolding and aggregation in neurodegenerative disease. ACS Chem Neurosci 10:2209–2221. https://doi.org/10.1021/acschemneuro.9b00143
Bluhm A, Schrempel S, von Horsten S, Schulze A, Rossner S (2021) Proteolytic alpha-synuclein cleavage in health and disease. Int J Mol Sci. https://doi.org/10.3390/ijms22115450
Nonaka T, Iwatsubo T, Hasegawa M (2005) Ubiquitination of alpha-synuclein. Biochemistry 44:361–368. https://doi.org/10.1021/bi0485528
Burmann BM, Gerez JA, Matecko-Burmann I, Campioni S, Kumari P, Ghosh D, Mazur A, Aspholm EE, Sulskis D, Wawrzyniuk M, Bock T, Schmidt A, Rudiger SGD, Riek R, Hiller S (2020) Regulation of alpha-synuclein by chaperones in mammalian cells. Nature 577:127–132. https://doi.org/10.1038/s41586-019-1808-9
Sulzer D, Edwards RH (2019) The physiological role of alpha-synuclein and its relationship to Parkinson’s disease. J Neurochem 150:475–486. https://doi.org/10.1111/jnc.14810
Vasili E, Dominguez-Meijide A, Outeiro TF (2019) Spreading of alpha-synuclein and Tau: a systematic comparison of the mechanisms involved. Front Mol Neurosci 12:107. https://doi.org/10.3389/fnmol.2019.00107
Ozansoy M, Basak AN (2013) The central theme of Parkinson’s disease: alpha-synuclein. Mol Neurobiol 47:460–465. https://doi.org/10.1007/s12035-012-8369-3
Kiely AP, Asi YT, Kara E, Limousin P, Ling H, Lewis P, Proukakis C, Quinn N, Lees AJ, Hardy J, Revesz T, Houlden H, Holton JL (2013) alpha-Synucleinopathy associated with G51D SNCA mutation: a link between Parkinson’s disease and multiple system atrophy? Acta Neuropathol 125:753–769. https://doi.org/10.1007/s00401-013-1096-7
Konno T, Ross OA, Puschmann A, Dickson DW, Wszolek ZK (2016) Autosomal dominant Parkinson’s disease caused by SNCA duplications. Parkinsonism Relat Disord 22(Suppl 1):S1-6. https://doi.org/10.1016/j.parkreldis.2015.09.007
Tofaris GK, Kim HT, Hourez R, Jung JW, Kim KP, Goldberg AL (2011) Ubiquitin ligase Nedd4 promotes alpha-synuclein degradation by the endosomal-lysosomal pathway. Proc Natl Acad Sci U S A 108:17004–17009. https://doi.org/10.1073/pnas.1109356108
Park GH, Park JH, Chung KC (2021) Precise control of mitophagy through ubiquitin proteasome system and deubiquitin proteases and their dysfunction in Parkinson’s disease. BMB Rep 54:592–600. https://doi.org/10.5483/BMBRep.2021.54.12.107
Rott R, Szargel R, Haskin J, Bandopadhyay R, Lees AJ, Shani V, Engelender S (2011) alpha-synuclein fate is determined by USP9X-regulated monoubiquitination. Proc Natl Acad Sci U S A 108:18666–18671. https://doi.org/10.1073/pnas.1105725108
Kumari R, Kumar R, Kumar S, Singh AK, Hanpude P, Jangir D, Maiti TK (2020) Amyloid aggregates of the deubiquitinase OTUB1 are neurotoxic, suggesting that they contribute to the development of Parkinson’s disease. J Biol Chem 295:3466–3484. https://doi.org/10.1074/jbc.RA119.009546
Acknowledgements
We thank Dr. X. Luo for collecting cells from PD patients and healthy siblings, Drs. Z. Huang and J. Wang for helping the analysis of sequencing data.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 81930084, U1601222), National Key Research and Development Program of China (2022YFC3401600), the Fundamental Research Funds for the Central Universities (No. 2021FZZX001-42), Ministry of Science and Technology of China (2103ZX10002008002), the Key Research and Development Program of Guangdong Province (2019B020235003), Department of Science and Technology of Guangdong Province (2020A1515110450), Traditional Chinese Medicine Bureau Of Guangdong Province of China (20211183, 20225009), the Special Project of State Key Laboratory of Dampness Syndrome of Chinese Medicine (SZ2021KF15) and the Specific Research Fund for TCM Science and Technology of Guangdong Provincial Hospital of Chinese Medicine (YN2015QN04 and 2020KT1079).
Author information
Authors and Affiliations
Contributions
YX conceptualized, designed, and supervised the research; SW and TL conducted the experiments.; YX, SW analyzed the data; YX and SW wrote the manuscript. All authors revised and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethics approval and consent to participate
The study was approved by the Guangdong Provincial Hospital of Traditional Chinese Medicine (TCM) Board of Ethics.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, S., Lin, T. & Xu, Y. Polymorphic USP8 allele promotes Parkinson’s disease by inducing the accumulation of α-synuclein through deubiquitination. Cell. Mol. Life Sci. 80, 363 (2023). https://doi.org/10.1007/s00018-023-05006-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-023-05006-0