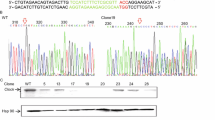
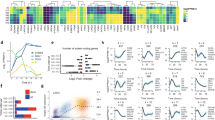
Abstract
Understanding the physiology of human-induced pluripotent stem cells (iPSCs) is necessary for directed differentiation, mimicking embryonic development, and regenerative medicine applications. Pluripotent stem cells (PSCs) exhibit unique abilities such as self-renewal and pluripotency, but they lack some functions that are associated with normal somatic cells. One such function is the circadian oscillation of clock genes; however, whether or not PSCs demonstrate this capability remains unclear. In this study, the reason why circadian rhythm does not oscillate in human iPSCs was examined. This phenomenon may be due to the transcriptional repression of clock genes resulting from the hypermethylation of histone H3 at lysine 27 (H3K27), or it may be due to the low levels of brain and muscle ARNT-like 1 (BMAL1) protein. Therefore, BMAL1-overexpressing cells were generated and pre-treated with GSK126, an inhibitor of enhancer of zest homologue 2 (EZH2), which is a methyltransferase of H3K27 and a component of polycomb repressive complex 2. Consequently, a significant circadian rhythm following endogenous BMAL1, period 2 (PER2), and other clock gene expression was induced by these two factors, suggesting a candidate mechanism for the lack of rhythmicity of clock gene expression in iPSCs.








Similar content being viewed by others
Availability of data
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Umemura Y, Maki I, Tsuchiya Y et al (2019) Human circadian molecular oscillation development using induced pluripotent stem cells. J Biol Rhythms 34:525–532. https://doi.org/10.1177/0748730419865436
Dierickx P, Vermunt MW, Muraro MJ et al (2017) Circadian networks in human embryonic stem cell-derived cardiomyocytes. EMBO Rep 18:1199–1212. https://doi.org/10.15252/embr.201743897
Vilchez D, Boyer L, Morantte I et al (2012) Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature 489:304–308. https://doi.org/10.1038/nature11468
Lee HJ, Gutierrez-Garcia R, Vilchez D (2017) Embryonic stem cells: a novel paradigm to study proteostasis? FEBS J 284:391–398. https://doi.org/10.1111/febs.13810
Banito A, Gil J (2010) Induced pluripotent stem cells and senescence: learning the biology to improve the technology. EMBO Rep 11:353–359. https://doi.org/10.1038/embor.2010.47
Sancar G, Brunner M (2014) Circadian clocks and energy metabolism. Cell Mol Life Sci 71:2667–2680. https://doi.org/10.1007/s00018-014-1574-7
Takahashi JS (2017) Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet 18:164–179. https://doi.org/10.1038/nrg.2016.150
Papazyan R, Zhang Y, Lazar MA (2016) Genetic and epigenomic mechanisms of mammalian circadian transcription. Nat Struct Mol Biol 23:1045–1052. https://doi.org/10.1038/nsmb.3324
Saini C, Morf J, Stratmann M et al (2012) Simulated body temperature rhythms reveal the phase-shifting behavior and plasticity of mammalian circadian oscillators. Genes Dev 26:567–580. https://doi.org/10.1101/gad.183251.111
Paulose JK, Rucker EB, Cassone VM (2012) Toward the Beginning of time: circadian rhythms in metabolism precede rhythms in clock gene expression in mouse embryonic stem cells. PLoS ONE 7:e49555. https://doi.org/10.1371/journal.pone.0049555
Yagita K, Horie K, Koinuma S et al (2010) Development of the circadian oscillator during differentiation of mouse embryonic stem cells in vitro. Proc Natl Acad Sci USA 107:3846–3851. https://doi.org/10.1073/pnas.0913256107
Umemura Y, Koike N, Matsumoto T et al (2014) Transcriptional program of Kpna2/Importin-α2 regulates cellular differentiation-coupled circadian clock development in mammalian cells. Proc Natl Acad Sci USA 111:E5039–E5048. https://doi.org/10.1073/pnas.1419272111
Umemura Y, Koike N, Ohashi M et al (2017) Involvement of posttranscriptional regulation of Clock in the emergence of circadian clock oscillation during mouse development. Proc Natl Acad Sci USA 114:E7479–E7488. https://doi.org/10.1073/pnas.1703170114
Li M, Liu G-H, Belmonte JCI (2012) Navigating the epigenetic landscape of pluripotent stem cells. Nat Rev Mol Cell Biol 13:524–535. https://doi.org/10.1038/nrm3393
Guenther MG, Frampton GM, Soldner F et al (2010) Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell 7:249–257. https://doi.org/10.1016/j.stem.2010.06.015
Kaneko H, Kaitsuka T, Tomizawa K (2020) Response to stimulations inducing circadian rhythm in human induced pluripotent stem cells. Cells 9:620. https://doi.org/10.3390/cells9030620
Bunger MK, Wilsbacher LD, Moran SM et al (2000) Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103:1009–1017. https://doi.org/10.1016/S0092-8674(00)00205-1
Stratmann M, Suter DM, Molina N et al (2012) Circadian Dbp transcription relies on highly dynamic BMAL1-CLOCK interaction with E boxes and requires the proteasome. Mol Cell 48:277–287. https://doi.org/10.1016/j.molcel.2012.08.012
Chan HL, Beckedorff F, Zhang Y et al (2018) Polycomb complexes associate with enhancers and promote oncogenic transcriptional programs in cancer through multiple mechanisms. Nat Commun 9:3377. https://doi.org/10.1038/s41467-018-05728-x
Schuettengruber B, Bourbon H-M, Di Croce L, Cavalli G (2017) Genome regulation by polycomb and trithorax: 70 years and counting. Cell 171:34–57. https://doi.org/10.1016/j.cell.2017.08.002
Atlasi Y, Stunnenberg HG (2017) The interplay of epigenetic marks during stem cell differentiation and development. Nat Rev Genet 18:643–658. https://doi.org/10.1038/nrg.2017.57
Yokobayashi S, Yabuta Y, Nakagawa M et al (2021) Inherent genomic properties underlie the epigenomic heterogeneity of human induced pluripotent stem cells. Cell Rep 37:109909. https://doi.org/10.1016/j.celrep.2021.109909
Yan P, Liu Z, Song M, Belmonte JCI, Xie W, Ren J, Zhang W, Sun Q, Qu J, Liu GH (2020) Genome-wide R-loop landscapes during Cell differentiation and reprogramming. Cell Rep 32:107870. https://doi.org/10.1016/j.celrep.2020.107870
Vollmers C, Panda S, DiTacchio L (2008) A high-throughput assay for siRNA-based circadian screens in human U2OS cells. PLoS One 3:e3457. https://doi.org/10.1371/journal.pone.0003457
McCabe MT, Ott HM, Ganji G et al (2012) EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492:108–112. https://doi.org/10.1038/nature11606
Takahashi K, Tanabe K, Ohnuki M et al (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872. https://doi.org/10.1016/j.cell.2007.11.019
Nakagawa M, Taniguchi Y, Senda S et al (2014) A novel efficient feeder-free culture system for the derivation of human induced pluripotent stem cells. Sci Rep 4:3594. https://doi.org/10.1038/srep03594
Chin MH, Mason MJ, Xie W et al (2009) Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell 5:111–123. https://doi.org/10.1016/j.stem.2009.06.008
Bernstein BE, Mikkelsen TS, Xie X et al (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125:315–326. https://doi.org/10.1016/j.cell.2006.02.041
Margueron R, Reinberg D (2011) The Polycomb complex PRC2 and its mark in life. Nature 469:343–349. https://doi.org/10.1038/nature09784
Pan G, Tian S, Nie J et al (2007) Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell 1:299–312. https://doi.org/10.1016/j.stem.2007.08.003
Harikumar A, Meshorer E (2015) Chromatin remodeling and bivalent histone modifications in embryonic stem cells. EMBO Rep 16:1609–1619. https://doi.org/10.15252/embr.201541011
Etchegaray J-P, Yang X, DeBruyne JP et al (2006) The polycomb group protein EZH2 is required for mammalian circadian clock function. J Biol Chem 281:21209–21215. https://doi.org/10.1074/jbc.M603722200
Margueron R, Li G, Sarma K et al (2008) Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell 32:503–518. https://doi.org/10.1016/j.molcel.2008.11.004
Lee SH, Li Y, Kim H et al (2022) The role of EZH1 and EZH2 in development and cancer. BMB Rep 55:595–601. https://doi.org/10.5483/BMBRep.2022.55.12.174
Mayran A, Drouin J (2018) Pioneer transcription factors shape the epigenetic landscape. J Biol Chem 293:13795–13804. https://doi.org/10.1074/jbc.R117.001232
Ameneiro C, Moreira T, Fuentes-Iglesias A et al (2020) BMAL1 coordinates energy metabolism and differentiation of pluripotent stem cells. Life Sci Alliance 3:e201900534. https://doi.org/10.26508/lsa.201900534
Gallardo A, Molina A, Asenjo HG et al (2020) The molecular clock protein Bmal1 regulates cell differentiation in mouse embryonic stem cells. Life Sci Alliance 3:e201900535. https://doi.org/10.26508/lsa.201900535
Thakur S, Storewala P, Basak U et al (2020) Clocking the circadian genes in human embryonic stem cells. Stem Cell Investig 7:9. https://doi.org/10.21037/sci-2020-014
Koronowski KB, Sassone-Corsi P (2021) Communicating clocks shape circadian homeostasis. Science 371:eabd0951. https://doi.org/10.1126/science.abd0951
Balsalobre A, Brown SA, Marcacci L et al (2000) Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289:2344–2347. https://doi.org/10.1126/science.289.5488.2344
Yagita K, Okamura H (2000) Forskolin induces circadian gene expression of rPer1, rPer2 and dbp in mammalian rat-1 fibroblasts. FEBS Lett 465:79–82. https://doi.org/10.1016/S0014-5793(99)01724-X
Verlande A, Masri S (2019) Circadian clocks and cancer: timekeeping governs cellular metabolism. Trends Endocrinol Metab 30:445–458. https://doi.org/10.1016/j.tem.2019.05.001
Kiessling S, Beaulieu-Laroche L, Blum ID et al (2017) Enhancing circadian clock function in cancer cells inhibits tumor growth. BMC Biol 15:13. https://doi.org/10.1186/s12915-017-0349-7
Ogino T, Matsunaga N, Tanaka T et al (2021) Post-transcriptional repression of circadian component CLOCK regulates cancer-stemness in murine breast cancer cells. Elife 10:e66155. https://doi.org/10.7554/eLife.66155
Seron-Ferre M, Valenzuela GJ, Torres-Farfan C (2007) Circadian clocks during embryonic and fetal development. Birth Defect Res C 81:204–214. https://doi.org/10.1002/bdrc.20101
Serón-Ferré M, Mendez N, Abarzua-Catalan L et al (2012) Circadian rhythms in the fetus. Mol Cell Endocrinol 349:68–75. https://doi.org/10.1016/j.mce.2011.07.039
van der Horst GTJ, Muijtjens M, Kobayashi K et al (1999) Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398:627–630. https://doi.org/10.1038/19323
Zheng B, Albrecht U, Kaasik K et al (2001) Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105:683–694. https://doi.org/10.1016/S0092-8674(01)00380-4
Kondratov RV, Kondratova AA, Gorbacheva VY et al (2006) Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev 20:1868–1873. https://doi.org/10.1101/gad.1432206
DeBruyne JP, Weaver DR, Reppert SM (2007) CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci 10:543–545. https://doi.org/10.1038/nn1884
Hanna J, Cheng AW, Saha K et al (2010) Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci USA 107:9222–9227. https://doi.org/10.1073/pnas.1004584107
Ware CB, Nelson AM, Mecham B et al (2014) Derivation of naive human embryonic stem cells. Proc Natl Acad Sci USA 111:4484–4489. https://doi.org/10.1073/pnas.1319738111
Okita K, Yamakawa T, Matsumura Y et al (2013) An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells 31:458–466. https://doi.org/10.1002/stem.1293
Acknowledgements
This research was funded by Japan Society for the Promotion of Science, grant number KAKENHI 18K06876, and Adaptable and Seamless Technology transfer Program through Target-driven R&D (A-STEP) from Japan Science and Technology Agency Grant Number JPMJTM19GL. The authors would like to thank Enago (www.enago.jp) for the English language review.
Funding
This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Number JP18K06876 and by Adaptable and Seamless Technology transfer Program through Target-driven R&D (A-STEP) from Japan Science and Technology Agency Grant Number JPMJTM19GL.
Author information
Authors and Affiliations
Contributions
HK performed experiments. HK, TK, and KT designed experiments and analyzed data. HK and TK wrote the paper. TK and KT oversaw the project.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kaneko, H., Kaitsuka, T. & Tomizawa, K. Artificial induction of circadian rhythm by combining exogenous BMAL1 expression and polycomb repressive complex 2 inhibition in human induced pluripotent stem cells. Cell. Mol. Life Sci. 80, 200 (2023). https://doi.org/10.1007/s00018-023-04847-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-023-04847-z