Abstract
Peroxisomes are essential for mitochondrial health, as the absence of peroxisomes leads to altered mitochondria. However, it is unclear whether the changes in mitochondria are a function of preserving cellular function or a response to cellular damage caused by the absence of peroxisomes. To address this, we developed conditional hepatocyte-specific Pex16 deficient (Pex16 KO) mice that develop peroxisome loss and subjected them to a low-protein diet to induce metabolic stress. Loss of PEX16 in hepatocytes led to increased biogenesis of small mitochondria and reduced autophagy flux but with preserved capacity for respiration and ATP capacity. Metabolic stress induced by low protein feeding led to mitochondrial dysfunction in Pex16 KO mice and impaired biogenesis. Activation of PPARα partially corrected these mitochondrial disturbances, despite the absence of peroxisomes. The findings of this study demonstrate that the absence of peroxisomes in hepatocytes results in a concerted effort to preserve mitochondrial function, including increased mitochondrial biogenesis, altered morphology, and modified autophagy activity. Our study underscores the relationship between peroxisomes and mitochondria in regulating the hepatic metabolic responses to nutritional stressors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatic peroxisomes are critical for the anabolic and catabolic processing of lipids, including the oxidation of very long-chain fatty acids and branched fatty acids, and the synthesis of plasmalogens, bile acids and sterol precursors. These lipid biochemical reactions are not entirely completed in peroxisomes as they require coordination with other organelles, including the ER and mitochondria [1, 2]. Peroxisomes are important for mitochondrial health, as genetic diseases such as Zellweger Spectrum Disorders, which result in the loss of functional peroxisomes, are associated with hepatic mitochondrial dysfunction [3, 4]. However, our understanding of the underlying mechanisms is limited.
Peroxisome loss in Zellweger Spectrum Disorders results from defects in one of 14 peroxins (PEX) responsible for peroxisome biogenesis and homeostasis [1, 5]. Some peroxins, such as PEX11, are essential for peroxisomal growth and division. Others, including PEX5, PEX7, PEX14, and PEX13 regulate peroxisomal matrix protein import, while PEX3, PEX16, and PEX19 are essential in initiating peroxisome biogenesis [5]. The loss of peroxins such as PEX5 results in ghost peroxisomes with membrane structures devoid of matrix proteins [6], whereas the loss of PEX16 completely abolishes peroxisomes [7]. Because peroxisomes interact with mitochondria [1, 2], the complete abolishment of peroxisomes may help us understand the functional interrelationship between these two organelles.
Mitochondria continuously undergo fission and fusion to maintain dynamic homeostasis. These processes are mediated primarily by Dynamin-related protein 1 (DRP1) and Mitofusin-2 (MFN2), respectively [8]. Impaired mitochondrial function can lead to reduced ATP production, excessive reactive oxygen species (ROS), altered metabolism, changes in calcium storage and apoptosis. The accumulation of damaged mitochondria increases cellular oxidative stress and the likelihood of cell death. Therefore impaired mitochondria are turned over by a selective autophagy called mitophagy [9,10,11].
Various peroxisome deficient mouse models have provided insight into hepatic mitochondrial homeostasis and function. PEX5 deficiency leads to the proliferation of mitochondria with altered morphology and function, and reduced respiration mainly due to a decrease in complex I [12, 13]. A similar phenotype was observed in PEX2 and PEX13 deficient mice [14, 15]. Similar mitochondria defects were also observed in metabolic stress conditions that cause peroxisome loss in the hepatocytes of non-PEX mutant animals. We recently reported that young mice and rats fed a very low protein diet had dramatically reduced hepatic peroxisomes due to the upregulation of peroxisome-specific autophagy, i.e., pexophagy. This reduction of peroxisomes was also associated with damaged and dysfunctional mitochondria [16, 17]. Interestingly, mitochondrial volume increases in most PEX deficient animal models and it has been suggested that this may be a consequence of increased mitochondrial biogenesis [11]. Alternatively, the accumulation of damaged and dysfunctional mitochondria may also result from defects in mitochondria quality control. Thus, it remains unclear whether these changes in mitochondria in hepatocytes without peroxisomes reflect an adaptive response to preserve cellular function or indicate cellular damage due to the loss of peroxisomes.
In this study, we aimed to uncover the role of peroxisomes in mitochondrial homeostasis. To understand the connection between peroxisome loss and mitochondrial function, we generated a liver-specific Pex16 knockout mouse and analyzed mitochondrial morphology, dynamics, and function. These mice were studied under normal nutritional conditions (i.e., normal diet) and nutritional stress (i.e., low protein diet feeding). We found that the loss of PEX16 increased the formation of small mitochondria through the upregulation of genes controlling their fission and fusion. Surprisingly, autophagy flux was reduced in the peroxisome-deficient model, potentially due to a reduction in the degradation of damaged mitochondria. The nutritional stressor led to mitochondrial dysfunction associated with the repression of their biogenesis despite an increase in autophagy. Upregulating mitochondria biogenesis and β-oxidation through PPARα activation improved mitochondrial homeostasis and function independently of the presence of peroxisomes.
Materials and methods
Mouse generation
The Albumin-Cre Pex16 flox/flox (Pex16f/f) mice were generated from Pex16tm1a(EUCOMM)Hmgu, which were obtained from the EUCOMM repository. The Pex16tm1a(EUCOMM)Hmgu were maintained on a C57BL/6J genetic background, and the Pex16 flox/flox mice were generated by crossing with mice expressing Flp recombinase under an actin promoter. To generate the hepatocyte specific Pex16 deficient mice (Pex16 KO), Albumin-Cre males were crossed with Pex16f/f females. Male Pex16f/+;Alb-cre mice were further crossed with female Pex16f/f to generate homozygous Pex16f/f;Alb-cre (aka Pex16 KO) and wild-type control mice. Only wild-type controls and homozygous Pex16f/f;Alb-cre males were used in the experiments. Genotypes were confirmed by PCR (Table S1). Procedures followed the Canadian Council for Animal Care guidelines and were approved by the Animal Care Committee at the Hospital for Sick Children.
Low protein diet mouse model and fenofibrate treatment
Pex16 KO and wild-type control mice were randomized at weaning (i.e., age 3 weeks) into two groups: (1) fed a normal 18% protein chow or (2) fed a low protein diet (LPD; 1% protein diet). The diets were isocaloric but differed in protein content (Fig. S2). Diets were continued for 14 days when animals were fasted for 5 h before being sacrificed. For the fenofibrate treatment experiments, Pex16 KO and wild-type control mice were randomized at weaning into four groups (1) fed a normal 18% protein chow or (2) fed an 18% protein chow and receiving fenofibrate treatment, (3) fed LPD 1% protein diet and (4) LPD and receiving fenofibrate. Fenofibrate (Sigma, F6020) was dissolved in dimethyl sulfoxide (DMSO) (concentration of 100 μg/μL) and given by gavage at a dose of 100 µg/g body weight. Fenofibrate treatment started 7 days after initiation of dietary interventions and continued for 7 days until sacrifice. DMSO was used for the control vehicle group.
Modulating autophagy flux
Pex16 KO and wild-type control mice were fed their attributed diets for 2 weeks and chloroquine (CQ) (Abcam, Cambridge, UK) treatment was delivered by intraperitoneal (IP) injection with a dose of 50 mg/kg of body weight 8 h prior to sacrifice. Mice were fasted for 4 h before sacrifice, and livers were harvested for various assays.
Histology and immunofluorescence
Livers were fixed, embedded in paraffin, sectioned (5 µm) and stained with H&E. Frozen sections (10 µm) were stained with Oil Red O or immunofluorescent antibodies. For this procedure, frozen sections were blocked with 3% bovine serum albumin (BSA) and 0.1% Tween-20 in phosphate-buffered saline (PBS) for 1 h at room temperature. Antibodies (Table S2) were diluted in 1% BSA and 0.1% Tween-20 in PBS. Slides were incubated in primary antibodies overnight at 4 °C, washed and then incubated with secondary antibodies (Table S2). Slides were stained with 4′,6-diamidino-2-phenylindole (DAPI) for 1 h at room temperature. Liver cryosections with 4-μm thickness were stained with 1:1000 fluorinated boron-dipyrromethene (BODIPY) to visualize fat droplets.
Western blotting
In brief, livers were lysed through sonication of tissue with an extraction buffer and protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, USA). Protein concentration was measured by Bicinchoninic Acid assay as per manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA, USA). Samples were then denatured by boiling in sodium dodecyl sulfate (SDS) and generally, 20 μg of protein was loaded onto SDS-PAGE gels. Proteins were transferred onto polyvinylidene fluoride membranes with Trans-Blot system (Thermo Fisher Scientific). Membranes were blocked with 3% BSA and 0.1% Tween-20 in tris-buffered saline (TBS) for one hour at room temperature. Antibodies (Table S2) were diluted in 1% BSA and 0.1% Tween-20 in TBS. Membranes were incubated in primary antibodies overnight at 4 °C, followed by secondary antibodies incubation for 1 h at room temperature. Membranes were visualized using Pierce enhanced chemiluminescence (Invitrogen, Carlsbad, CA, USA).
qPCR
Total RNA was extracted from the liver using Direct-zol RNA Miniprep Kit (ZYMO Research Inc., USA). cDNA was synthesized from 500 ng of total RNA using the SuperScript VILO cDNA Synthesis Kit (Thermo Fisher Scientific, Rockford, IL, USA). For qPCR, 0.1 μl of cDNA was used with Advanced qPCR Mastermix with SuperG Dye (Wisent, Bioproducts, Saint-Jean-Baptiste, QC, Canada). mtDNA was measured using 500 ng of genomic DNA in each qPCR reaction. qPCR was performed using a CFX384 Touch Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA) using the primers listed in Table S3.
Hepatic triglyceride assay
Briefly, 50 mg of wet liver tissue was homogenized in cold PBS and tissue lipids were extracted with methanol-chloroform, dried and dissolved for triglyceride analysis. Liver and serum triglyceride concentrations were determined with a commercial kit (Randox Laboratories, Crumlin, UK). Photometric absorbances were read at 500 nm. Triglyceride values were normalized to protein concentration calculated from the same liver tissue by BCA assay as described above.
Hepatic ATP assay
Liver tissue (~ 10 mg) was homogenized using a dounce homogenizer in perchloric acid (PCA). Excess PCA was precipitated by adding ice-cold 2 M KOH to neutralize the samples (pH 6.5–8). The deproteinized samples were then assayed. ATP concentrations were measured using a commercial ATP (Colorimetric/Fluorometric) assay kit (Abcam). Fluorescence was measured at Excitation/Emission = 537/578 nm with Skanlt software by Varioskan Lux Multimode Microplate Reader (Thermo Fisher Scientific). ATP concentration was established by comparing fluorescence readings from samples to standards provided within the kit.
Transmission electron microscopy (TEM)
For transmission electron microscopy (TEM), fresh liver samples (~ 1 mm2) were fixed in 2% glutaraldehyde in 0.1 M sodium cacodylate buffer, rinsed in cacodylate buffer, post-fixed in 1% osmium tetroxide in cacodylate buffer. Samples were then dehydrated through a series of graded ethanol followed by incubation in propylene oxide, and then embedded in Quetol-Spurr resin. Sections (90 nm) were cut on a Leica Ultracut ultramicrotome (Leica Microsystem, Wetzlar, Germany), stained with uranyl acetate and lead citrate, and viewed in an FEI Tecnai 20 transmission electron microscope (Field Electron and Ion Company, Hillsboro, Oregon, USA). Mitochondrial morphology was measured using Image J software.
Central carbon metabolism assay
To quantify metabolites of central carbon metabolism (CCM) in mouse liver, we used UPLC-MRM/MS, a Dionex 3400 UHPLC system (Dionex Sunnyvale, CA, USA) coupled to a 4000 QTRAP mass spectrometer (SCIEX, Framingham, MA, USA). Liver metabolites were extracted and quantified including those from tricarboxylic acid (TCA) cycle, glucose and selected sugar phosphates, and other phosphate-containing metabolites and nucleotides (Tables S4 and S5). Concentrations of the detected metabolites were calculated from internal linear-regression calibration curves.
Isolation of liver mitochondria and mitochondrial respiration
Mitochondria were isolated from liver tissue by differential centrifugation as described previously [18]. The livers were dissected and minced in ice-cold mitochondrial isolation buffer composed of 100 mM Tris-MOPS, 10 mM EGTA/Tris and 90 mM sucrose, adjusted pH to 7.4. The suspension was transferred to a glass dounce homogenizer and homogenized. Following a series of centrifugations at different speeds at 4 °C, the pellet containing mitochondria was resuspended in an approximate volume in the mitochondrial isolation buffer. Total mitochondrial protein was determined using Pierce BCA Protein Assay kit (Thermo Fisher Scientific). Finally, the mitochondrial preparations (5 mg/well) were used for the coupling assay by the Agilent Seahorse XFe/XF96 analyzer (Agilent Technologies, Santa Clara, CA, USA). The mitochondrial respiration was measured during exposure to different substrates (glutamate, malate, succinate, and pyruvic acid). The final concentration of the injections was as follows: ADP was 1 mM, oligomycin was 2.5 mg/ml, FCCP was 2.2 mM/ml and rotenone was 2 mM/ml [19].
Statistics
Data analyses were performed with GraphPad Prism 8.0 and Image J [20]. CCM data were processed by mixOmics packages with R software [21]. A Partial Least Squared discriminant analysis (PLS-DA) was performed to visualize general clustering and trends. PLS-DA scored plots of components one and two, comparing different diet groups were generated. KEGG pathway analysis was conducted in MetaboAnalyst 4.0. For non-CCM data, two group comparisons were performed using a paired two-tailed Student’s t test. The data of more than two groups were analyzed using one-way ANOVA followed by Turkey’s post-hoc t tests to compare all possible pairs of experimental groups. A value of P < 0.05 was considered significant.
Results
Loss of PEX16 leads to disappearance of peroxisomal membrane and hepatic steatosis
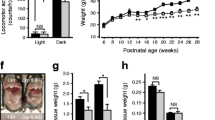
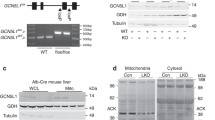
We generated a liver specific Pex16 knockout (Pex16 KO) mouse model by Albumin-Cre-mediated homologous recombination of a floxed allele (Fig. 1a). Western blot analysis of liver tissue showed the loss of PEX16 and peroxisomal membrane protein 70 (PMP70) at postnatal week 3, indicating efficient Pex16 deletion and peroxisome loss (Fig. 1b). Decreased immunofluorescence of PMP70 confirmed peroxisome ablation in hepatocytes (Fig. 1c, Fig. S1a). Pex16 KO mice had a decreased body weight compared to wild-type control animals at postnatal week 3 (Fig. 1d, e). However, the liver weight adjusted for the body weight of Pex16 KO mice was significantly increased (Fig. 1d, f). Although the structure and morphology of their liver tissue appeared normal (Fig. 1g), the Oil Red O staining revealed an increased presence of oil droplets in the livers of Pex16 KO mice at 3 weeks (Fig. 1h) and 5 weeks of age (Fig. S1b). Together, these results show that hepatic loss of PEX16 leads to the disappearance of peroxisomes and hepatic steatosis.
Hepatic steatosis in 3-week weanlings with hepatocyte specific deletion of Pex16. a Schematic of Albumin-Cre-mediated Pex16 gene deletion. b Immunoblotting of PEX16 and PMP70 in wild-type and Pex16 KO liver, and quantification of band intensity relative to GAPDH. c Immunofluorescence of PMP70 in control and Pex16 KO liver. d Mouse and liver morphology of wild-type and Pex16 KO groups. e Quantification of body weight and f Liver weight and liver to body weight ratio of control mice and Pex16 KO mice. g H&E staining and h Oil red O staining of wild-type and Pex16 KO liver. Data are represented as mean ± SEM, n = 6 mice per group. Scale bar: c, g, h 50 µm, d 10 mm. *p < 0.05, **p < 0.01, ***p < 0.001. See also Fig. S1
Nutritional stress induced by a low protein diet exacerbates hepatic steatosis caused by the loss of peroxisomes
To determine susceptibility to nutritional stress, we subjected Pex16 KO mice to a low protein diet (LPD) (Fig. S2) for 2 weeks starting at weaning (age 3 weeks) (Fig. 2a). As expected, body weight significantly decreased during LPD feeding in both Pex16 KO mice and wild-type controls, whereas caloric intake was similar when corrected for body weight (Fig. 2b, c). The size and weight of the liver relative to body weight also decreased in mice fed an LPD (Fig. 2d, e). However, the liver enlargement seen in the Pex16 KO mice was no longer observed when these animals were fed LPD (Figs. 1d, f, 2d, e). Histological analysis showed more vacuoles in the livers of Pex16 KO mice, an effect exacerbated by the LPD (Fig. 2f). Oil Red O staining confirmed that these vacuoles corresponded to lipid droplets (Fig. 2g). Lipid droplets were fewer and smaller in the Pex16 KO mice fed a regular diet compared to the wild-type mice fed an LPD (Fig. 2g, h). In contrast, there were more lipid droplets in the LPD-fed Pex16 KOs compared to the wild-type controls fed the LPD. Lipid staining with BODIPY further demonstrated that the Pex16 KOs fed LPD had the most severe hepatic steatosis (Fig. 2h, i).
Growth delay and severe hepatic steatosis in LPD fed Pex16 KO mice. a Schematic of experimental design. b Mouse morphology of wild-type and Pex16 KO mice fed a control diet or LPD. c Quantification of food intake, body weight and body weight change. d Liver morphology of mice after 2 weeks of control diet or LPD. e Quantification of liver weight and liver to body weight ratio. f Liver H&E staining of wild-type and Pex16 KO mice fed a control diet or LPD. g Oil red O staining. h Immunofluorescence of BODIPY staining. i Quantification of lipid droplet content from Oil Red O and BODIPY staining. Data are represented as mean ± SEM, n = 6–8 mice per group. Scale bar: b, d 10 mm, f, g 500 µm, h 50 µm. *p < 0.05, **p < 0.01, ***p < 0.001. See also Fig. S2
Loss of peroxisomes leads to disturbed hepatic energy homeostasis, which is exacerbated by nutritional stress
To determine the effect of peroxisome depletion on liver metabolism during normal feeding and nutritional stress, we measured fasting blood glucose levels in Pex16 KOs. During fasting, the liver becomes primarily responsible for gluconeogenesis, a process that consumes ATP in the liver. Fasting glucose was not affected in the Pex16 KOs compared to the wild-type mice (Fig. 3a, Fig. S3a). However, it was reduced by LPD and exacerbated by the loss of PEX16, indicating reduced hepatic glucose production. Similarly, ATP levels decreased after LPD feeding in Pex16 KO and wild-type mice (Fig. 3b). We further analyzed the hepatic intermediate metabolome targeting the central carbon metabolism. A total of 53 metabolites were detected, and Pex16 KOs showed a distinct metabolic profile from wild-type controls (Fig. S3b). With LPD feeding, these differences between Pex16 KOs and wild-type controls were still apparent, but the overall differences in metabolite profile appeared to be linked to the diet. Levels of each metabolite are shown in the heat map and the tables (Fig. 3c, and Tables S4, S5). Pathway enrichment analysis revealed that the TCA cycle was affected by both the loss of PEX16 and by LPD feeding (Fig. 3d). Specifically, entry into the TCA cycle appeared to be blocked in wild-type mice fed an LPD as shown by an increase in citrate and a concomitant decrease in acetyl-CoA. This change was more pronounced in the Pex16 KOs fed an LPD associated with a substantial reduction in NAD+ content. These results indicate that loss of PEX16 affects overall energy homeostasis and induces a metabolic stress pattern that shows some similarities to that induced by LPD feeding.
Loss of Pex16 leads to changes in hepatic central carbon metabolism (CCM) exacerbated by LPD feeding in mice. a Blood glucose test in mice fasted for 15 h. Grey asterisks indicate significance between wild-type fed either control or LPD diets, pink asterisks indicate significance between wild-type fed LPD vs Pex16 KO fed LPD. b Hepatic ATP levels. c Heat map presenting cluster analysis of CCM data. High level is in red, and low level is in blue as per legend. d KEGG pathways enriched for hepatic metabolism related pathways. The most affected pathways are related to citric acid cycle (TCA cycle) and fatty acid degradation/elongation (highlighted in red). e Hepatic triglycerides (TG) assay. f Western blots of hepatic PPARα, AMPKα and p-AMPKα, and quantification of band intensity relative to β-actin and ratio of p-AMPKα/AMPKα. Data are represented as mean ± SEM, n = 6–8 mice per group. *p < 0.05, **p < 0.01, ***p < 0.001. See also Fig. S3
Pex16 KO mice show increased triglyceride content despite the increase in the expression of β-oxidation genes
To better understand the etiology of hepatic steatosis caused by the loss of PEX16, we assessed relevant lipid pathways in the liver. We found that the loss of peroxisomes led to increased hepatic triglycerides, which was exacerbated by an LPD (Fig. 3e). This observation was consistent with our histological findings. As hepatic steatosis can be caused by increased lipogenesis, we examined the expression levels of lipogenesis regulatory genes. qPCR of acetyl-CoA carboxylase alpha (Acaca), considered a rate-limiting enzyme for lipogenesis, and fatty acid synthase (Fas) were decreased in the Pex16 KO mice, as well as in both the wild-type and Pex16 KO mice that were fed an LPD (Fig. S3c). We then evaluated the inhibition of the lipogenic pathway by assessing the phosphorylation status of AMP-activated protein kinase alpha (AMPKα) in the liver. Phosphorylated AMPKα protein levels and pAMPKα/ AMPkα were increased by both loss of PEX16 and LPD feeding, but the effect of diet was more modest (Fig. 3f). Consistent with increased AMPKα phosphorylation, the expression of genes regulating mitochondrial β-oxidation, peroxisomal β-oxidation and CoA formation were increased in Pex16 KO mice fed a control diet (Fig. S3d–f). However, these increases in β-oxidation genes did not occur in Pex16 KO mice fed an LPD. Instead, both wild-type mice and Pex16 KO mice on an LPD showed a significant decrease in the expression of β-oxidation genes. The protein level of nuclear hormone receptor peroxisome proliferator-activated receptor alpha (PPARα), an important transcriptional regulator of β-oxidation, was reduced in the Pex16 KO animals, and it was greatly decreased by LPD feeding in both strains (Fig. 3f). This result suggests that the increase in steatosis in the LPD mice may be caused by a decrease in β-oxidation. However, for the Pex16 KO mice in control diet conditions, the increase in triglyceride levels and lipid droplets was observed in their livers despite an increase in β-oxidation gene expression, suggesting a potentially altered mitochondrial function.
Hepatic mitochondrial dynamics are differentially altered in response to loss of peroxisomes and nutritional stress
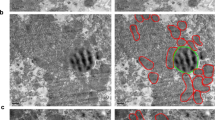
Mitochondria are dynamic organelles. They elongate and divide to meet the metabolic needs of the cells, thus their morphology reflects the bioenergetic status of the cell [8]. For instance, acute nutrient deprivation leads to mitochondria elongation, which promotes increased oxidative phosphorylation and ATP production, and simultaneously prevents non-specific bulk degradation of mitochondria [22,23,24]. Conversely, extensive mitochondrial fission occurs during oxidative stress caused by oxygen deprivation or chemical inhibition of the mitochondrial respiration change complexes [25]. To assess the mechanism of the disruption of hepatic energy homeostasis in the Pex16 KO mice, we examined their mitochondria in normal conditions and during metabolic stress. Immunofluorescent microscopy imaging of hepatocytes stained for the mitochondrial heat shock protein 60 (HSP60) (Fig. 4a) and transmission electron microscopy (Fig. 4b, Fig. S4a) were performed to compare mitochondrial morphology between the different mouse strains and conditions.
Loss of Pex16 and LPD feeding differentially affect mitochondrial stress. a Immunofluorescence of HSP60 in hepatocytes of liver sections. b Transmission electron microscopy (TEM) images (× 20,000), hepatocyte mitochondria are outlined in white, arrows indicate hepatocyte peroxisomes, asterisks indicate fat granules. c Quantification of mitochondrial area, mitochondrial aspect ratio (length/width) and mitochondrial number from TEM images. d qPCR analysis of hepatocyte mitochondrial number. e Western blots of hepatic mitochondrial markers HSP60 and TOM20, and quantification of band intensity relative to β-actin. f Western blots of hepatic MFN2, DRP1, pDRP1, and PINK1 of liver tissue and isolated mitochondria, quantification of band intensity relative to β-actin, TOM20 and COX IV respectively and the ratio of pDRP1/DRP1. g Hepatocyte mitochondrial respiration/function was determined by oxygen consumption rate (OCR). Red asterisks indicate significance between wild-type vs Pex16KO, grey asterisks indicate significance between wild-type vs wild-type fed an LPD, pink asterisks indicate significance between wild-type + LPD vs Pex16 KO + LPD liver. h Western blots of mitochondrial electron transport chain (ETC) complexes subunits, NDUFB8 (Complex I), SDHB (Complex II), UQCRC2 (Complex III), MTCO1 (Complex IV) and ATP5A (Complex V) from isolated hepatic mitochondria, quantification of band intensity relative to COX IV. Data are represented as mean ± SEM, n = 6–8 mice per group. Scale bar: a 10 µm and b 1 µm. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See also Fig. S4
In comparison to the wild-type mice, there were more but smaller mitochondria in the Pex16 KO mice hepatocytes in both metabolic conditions (Fig. 4a, b, circles with dashed lines). Quantification of the mitochondria size (area) and number verified their differences (Fig. 4c). The increase in mitochondria in the Pex16 KO animals was further corroborated by both an increase in the mitochondrial DNA (mtDNA) copy number (Fig. 4d) and by higher levels of the outer membrane translocase 20 (TOM20) and HSP60 (Fig. 4e) compared to wild-type controls.
Under LPD-induced metabolic stress, both wild-type and Pex16 KO mice had larger but fewer hepatic mitochondria (Fig. 4a–c), as assessed by protein levels and mtDNA. This effect was the strongest in the wild-type mice, as Pex16 KO mice fed an LPD had smaller and more numerous mitochondria than the wild-type mice fed an LPD (Fig. 4a–e). We also assessed levels of hepatic peroxisomal proteins and found that wild types fed an LPD had substantially decreased levels of PMP70 and PEX16, which reflected the results shown in livers of amino acid starved rats [17] (Fig. S4b).
We next assessed whether changes in mitochondrial morphometrics were related to mitochondrial fusion and fission by assessing the level of MFN2 and DRP1 respectively. We showed by western blot that MFN2, total DRP1, and phospho-DRP1-S637 (pDRP1) were increased in the Pex16 KO mice compared to the wild types (Fig. 4f). However, as mitochondria are more numerous in the hepatocytes of Pex16 KO mice (Fig. 4a–d), we normalized MFN2 and DRP1 expression to that of TOM20 and found that these proteins were lower in mitochondria of Pex16 KO compared to those in the wild-type animals. However, the pDRP1/DRP1 ratio did not differ between strains fed LPD. The dynamics of mitochondrial fission and fusion are complex, and our data are inconclusive regarding the link between increased fission and smaller mitochondrial size in Pex16 KO hepatocytes. The complexity may derive from mitochondria incurring different or compounded damage in response to the loss of peroxisomes in normal versus LPD feeding, thus, the insults could induce a different combination of repair responses leading to an intermediate phenotype in mitochondrial morphology.
Loss of peroxisomes exacerbates nutritional stress-induced mitochondrial dysfunction
To assess whether the mitochondrial morphological changes were associated with impaired function, we evaluated mitochondrial health by measuring the oxygen consumption rate (OCR) of isolated hepatic mitochondria. Their basal and maximal respiration was assessed in the different strains fed a normal diet or LPD. Baseline OCR did not differ between groups, but the Pex16 KO mice fed a normal diet had a higher reserve respiration capacity (maximal respiration) compared to wild-type controls (Fig. 4g). Interestingly, the hepatic mitochondria of the Pex16 KO mice showed a gradual decrease in OCR following FCCP treatment, which was not observed in wild-type mice. In both LPD-fed groups, mitochondria showed very little respiration reserve as their maximal respiration did not significantly increase over their basal OCR. These results reveal that hepatic mitochondrial function is not impaired by loss of peroxisomes, which is consistent with the unchanged levels of ATP at basal conditions. Instead, the higher reserve respiration capacity in the peroxisome-deficient mice suggests that their mitochondria may have adapted to the changed cellular condition and resulted in mitochondria with a higher capacity to produce ATP. However, under nutritional stress, the mitochondria of Pex16 KO animals have lost their increased reserve respiration capacity.
Next, we assessed the complexes of the electron transport chain (ETC) in isolated mitochondria. We normalized mitochondria complex proteins detected with immunoblotting to cytochrome c oxidase subunit 4 (COX IV), the terminal enzyme in the respiratory chain to allow comparison of relative changes in these complex proteins as described by others [26]. When normalized to COX IV, mitochondrial complex II (SDHB), III (UQC RC2) and V (ATP5A) were modestly increased in the mitochondria isolated from Pex16 KOs compared to wild types, while complex I (NDUFB8) and IV (MTCO1) were modestly decreased (Fig. 4h). LPD feeding decreased all the complexes compared to normal-fed animals, but no differences were observed between the two LPD-fed groups except for complex V. In addition, the normalization protein COX IV differed across conditions where levels appeared to increase in the mitochondria of Pex16 KO mice, which reflects an increase in mitochondria mass observed in these mice compared to the wild-type mice (Fig. 4c). Together with the mitochondria stress test, these results suggest that the Pex16 KO animals are able to adapt to peroxisomal loss under normal conditions by increasing mitochondria numbers and the reserve capacity of the mitochondria. However, the increased reserve capacity is lost under metabolic stress.
PEX16 deficiency alters mitochondria quality control related to a decrease in autophagy
In both normal and LPD-fed conditions, the mitochondria of the Pex16 KO animals were more fragmented and more numerous but showed no difference in ATP production or basal respiration compared to their wild-type counterpart. As these observations could suggest altered mitochondrial quality control, we next examined for defects in mitophagy, the autophagic degradation of mitochondria [9]. An indicator of mitochondria damage is the accumulation of PTEN-induced kinase 1 (PINK1). PINK1 is targeted to the mitochondria and is readily cleaved upon entry by the inner membrane protease PARL, the cleaved products are retro-translocated and degraded by proteasomes in healthy mitochondria, whereas full-length PINK1 accumulates in damaged mitochondria [27]. This accumulation initiates PARKIN-dependent mitophagy, which induces mitochondria fragmentation and the recruitment of autophagic factors to the damaged mitochondria [28]. Therefore, we first assessed whether PINK1 accumulated in Pex16 KO livers. Full-length PINK1 was barely detectable in all conditions (Fig. 4f). When normalized to β-actin, we found that, in wild-type mice, LPD feeding led to a small but significant decrease in PINK1 compared to normal diet feeding. However, we saw an increase in cleaved PINK1 (42 kDa fragment) in the Pex16 KO mice fed a control diet or LPD (Fig. 4f). Similar results were observed when cleaved PINK1 was measured in isolated mitochondria and adjusted for COX IV (Fig. 4f). The significance of the accumulation of cleaved PINK1 is unclear, but it is known that in healthy mitochondria, cleaved PINK1 is efficiently removed from the mitochondria by retro-translocation and degradation [29, 30]. However, we did not observe an accumulation of full-length PINK1, which would be indicative of significant mitochondrial damage, typically observed during chemically induced mitochondria damage [31].
We next examined whether autophagy was downregulated in Pex16 KO livers. We first analyzed the autophagy receptor protein sequestosome-1 (p62) and microtubule-associated protein 1A/1B light chain 3B (LC3B). p62 is turned over by autophagy, thus it accumulates in cells when autophagy or autophagy flux is defective [32]. LC3B-I is the cleaved version of the proLC3B before it is conjugated to phosphatidylethanolamine. The LC3B-PE, also known as LC3B-II, has increased mobility on SDS-PAGE due to lipidation (Fig. 5b), and it can be used to measure the amount of autophagosomes in the cell. We found that both p62 and LC3B-II were higher in the Pex16 KO mice on regular feed, whereas LC3B-II was observed to be higher in the LPD-fed Pex16 KO mice compared to LPD-fed wild types (Fig. 5b). However, p62 and LC3B-II levels were lower in the LPD mice than the mice with regular feed, suggesting a difference in autophagy induction. Since a higher p62 level and LC3B-II can suggest a defect in autophagy flux (the rate of autophagy substrate degraded in lysosomes), we measured the LC3B-II in mice treated with the lysosomotropic agent chloroquine to inhibit lysosome degradation. Acute administration of chloroquine to inhibit autophagic flux led to a moderate increase in p62 levels for all conditions compared to the untreated animals as expected (Fig. 5c, d). The wild-type control-fed mice showed a significant increase in LC3B-II content after chloroquine treatment (Fig. 5c). However, the Pex16 KO fed the control diet actually showed a modest decrease in their LC3B-II content upon chloroquine treatment compared to their untreated counterpart. The LPD groups showed no increase after chloroquine (Fig. 5c). These experiments suggest that the rate of autophagy flux is slowed in the livers of Pex16 KO mice compared to wild-type animals, which may contribute to the accumulation of mitochondria in the liver of Pex16 KO mice (Fig. 4a–e).mTORC1 is the main regulator of autophagy and the inhibition of mTORC1 kinase activity induces autophagy [33]. To further investigate whether the impact of functional loss of PEX16 on autophagy is related to changes in mTORC1 activation, we analyzed phosphorylated p70S6K (p-p70S6K) and phosphorylated 4EBP1 (p-4EBP1), two downstream substrates of mTORC1 kinase activity. p-p70S6K/p70S6K ratio was down-regulated in the Pex16 KO mice consistent with inhibition of mTORC1, but p-4EBP1/4EBP1 ratio was not changed (Fig. 5e, f). Both strains fed LPD showed a decrease in both p-p70S6K/p70S6K and p-4EBP1/4EBP1 ratios supporting that there is an increase in autophagy. Interestingly, loss of functional peroxisomes increased total protein levels of 4EBP1 and p70S6K, with mRNA levels p70S6K most dramatically upregulated in Pex16 KO mice (Fig. S5a, S5b). Increased ROS has been shown to upregulate 4EBP and S6K transcription [34] and could be a mechanism that explains the gene and protein expression changes in our model. While the increase in 4EBP1 and p70S6K levels in the Pex16 KOs limits our ability to draw firm conclusions on mTORC1 activation, this effect may explain the lack of a difference in the p-4EBP1/4EBP1 ratio in comparison to wild types. In summary, the accumulation of altered mitochondria in the Pex16 KO mice may result from a downstream impairment in overall autophagic flux, despite indications that mTORC1 is inhibited.
Loss of Pex16 and LPD feeding differentially affect autophagy and mitochondrial biogenesis. a Schematic of mice treated with chloroquine (CQ) for 8 h under wild-type control and Pex16 KO mice fed a control diet or LPD. b Western blots of hepatic p62, LC3B-I and II, and quantification of band intensity relative to β-actin of wild-type and Pex16 KO mice fed a control diet or LPD. c Western blots of hepatic p62 and LC3B-I and II, and quantification of band intensity relative to β-actin of wild-type and Pex16 KO mice fed a control diet and treated by CQ. d Western blots of hepatic p62 and LC3B-I and II, and quantification of band intensity relative to β-actin of wild-type and Pex16 KO mice fed an LPD diet and treated by CQ. e Western blot of mTOR downstream effector p-p70S6K and p70S6K, and quantification of band intensity relative to β-actin and the ratio of p-p70S6K/p70S6K. f Western blot of mTOR downstream effector p4EBP1, 4EBP1, and quantification of band intensity relative to β-actin and the ratio of p4EBP1/4EBP1. g Western blot of mitochondrial biogenesis marker PGC-1α and NRF1, and quantification of band intensity relative to β-actin. Data are represented as mean ± SEM, n = 6–8 mice per group. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. CQ chloroquine. See also Fig. S5
Pex16 KO mice show increased mitochondrial biogenesis
To determine whether the changes in mitochondrial content were related to mitochondrial biogenesis, we analyzed the levels of peroxisome-proliferator-activated receptor coactivator-1α (PGC-1α) and its downstream target nuclear respiratory factor 1 (NRF1) [35]. PGC-1α and NRF1 protein (Fig. 5g) and Pgc-1α and Nrf1 mRNA (Fig. S5c, S5d) were elevated in the liver of Pex16 KO compared to the wild types, consistent with the increase in mitochondrial number and mass. In contrast, PGC-1α content was decreased by LPD feeding to similar levels in the wild type mice and Pex16 KOs to the normal feeding (Fig. 5g, Fig. S5c). Our results indicate that increased mitochondrial content in Pex16 KO is related to both reduced autophagy and increased mitochondrial biogenesis. In contrast, the reduced mitochondrial content related to LPD feeding appears to be caused by reduced mitochondrial biogenesis.
Activation of PPARα improves hepatic steatosis caused by loss of functional peroxisomes and low protein diet feeding
PPARα is a nuclear receptor that enhances mitochondrial biogenesis and β-oxidation, and it was recently found to induce autophagy [36]. Therefore, we next assessed whether PPARα activation with fenofibrate could improve hepatic metabolism in Pex16 KO mice fed either a control diet or an LPD (Fig. 6a). Fenofibrate did not affect body weight, liver weight, food intake, or fasting glucose concentrations in Pex16 KO or wild-type mice fed a control diet or an LPD (Fig. 6b, Fig. S6a, b, S6g, h). However, fenofibrate treatment reduced hepatic steatosis in Pex16 KO mice fed either a control diet or an LPD (Fig. 6c–g, Fig. S6f–h). Fenofibrate had a similar effect in wild types fed LPD, whereas vehicle treatment did not improve hepatic lipid accumulation (Fig. S6c–f, S6h).
Fenofibrate (FF) treatment reduces hepatic steatosis in Pex16 KO and wild-type mice fed an LPD. a Schematic of experiments where mice fed an LPD were treated with fenofibrate through gavage for 1 week. b Blood glucose test in mice fasted for 15 h, and liver weight. Grey asterisks indicate significance between wild-type + LPD vs wild-type + LPD + FF, pink asterisks indicate significance between Pex16 KO + LPD vs Pex16 KO + LPD + FF. c Hepatic triglycerides (TG) assay. d Quantification of lipids from Oil Red O staining sections shown in g. e Liver morphology of LPD + FF treated wild-type and Pex16 KO mice. f Liver H&E staining. g Liver Oil red O staining. Data are represented as mean ± SEM, n = 6–8 mice per group. Scale bar: e 10 mm, f, g 500 µm. *p < 0.05, **p < 0.01. FF fenofibrate. See also Fig. S6
PPARα activation improves hepatic mitochondrial function independently of functional peroxisomes
We next examined the effect of activating PPARα on mitochondrial fitness in Pex16 KOs fed a control diet or an LPD. Fenofibrate treatment increased peroxisome numbers in wild types fed LPD (Fig. 7a) consistent with an increase in PMP70 (Fig. S7e). Peroxisomal structures were not observed in the EM micrographs of the Pex16 KO livers (Fig. 7a), although we did not perform alkaline 3,3′-diaminobenzidine staining to directly visualize peroxisomes. We therefore further validated our EM findings using immunofluorescence staining of PMP70, which showed an increase with fenofibrate treatment, including small increases in Pex16 KO livers (Fig. S7e). However, this PMP70 staining co-localized with endothelial cells as indicated by co-staining with CD31, an endothelial marker (Fig. S7f). This indicates that fenofibrate treatment can stimulate non-hepatocyte peroxisome biogenesis. Mitochondria were smaller after fenofibrate treatment in Pex16 KOs fed an LPD (Fig. 7a, b). However, their numbers were not changed (Fig. 7b). In contrast, the mitochondria of Pex16 KOs on a control diet treated with fenofibrate showed no clear change in size but were reduced in number compared to untreated mice (Fig. S7a–c). In wild types fed LPD, fenofibrate treatment increased mitochondrial numbers, but these mitochondria were smaller and less elongated (Fig. 7a–c). Consistently, TOM20 protein levels and mtDNA were not significantly increased with fenofibrate treatment in the Pex16 KOs fed an LPD but were increased in wild-type animals (Fig. 7c, d). These changes in mitochondria were associated with a fenofibrate-induced decrease in p-AMPKα and increase in PPARα protein levels (Fig. 7e). Also, mitochondrial content was not impacted by DMSO treatment in wild types fed a control diet (Fig. S7a–e).
Fenofibrate (FF) treatment improves mitochondrial respiration in Pex16 KO and wild-type mice fed an LPD. a Transmission electron microscopy (TEM) images (× 20,000), hepatocyte mitochondria are outlined in white, arrows indicate hepatocyte peroxisomes, asterisks indicate lipids. b Measurement of mitochondrial area, mitochondrial aspect ratio (length/width), and mitochondrial number from TEM images. c qPCR analysis of hepatocyte mitochondrial number. d Western blots of hepatic TOM20 and quantification of band intensity relative to β-actin. e Western blots of hepatic PPARα, p-AMPKα, AMPKα, quantification of band intensity relative to β-actin and the ratio of p-AMPKα/AMPKα. f Western blots of mitochondrial electron transport chain (ETC) complexes subunits, NDUFB8 (Complex I), SDHB (Complex II), UQCRC2 (Complex III), MTCO1 (Complex IV) and ATP5A (Complex V) from isolated hepatic mitochondria, quantification of band intensity relative to COX IV. g Hepatic ATP levels. h Hepatocyte mitochondrial respiration/function was determined by oxygen consumption rate (OCR). Grey asterisks indicate significance between wild-type + LPD vs wild-type + LPD + FF, pink asterisks indicate significance between Pex16 KO + LPD vs Pex16 KO + LPD + FF. Data are represented as mean ± SEM, n = 6–8 mice per group. Scale bar: a 1 µm, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. FF fenofibrate. See also Fig. S7
As expected, fenofibrate treatment upregulated mRNA levels of most genes involved in mitochondrial β-oxidation, peroxisomal β-oxidation and CoA synthesis in Pex16 KO and wild-type mice fed an LPD (Fig. S7g). Gene expression of lipogenesis markers Acaca and Fasn were downregulated in fenofibrate-treated mice but this response to treatment was absent in Pex16 KO mice fed a control diet (Fig. S7h).
Mitochondrial protein complexes isolated from mitochondria were increased by fenofibrate in wild-type mice fed an LPD compared to untreated mice, although the effect appeared less pronounced in the Pex16 KOs (Fig. 7f). No substantial effects of fenofibrate treatment on mitochondrial complex proteins were observed in the Pex16 KOs fed a control diet, with a modest increase in complex IV and a small decrease in complex II and V (Fig. S7i). This data suggests that PPARα activation improves mitochondrial function in Pex16 KOs fed an LPD through an increase in mitochondrial complex content without an overall effect on mitochondrial mass.
In line with these results, the ATP content was similarly increased in Pex16 KOs fed an LPD after fenofibrate treatment compared to wild types (Fig. 7g). Finally, mitochondria showed improved respiration in the Pex16 KOs with or without LPD feeding (Fig. 7h, Fig. S7j). These results indicate that PPARα activation improves mitochondrial fitness in mice subjected to LPD-induced nutritional stress despite an absence of functional peroxisomes.
PPARα activation stimulates mitochondrial biogenesis and autophagy in Pex16 KO mice
We next assessed the effects of PPARα activation on markers of mitochondrial dynamics, biogenesis, and autophagy. Both MFN2 protein level and pDRP1/DRP1 ratio were increased in the fenofibrate-treated Pex16 KOs fed a control diet or an LPD compared to untreated animals (Fig. 8a, Fig. S8a). Both Pex16 KOs and the wild-type animals fed an LPD and treated with fenofibrate showed an increase in autophagosome formation (ratio LC3B-II/I) and a decrease in p62 (Fig. 8b), which suggests an increase in autophagic flux. Similar changes in the autophagic factors were also observed in fenofibrate-treated Pex16 KO mice fed the control diet (Fig. S8b).
Fenofibrate (FF) treatment stimulates mitochondrial biogenesis and autophagy in Pex16 KO and wild-type mice fed an LPD. a Western blots of hepatic MFN2, DR1, pDRP1, and quantification of band intensity relative to β-actin and TOM20, and the ratio of pDRP1/DRP1. b Western blot of hepatic autophagy and mitophagy markers p62, LC3B and PINK1 in liver lysates, and PINK1 in isolated hepatic mitochondria, quantification of band intensity relative to β-actin and COX IV. c Western blot of autophagy marker p-p70S6K and p70S6K, and quantification of band intensity relative to β-actin and the ratio of p-p70S6K/p70S6K. d Western blot of autophagy marker p4EBP1, 4EBP1, and quantification of band intensity relative to β-actin and the ratio of p4EBP1/4EBP1. e Western blot of mitochondrial biogenesis marker PGC-1α and NRF1, and quantification of band intensity relative to β-actin. Data are represented as mean ± SEM, n = 6–8 mice per group. *p < 0.05, **p < 0.01, ***p < 0.001. FF fenofibrate. See also Fig. S8
The effects on autophagy were associated with a decrease in p-p70S6K/p70S6K indicating inhibition of mTOR (Fig. 8c). However, fenofibrate treatment increased p-4EBP1/4EBP1 in all mouse groups (Fig. 8d). mRNA levels of these factors showed a similar pattern (Fig. S8d). PGC-1α and NRF1 were upregulated in Pex16 KOs as well as wild-type animals after fenofibrate treatment (Fig. 8e, Fig. S8e). Together, these data suggest that the improved mitochondrial health observed after PPARα activation in Pex16 KOs fed an LPD is related to increased mitochondrial biogenesis and enhanced autophagy.
Discussion
In this study, we describe the metabolic adaptation that occurs in a peroxisome-deficient hepatocytes model. The knockout of the peroxisomal biogenesis gene PEX16, which results in the loss of peroxisomal membrane, causes changes in mitochondria morphology and function. However, the hepatocytes of the Pex16 KO mice still produced ATP at levels comparable to wild-type animals (Fig. 3e), suggesting the observed mitochondria changes may act as a compensatory response in livers with impaired peroxisomal function. Functionally, mitochondria from Pex16 KO hepatocytes have increased reserve respiration capacity. Further, the hepatocytes showed upregulation in mitochondrial biogenesis and reduction in autophagy. However, Pex16 KO mice were less able to adapt to metabolic stress induced through LPD feeding. However, activating PPARα improved mitochondrial and overall metabolic function in the absence of functional peroxisomes in hepatocytes by stimulating mitochondrial biogenesis and autophagy. Our work not only provides further support to the importance of peroxisomes in maintaining mitochondria health, but it also reveals a certain adaptive capacity of hepatocytes to maintain mitochondrial function without functional peroxisomes.
Loss of PEX16 and peroxisomes affects mitochondrial homeostasis and function
PEX16 is essential for peroxisome biogenesis as its absence results in the loss of peroxisomal membrane [37] and is associated with Zellweger syndrome [7] and leukodystrophy [38]. We developed a liver-specific Pex16 KO mouse model as demonstrated by the loss of PEX16 and the peroxisome membrane protein, PMP70. These mice developed hepatic steatosis similar to other models of loss of peroxisome function, such as in PEX2 and PEX5 deficiency [12,13,14]. Mitochondrial abnormalities are found in patients with Zellweger syndrome [39] and mouse models with deletion of Pex2 and Pex5 [13, 14, 40], underscoring the role of peroxisomes in mitochondrial health. In this study, we showed that loss of Pex16 leads to the accumulation of small mitochondria, but with only minor functional disruption (Fig. 3). Mitochondria morphological changes without ATP disruption were also reported in Pex5 KO mice at 10 weeks of age [13] compared to our mice sacrificed at 5 weeks of age. The more severe hepatic mitochondrial changes found early in life were in whole body Pex2 and Pex5 KO models. It should be noted that these mice have very different peroxisome content. The Pex16 KO livers are absent of any peroxisomal structures owing to the role of PEX16 in peroxisome membrane formation. At the same time, the KO of Pex5 in hepatocytes leads to fewer but larger peroxisomal structures than in wild-type hepatocytes. The peroxisomal membrane proteins may have functions that affect mitochondria. Finally, the more severe phenotype in the whole body Pex2 and Pex5 KO models could have been related to extrahepatic pathophysiological effects secondarily affecting the liver [12, 14].
Interestingly, the stress test of mitochondria isolated from the Pex16 KO showed a rapid loss of oxygen consumption rate upon treatment with FCCP. This decrease is unlikely due to a limitation of FCCP as the concentration of mitochondria used was the same as the wild-type mitochondria, which did not show this loss in its OCR upon FCCP treatment (Fig. 4g and Fig S7j). However, we cannot rule out a possible issue with substrate concentration. Further, we observed an accumulation in cleaved PINK1 in the Pex16 KO liver, but the cause of this accumulation is not known. Given that PINK1 is readily cleaved by the inner membrane protease PARL, and retro-translocated out to the cytosol for degradation by the proteasome [31], an accumulation of the cleaved form of PINK1 may indicate a mild defect in the mitochondria. Finally, we also found that the Pex16 KO mice fed an LPD were more susceptible to mitochondrial damage compared to the wild-type mice. Therefore, we propose that the Pex16 KO mitochondria are more susceptible to metabolic stress-induced damage.
Peroxisomes have been proposed to act in regulating mitochondria redox homeostasis [2]. Smaller or fragmented mitochondria have decreased potential [8]. The smaller mitochondria observed in the Pex16 KO mice are likely a compensation mechanism to prevent mitochondria-induced oxidative stress in the absence of peroxisomes. In fact, similar fragmentation in mitochondria is observed during hypoxia where DRP1 is activated to promote mitochondria fragmentation [41]. However, since a decreased potential is correlated with decreased ATP production, the Pex16 KO mice appear to compensate by increasing the amount of mitochondria (Fig. 4). Finally, we observed that the Pex16 KO mice have a higher maximum respiration capacity compared to wild-type mice and higher levels of ETC proteins, yet they do not produce more ATP. This suggests the Pex16 KO hepatocyte mitochondria function is downregulated to minimize the production of ROS. Therefore, we propose that hepatocyte mitochondrial function in the Pex16 KO mice may be maintained through a compensatory response. However, these changes make their mitochondria more susceptible to damage during metabolic stress.
The loss of PEX16 did lead to mild hepatic steatosis, even on a control diet, despite having preserved mitochondrial function. Hepatic steatosis can be caused by increased hepatic lipogenesis or extrahepatic lipolysis, with the latter leading to a higher flux of fatty acids to the liver. Alternatively, decreased β-oxidation or very-low-density lipoprotein secretion could contribute to hepatic lipid accumulation. Lipogenesis was unlikely to be responsible, given the reduced expression of lipogenic genes in the Pex16 KO compared to wild-type mice. Increased lipolysis through enhanced lipoprotein or hormone-sensitive lipase activity was also less likely, given that this was a liver-specific KO model. Limited data suggest that very-low-density lipoprotein secretion is not altered in patients with peroxisome biogenesis disorders [42]. Although expression of genes encoding for enzymes controlling β-oxidation was increased in Pex16 KO mice, we cannot exclude reduced β-oxidation in these mice. As mitochondrial function was decreased in the Pex16 KO mice fed an LPD, this is likely the most critical contributor to the exacerbated steatosis in these mice.
Decreased autophagy and induction of mitochondria biogenesis contribute to increased mitochondrial mass in Pex16 KO mice
Our studies suggest that the increase in mitochondrial mass might be caused by two distinct mechanisms. First, the activation of AMPK by functional loss of peroxisome (Fig. 3) could induce mitochondrial biogenesis via the AMPK-dependent activation of PGC-1α [43]. The increase in activated pAMPK also inhibits mTORC1 and activates autophagy [44]. We found pAMPK to be increased in livers of Pex16 KO, a result similar to that described in liver-specific PEX5 deficiency [45]. Second, our autophagy assays suggest that hepatocyte autophagic flux is decreased in Pex16 KO mice. We speculate that this increased mitochondrial biogenesis and reduced autophagic flux are compensatory mechanisms that preserve mitochondrial function by increasing mitochondrial mass. Down-regulating β-oxidation could also prevent further mitochondrial damage by limiting the production of ROS.
Loss of functional peroxisomes exacerbates low protein diet induced mitochondrial changes through separate pathways
Nutritional stress induced by LPD in weanlings exacerbated the hepatic steatosis caused by Pex16 deficiency. Interestingly, cold-induced stress also exacerbates mitochondrial damage in brown adipocytes in adipose-specific Pex16 KO mice [46]. The small mitochondria seen in the livers of the Pex16 KO mice were not described in those of the Pex5 deficient model [12], and this might be related to potential differential effects on mitochondrial homeostasis where PEX16 is involved in the early steps of peroxisome biogenesis, whereas PEX5 imports peroxisomal proteins. Interestingly, LPD feeding led to a fewer and smaller mitochondria compared to normal chow diet animals, associated with signs of reduced biogenesis and despite possibly lower autophagy activation. These findings are contrary to those described in a rat model of LPD-induced loss of functional peroxisomes and hepatic mitochondrial dysfunction where an accumulation of damaged mitochondria was observed with LPD feeding [17]. This discrepancy might relate to differences in severity of the two model systems, or a temporal difference as the rats were placed on an LPD for 4 weeks instead of the 2 weeks in the mice.
Fibrates improve mitochondrial function in Pex16 KO low protein fed mice
PPARα induces peroxisome and mitochondrial biogenesis, increases β-oxidation, and has recently been shown to induce autophagy [36]. Previously, we demonstrated that activation of PPARα could reverse peroxisome loss and improve mitochondria fitness in the hepatocytes of rats subjected to LPD [17]. Here, we examined whether peroxisomes are necessary for the possible rescue of mitochondrial function by PPARα induction. Fibrate treatment up-regulated genes involved in peroxisome biogenesis (Fig. 7a) and peroxisomal and mitochondrial β-oxidation, and this was associated with reduced lipids in both LPD-fed Pex16 KOs and wild-type mice, suggesting a peroxisome-independent effect. Fibrates also increased mitochondrial biogenesis, which is consistent with recent observations [47]. Fibrate treatment also led to an induction of autophagy as indicated by decreased mTORC1 activity, increased LC3B-II, and decreased p62. Activation of PPARα has been shown to up-regulate autophagy transcriptionally [36], and this could account, in part, for the increased autophagy in our model. Of interest, fibrate treatment has been piloted in patients with Zellweger Syndrome but without clear clinical impact [48, 49]. The lack of effect in patients could be related to the possibility that the disease state was already too advanced for these patients with Zellweger syndrome to benefit from fibrate treatment, or it is also possible that mitochondrial damage may not be the sole pathology of Zellweger syndrome.
In conclusion, this study describes the phenotypical consequences of hepatic loss of Pex16, which are exacerbated by nutritional stress induced by LPD. We demonstrated that Pex16 deficiency leads to enhanced mitochondrial biogenesis and reduced autophagy resulting in an increase in mitochondrial mass (Fig. 9), in order to compensate for the loss of the peroxisomal protective role. These mitochondria do maintain their respiration potential but are more susceptible to metabolic stress-induced damage. PPARα treatment improved mitochondrial health by overcoming the reduction in autophagy flux caused by the loss of PEX16 and by enhancing mitochondrial biogenesis. This improvement is independent of the peroxisome function. Our finding that mitochondrial dysfunction can be uncoupled from peroxisome loss is of relevance for children with peroxisomal biogenesis disorders and suggest that interventions to restore mitochondrial homeostasis and preventing additional stressors such as malnutrition could improve their outcome.
Proposed mechanistic model by which Pex16 loss and LPD lead to damaged mitochondria and reduced autophagy. Loss of hepatic Pex16 leads to fragmented mitochondria. Feeding a low protein diet leads to elongated mitochondria. A low protein diet in mice with peroxisomal defects impacts mitochondrial biogenesis and autophagy. Activation of the PPARα pathway restores mitochondrial biogenesis and hepatic mitochondrial function in low protein-fed mice, even in the absence of functional peroxisomes
Data availability
The datasets generated during and/or analyzed during this study are available in the BioStudies repository, https://www.ebi.ac.uk/biostudies, accession number S-BSST100.
Abbreviations
- Acaa1a:
-
Acetyl-coenzyme A carboxylase carboxyl transferase subunit α
- Acaca:
-
Acetyl-CoA carboxylase α
- Acadm1:
-
Acyl-CoA dehydrogenase medium chain 1
- Acadv1:
-
Very long-chain specific acyl-CoA dehydrogenase 1
- Acadl:
-
Acyl-CoA dehydrogenase long chain
- Acca2:
-
Acetyl-CoA carboxylase α subunit
- Acot12:
-
Acyl-CoA thioesterase 12
- Acox1:
-
Peroxisomal acyl-coenzyme A oxidase 1
- Acsl1:
-
Acyl-CoA synthetase long chain family member 1
- ADP:
-
Adenosine diphosphate
- AMP:
-
Adenosine monophosphate
- AMPK:
-
AMP-activated protein kinase
- ATP:
-
Adenosine triphosphate
- BSA:
-
Bovine serum albumin
- CCM:
-
Central carbon metabolism
- CQ:
-
Chloroquine
- COX IV:
-
Cytochrome c oxidase subunit 4
- DAPI:
-
4′,6-Diamidino-2-phenylindole
- DRP1:
-
Dynamin-1-like protein
- DMSO:
-
Dimethyl sulfoxide
- Ech1:
-
Enoyl-CoA hydratase 1fabp1
- ER:
-
Endoplasmic reticulum
- ESCs:
-
Mouse embryonic stem cells
- ETC:
-
Electron transport chain
- 4EBP1:
-
Eukaryotic translation initiation factor 4E binding protein 1
- Fabp1:
-
Fatty acid binding protein 1
- Fasn:
-
Fatty acid synthase
- FCCP:
-
Carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone
- FF:
-
Fenofibrate
- GMP:
-
Guanosine monophosphate
- GDP:
-
Guanosine diphosphate
- GTP:
-
Guanosine triphosphate
- H&E:
-
Haemotoxylin and eosin
- HSP60:
-
Heat shock protein 60
- IP:
-
Intraperitoneal
- KO:
-
Knockout
- LC3B:
-
Microtubule-associated protein light chain 3B
- LPD:
-
Low protein diet
- MFN2:
-
Mitofusin-2
- mtDNA:
-
Mitochondrial DNA
- NAD:
-
Nicotinamide adenine dinucleotide
- NADH:
-
Nicotinamide adenine dinucleotide hydride
- NADPH:
-
Nicotinamide adenine dinucleotide phosphate
- NRF1:
-
Nuclear respiratory factor 1
- OCR:
-
Oxygen consumption rat
- PBS:
-
Phosphate-buffered saline
- PCA:
-
Perchloric acid
- PEP:
-
Prolyl endopeptidase
- PINK1:
-
PTEN induced kinase 1
- PMP70:
-
Peroxisomal membrane protein 70
- p62/SQSTM1:
-
Autophagy adaptor protein sequestosome 1
- p70S6K:
-
Ribosomal protein S6 kinase β-1
- PEX:
-
Peroxin
- PPARGC1A:
-
Peroxisome-proliferator-activated receptor coactivator-1α
- PPARα:
-
Peroxisome proliferator activated receptor α
- Rpl13a:
-
Ribosomal protein L13a
- TBS:
-
Tris-buffered saline
- TCA:
-
Tricarboxylic acid
- TEM:
-
Transmission electron microscopy
- TOM20:
-
Membrane translocase 20
- UMP:
-
Uridine monophosphate
- UDP:
-
Uridine diphosphate
- UTP:
-
Uridine triphosphate
References
Farré J, Mahalingam SS, Proietto M, Subramani S (2019) Peroxisome biogenesis, membrane contact sites, and quality control. EMBO Rep 20:e46864. https://doi.org/10.15252/embr.201846864
Fransen M, Lismont C, Walton P (2017) The peroxisome-mitochondria connection: how and why? Int J Mol Sci 18:1126. https://doi.org/10.3390/ijms18061126
Mooi WJ, Dingemans KP, Van Den Bergh Weerman MA et al (1983) Ultrastructure of the liver in the cerebrohepatorenal syndrome of Zellweger. Ultrastruct Pathol 5:135–144. https://doi.org/10.3109/01913128309141833
Trijbels JMF, Berden JA, Monnens LAH et al (1983) Biochemical studies in the liver and muscle of patients with Zellweger syndrome. Pediatr Res 17:514–517. https://doi.org/10.1203/00006450-198306000-00018
Germain K, Kim PK (2020) Pexophagy: a model for selective autophagy. Int J Mol Sci 21:578. https://doi.org/10.3390/ijms21020578
Soliman K, Göttfert F, Rosewich H et al (2018) Super-resolution imaging reveals the sub-diffraction phenotype of Zellweger Syndrome ghosts and wild-type peroxisomes. Sci Rep 8:7809. https://doi.org/10.1038/S41598-018-24119-2
Honsho M, Tamura S, Shimozawa N et al (1998) Mutation in PEX16 is causal in the peroxisome-deficient Zellweger syndrome of complementation group D. Am J Hum Genet 63:1622–1630. https://doi.org/10.1086/302161
Chan DC (2020) Mitochondrial dynamics and its involvement in disease. Annu Rev Pathol Mech Dis 15:235–259. https://doi.org/10.1146/annurev-pathmechdis-012419-032711
Ploumi C, Daskalaki I, Tavernarakis N (2017) Mitochondrial biogenesis and clearance: a balancing act. FEBS J 284:183–195. https://doi.org/10.1111/febs.13820
Kim I, Rodriguez-Enriquez S, Lemasters JJ (2007) Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys 264:245–253. https://doi.org/10.1016/j.abb.2007.03.034
Peeters A, Shinde AB, Dirkx R et al (2015) Mitochondria in peroxisome-deficient hepatocytes exhibit impaired respiration, depleted DNA, and PGC-1α independent proliferation. Biochim Biophys Acta Mol Cell Res 1853:285–298. https://doi.org/10.1016/j.bbamcr.2014.11.017
Baumgart E, Vanhorebeek I, Grabenbauer M et al (2001) Mitochondrial alterations caused by defective peroxisomal biogenesis in a mouse model for Zellweger syndrome (PEX5 knockout mouse). Am J Pathol 159:1477–1494. https://doi.org/10.1016/S0002-9440(10)62534-5
Dirkx R, Vanhorebeek I, Martens K et al (2005) Absence of peroxisomes in mouse hepatocytes causes mitochondrial and ER abnormalities. Hepatology 41:868–878. https://doi.org/10.1002/hep.20628
Keane MH, Overmars H, Wikander TM et al (2007) Bile acid treatment alters hepatic disease and bile acid transport in peroxisome-deficient PEX2 Zellweger mice. Hepatology 45:982–997. https://doi.org/10.1002/hep.21532
Maxwell M, Bjorkman J, Nguyen T et al (2003) Pex13 inactivation in the mouse disrupts peroxisome biogenesis and leads to a Zellweger syndrome phenotype. Mol Cell Biol 23:5947–5957. https://doi.org/10.1128/mcb.23.16.5947-5957.2003
Sargent G, van Zutphen T, Shatseva T et al (2016) PEX2 is the E3 ubiquitin ligase required for pexophagy during starvation. J Cell Biol 214:677–690. https://doi.org/10.1083/jcb.201511034
van Zutphen T, Ciapaite J, Bloks VW et al (2016) Malnutrition-associated liver steatosis and ATP depletion is caused by peroxisomal and mitochondrial dysfunction. J Hepatol 65:1198–1208. https://doi.org/10.1016/j.jhep.2016.05.046
Klionsky DJ, Abdelmohsen K, Abe A et al (2016) Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12:1–222. https://doi.org/10.1080/15548627.2015.1100356
Iuso A, Repp B, Biagosch C et al (2017) Assessing mitochondrial bioenergetics in isolated mitochondria from various mouse tissues using Seahorse XF96 Analyzer. Methods Mol Biol 1567:217–230. https://doi.org/10.1007/978-1-4939-6824-4_13
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. https://doi.org/10.1038/NMETH.2089
(2013) R: a language and environment for statistical computing
Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J (2011) Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci USA 108:10190–10195. https://doi.org/10.1073/PNAS.1107402108
Gomes LC, Di BG, Scorrano L (2011) During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol 13:589–598. https://doi.org/10.1038/NCB2220
Tondera D, Grandemange S, Jourdain A et al (2009) SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J 28:1589–1600. https://doi.org/10.1038/EMBOJ.2009.89
Galloway CA, Lee H, Yoon Y (2012) Mitochondrial morphology-emerging role in bioenergetics. Free Radic Biol Med 53:2218–2228. https://doi.org/10.1016/J.FREERADBIOMED.2012.09.035
Schlaak RA, Frei A, SenthilKumar G et al (2020) Differences in expression of mitochondrial complexes due to genetic variants may alter sensitivity to radiation-induced cardiac dysfunction. Front Cardiovasc Med 7:23. https://doi.org/10.3389/FCVM.2020.00023
Deas E, Plun-Favreau H, Gandhi S et al (2011) PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum Mol Genet 20:867–879. https://doi.org/10.1093/hmg/ddq526
Matsuda N, Sato S, Shiba K et al (2010) PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol 189:211–221. https://doi.org/10.1083/jcb.200910140
Takatori S, Ito G, Iwatsubo T (2008) Cytoplasmic localization and proteasomal degradation of N-terminally cleaved form of PINK1. Neurosci Lett 430:13–17. https://doi.org/10.1016/J.NEULET.2007.10.019
Sim CH, Gabriel K, Mills RD et al (2012) Analysis of the regulatory and catalytic domains of PTEN-induced kinase-1 (PINK1). Hum Mutat 33:1408–1422. https://doi.org/10.1002/HUMU.22127
Sekine S, Youle RJ (2018) PINK1 import regulation; a fine system to convey mitochondrial stress to the cytosol. BMC Biol 16:2. https://doi.org/10.1186/S12915-017-0470-7
Klionsky DJ, Abdel-Aziz AK, Abdelfatah S et al (2021) Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy 17:1–382. https://doi.org/10.1080/15548627.2020.1797280
Russell RC, Yuan H-X, Guan K-L (2014) Autophagy regulation by nutrient signaling. Cell Res 24:42–57. https://doi.org/10.1038/cr.2013.166
Toshniwal AG, Gupta S, Mandal L, Mandal S (2019) ROS inhibits cell growth by regulating 4EBP and S6K, independent of TOR, during development. Dev Cell 49:473-489.e9. https://doi.org/10.1016/J.DEVCEL.2019.04.008
Jornayvaz FR, Shulman GI (2010) Regulation of mitochondrial biogenesis. Essays Biochem 67:69–84. https://doi.org/10.1042/BSE0470069
Lee JM, Wagner M, Xiao R et al (2014) Nutrient-sensing nuclear receptors coordinate autophagy. Nature 516:112–115. https://doi.org/10.1038/nature13961
Honsho M, Hiroshige T, Fujiki Y (2002) The membrane biogenesis peroxin Pex16p: topogenesis and functional roles in peroxisomal membrane assembly. J Biol Chem 277:44513–44524. https://doi.org/10.1074/jbc.M206139200
Ebberink MS, Csanyi B, Chong WK et al (2010) Identification of an unusual variant peroxisome biogenesis disorder caused by mutations in the PEX16 gene. J Med Genet 47:608–615. https://doi.org/10.1136/jmg.2009.074302
Goldfischer S, Moore CL, Johnson AB et al (1973) Peroxisomal and mitochondrial defects in the cerebro-hepato-renal syndrome. Science 182:62–64
Baes M, Gressens P, Baumgart E et al (1997) A mouse model for Zellweger syndrome. Nat Genet 17:49–57. https://doi.org/10.1038/ng0997-49
Kim H, Scimia MC, Wilkinson D et al (2011) Fine-tuning of Drp1/Fis1 availability by AKAP121/Siah2 regulates mitochondrial adaptation to hypoxia. Mol Cell 44:532–544. https://doi.org/10.1016/J.MOLCEL.2011.08.045
van der Hoek YY, Wanders RJ, van den Ende AE et al (1997) Lipoprotein[a] is not present in the plasma of patients with some peroxisomal disorders. J Lipid Res 38:1612–1619. https://doi.org/10.1016/S0022-2275(20)37179-0
Zong H, Ren JM, Young LH et al (2002) AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proc Natl Acad Sci USA 99:15983–15987. https://doi.org/10.1073/pnas.252625599
Mihaylova MM, Shaw RJ (2011) The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol 13:1016–1023. https://doi.org/10.1038/ncb2329
Peeters A, Fraisl P, Van Den Berg S et al (2011) Carbohydrate metabolism is perturbed in peroxisome-deficient hepatocytes due to mitochondrial dysfunction, AMP-activated protein kinase (AMPK) activation, and peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) suppression. J Biol Chem 286:42162–42179. https://doi.org/10.1074/jbc.M111.299727
Park H, He A, Tan M et al (2019) Peroxisome-derived lipids regulate adipose thermogenesis by mediating cold-induced mitochondrial fission. J Clin Investig 129:694–711. https://doi.org/10.1172/JCI120606
Iershov A, Nemazanyy I, Alkhoury C et al (2019) The class 3 PI3K coordinates autophagy and mitochondrial lipid catabolism by controlling nuclear receptor PPARα. Nat Commun 10:1566. https://doi.org/10.1038/s41467-019-09598-9
Lazarow PB, Black V, Shio H et al (1985) Zellweger syndrome: biochemical and morphological studies on two patients treated with clofibrate. Pediatr Res 19:1356–1364. https://doi.org/10.1203/00006450-198512000-00030
Björkhem I, Blomstrand S, Glaumann H, Strandvik B (1985) Unsuccessful attempts to induce peroxisomes in two cases of Zellweger disease by treatment with clofibrate. Pediatr Res 19:590–593. https://doi.org/10.1203/00006450-198506000-00018
Acknowledgements
We would like to acknowledge Prof Barbara Bakker for the excellent technical support towards the mitochondrial function assays. We thank Michelle Huong (Department of Human Biology, University of Toronto) for help with genotyping, Dr. Abdalla Ahmed (The Hospital for Sick Children) for feedback and suggestions, Ms. Celine Bourdon (The Hospital for Sick Children) for feedback and editing, Dr. Dave Schibli and Dr. Jun Han (UVic Genome BC Proteomic Centre and The Metabolomics Innovation Centre) for conducting the CCM assay, Paul Paroutis and Kimberly Lau (Imaging Facility at The Hospital for Sick Children) for help with confocal microscopy and LAS (Lab Animal Services, The Hospital for Sick Children) for mouse husbandry and care.
Funding
This work was supported by Canadian Institutes of Health Research (Grant reference number 156307).
Author information
Authors and Affiliations
Contributions
LC and DL designed and performed the experiments and wrote the manuscript. SL generated and initially characterized the Albumin-Cre Pex16 KO mouse model and edited the manuscript. GH analyzed the central carbon metabolism assay, helped interpret this data and edited the manuscript. BW measured mitochondrial area from TEM images and edited the manuscript. PD provided expertise on interpretation of the mitochondrial data and edited the manuscript. RL and JB were responsible for TEM experiments with JB also providing input in interpretation of the autophagy data and both edited the manuscript. PK and RB conceived the project, designed the experiments, and wrote and edited the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval and consent to participate
Procedures followed the Canadian Council for Animal Care guidelines and were approved by the Animal Care Committee at the Hospital for Sick Children.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chi, L., Lee, D., Leung, S. et al. Loss of functional peroxisomes leads to increased mitochondrial biogenesis and reduced autophagy that preserve mitochondrial function. Cell. Mol. Life Sci. 80, 183 (2023). https://doi.org/10.1007/s00018-023-04827-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-023-04827-3