Abstract
Repair-supportive mesenchymal cells (RSMCs) have been recently reported in the context of naphthalene (NA)-induced airway injury and regeneration. These cells transiently express smooth muscle actin (Acta2) and are enriched with platelet-derived growth factor receptor alpha (Pdgfra) and fibroblast growth factor 10 (Fgf10) expression. Genetic deletion of Ctnnb1 (gene coding for beta catenin) or Fgf10 in these cells using the Acta2-Cre-ERT2 driver line after injury (defined as NA-Tam condition; Tam refers to tamoxifen) led to impaired repair of the airway epithelium. In this study, we demonstrate that RSMCs are mostly captured using the Acta2-Cre-ERT2 driver when labeling occurs after (NA-Tam condition) rather than before injury (Tam-NA condition), and that their expansion occurs mostly between days 3 and 7 following NA treatment. Previous studies have shown that lineage-traced peribronchial GLI1+ cells are transiently amplified after NA injury. Here, we report that Gli1 expression is enriched in RSMCs. Using lineage tracing with Gli1Cre−ERT2 mice combined with genetic inactivation of Fgf10, we show that GLI1+ cells with Fgf10 deletion fail to amplify around the injured airways, thus resulting in impaired airway epithelial repair. Interestingly, Fgf10 expression is not upregulated in GLI1+ cells following NA treatment, suggesting that epithelial repair is mostly due to the increased number of Fgf10-expressing GLI1+ cells. Co-culture of SCGB1A1+ cells with GLI1+ cells isolated from non-injured or injured lungs showed that GLI1+ cells from these two conditions are similarly capable of supporting bronchiolar organoid (or bronchiolosphere) formation. Single-cell RNA sequencing on sorted lineage-labeled cells showed that the RSMC signature resembles that of alveolar fibroblasts. Altogether, our study provides strong evidence for the involvement of mesenchymal progenitors in airway epithelial regeneration and highlights the critical role played by Fgf10-expressing GLI1+ cells in this context.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The bronchi of the lung represent important interfaces with the outside environment and are as such exposed to many toxins and pathogens. Therefore, the bronchial epithelium undergoes a process of injury repair. The relevant bronchial epithelial cells for the repair process in the bronchi are the non-ciliated secretory cells or club cells [1, 2]. These cells express secretoglobin family 1A member 1 (SCGB1A1, a.k.a. Clara cell 10 kDa secretory protein CC10 or CCSP), and they represent progenitors for both club and ciliated cells [3, 4]. A large subset of club cells expresses high levels of cytochrome P450 family 2 subfamily f polypeptide 2 (CYP2F2), which is involved in the transformation of naphthalene (NA) into trans-1R-hydroxy-2R-glutathionyl-1,2-dihydronaphthalene, a toxic intermediate that leads to the depletion of club cells in the lung [5,6,7]. In addition, club cells expressing low levels of CYP2F2, called variant club cells, escape the deleterious effect of NA and represent a pool of progenitors. These cells are present at proximity of neuroepithelial bodies (NEBs) and bronchioalveolar duct junctions (BADJs). These cells are critical for the repair process [8,9,10]. The NA injury model is therefore useful to investigate airway regeneration.
Epithelial–mesenchymal interactions orchestrated by fibroblast growth factors, which have been described in the embryonic lung [11], are also at play after injury in the adult lung. It has been proposed that following NA injury in adult mice, WNT7b expressed by surviving ciliated cells acts on airway smooth muscle cells (ASMCs) leading to the increased expression of Fgf10 [12]. FGF10 binds to and activates FGFR2b expressed in variant club cells to control their amplification [12]. Leucine-rich repeat-containing G-protein coupled receptor 6 (Lgr6)-positive cells represent an ASMC subpopulation that enhances repair of the bronchial epithelium following NA administration and also involves a WNT-FGF10 positive feedback loop [13]. Therefore, there is a consensus in the field that ASMCs are a mesenchymal niche for variant club cells following injury.
In a recently published paper, we showed that during lung repair after NA injury, a mesenchymal population of ACTA2+ PDGFRα+ cells emerges in the non-cartilaginous conducting airway niche, which is normally populated by ASMCs [14]. This cell population, which we termed “repair-supportive mesenchymal cells” (RSMCs), is distinct from conventional ASMCs, which have previously been proposed to contribute to epithelial repair [14]. Gene expression analysis on sorted lineage-labeled cells showed that RSMCs express low levels of ASMC marker genes, but high levels of the pro-regenerative marker Fgf10 [14]. Organoid co-cultures demonstrated an enhanced ability for ACTA2+ cell-derived RSMCs in supporting club cell growth in vitro [14, 15].
In this study, we carried out lineage tracing of ACTA2+ cells captured before (defined as Tam-NA condition) or after NA injury (defined as NA-Tam condition). We also analyzed the contribution of FGF10 derived from the GLI1+ lineage to the repair process. Bronchiolosphere cultures using GLI1+ mesenchymal cells isolated from non-injured or injured lungs co-cultured with SCGB1A1+ cells were also carried out. Our study demonstrates the critical role played by Fgf10-expressing GLI1+ cells in the context of repair of the bronchial epithelium.
Materials and methods
Mice, tamoxifen administration and naphthalene treatment
Acta2-Cre-ERT2 mice (STOCK Tg(Acta2-cre/ERT2)12Pcn) [16] were kindly provided by Dr. Pierre Chambon (University of Strasbourg, France). Fgf10flox mice (B6;129-Fgf10tm1.2Sms /J) [17, 18] were a generous gift from Dr. Suzanne L. Mansour (University of Utah, USA). tm9 tdTomatoflox (hereafter referred to as tdTomatoflox) (B6;129S6-Gt(ROSA)26Sortm9(CAG−tdTomato)Hze /J, JAX stock number 007905), tm14 tdTomatoflox (B6.Cg-Gt(ROSA)26Sortm14(CAG−tdTomato)Hze /J, JAX stock number 007914), Scgb1a1Cre−ERT (B6N.129S6(Cg)-Scgb1a1tm1(cre/ERT)Blh /J, JAX stock number 016225) and Gli1Cre−ERT2 (STOCK Gli1tm3(cre/ERT2)Alj /J, JAX stock number 007913) were purchased from the Jackson laboratory. The tm14 tdTomatoflox line was used in combination with the Scgb1a1Cre−ERT line, while the tm9 tdTomatoflox line was used in combination with the Acta2-Cre-ERT2, Gli1Cre−ERT2 and Gli1Cre−ERT2; Fgf10flox lines. To induce Cre-ERT2-mediated genetic recombination, Acta2-Cre-ERT2 and Gli1Cre−ERT2 mice were fed tamoxifen-containing food (0.4 g of tamoxifen per kg of food) (Altromin, Germany). Tamoxifen powder was purchased from Sigma-Aldrich. For naphthalene treatment, 8–12-week-old female mice were used. Naphthalene (Sigma) was dissolved in corn oil (20 mg/mL) and was administered intraperitoneally at a dose of 0,275 mg naphthalene per g of body weight.
Immunofluorescence
Lungs were flushed by transcardiac perfusion with PBS and then, either fixed with 4% paraformaldehyde for paraffin embedding and 5 μm sectioning using a microtome or inflated with 1.7% low melting agarose for 200 μm sectioning using a vibratome. Immunofluorescence (IF) was performed using monoclonal anti-ACTA2 (Sigma, 1:200), monoclonal anti-SCGB1A1 (Santa Cruz Biotechnology, 1:200), polyclonal anti-KI67 (Thermo Fisher Scientific, 1:200) and polyclonal anti-RFP antibodies (Thermo Fisher Scientific, 1:200; used only when combined with KI67 staining which requires antigen retrieval). Nuclei were stained using DAPI (Life Technologies). Tile scans of thin sections were acquired using EVOS FL2 Imaging System (Thermo Fischer Scientific). Other fluorescent images were acquired using a Leica DM550 B fluorescence microscope, an SP5 or SP8 confocal microscope, both equipped with a white-light laser and hybrid detectors (Leica Microsystems). Where applicable, three-dimensional (3D) reconstruction of z-stacks was performed using LAS X software (version 3.5; Leica Microsystems). For quantification of stained control and experimental lungs, sections from at least 3 independent lungs were analyzed and multiple images were used (n > 8).
Flow cytometry analysis and cell sorting
Flow cytometry and cell sorting were carried out as we previously described [14]. In brief, lungs were isolated in Hank’s Balanced Salt Solution (HBSS, Gibco), minced into small pieces and incubated with 0.5% collagenase type IV in HBSS (Life Technologies) at 37 °C for 45 min. Lung homogenates were then passed through 70 and 40 μm cell strainers (BD Biosciences) to obtain single-cell suspensions. Cells were centrifuged at 4 °C at 1000 rpm for 5 min and then resuspended in MACS buffer and stained with anti-PDGFRα (APC-conjugated, 1:100), anti-EpCAM (APC-Cy7-conjugated, 1:100), anti-CD31 (Pacific Blue-conjugated, 1:100) and anti-CD45 (Pacific Blue-conjugated, 1:100) antibodies (all from Biolegend) for 20 min on ice in the dark. Then, cells were washed with MACS buffer. Flow cytometry and cell sorting were done using FACSAria III cell sorter (BD Biosciences).
Quantitative real-time PCR
For bulk RNA extraction from lungs, accessory lobes were lysed and subjected to RNeasy Mini kit (Qiagen) as we previously described [14]. Extraction of RNA from sorted cells was performed using RNeasy Micro kit (Qiagen). Primers were designed using the Universal Probe Library Assay Design center program (Roche Applied Science), and quantitative real-time PCR (qPCR) was performed using Light Cycler 480 II (Roche Applied Science). Hypoxantine-guanine phosphoribosyltransferase (Hprt) was used as a reference gene.
Single-cell RNA sequencing
Single-cell RNA-seq of tdTomato+ (hereafter referred to as tdTom+) cells isolated from Acta2-Cre-ERT2; tdTomatoflox lungs was performed using the ICELL8 platform (Takara Bio) using the V2 protocol with in-chip preamplification. Samples were presorted into HBSS (Gibco) at a concentration of 20 cells/µL and stained with Hoechst dye (Invitrogen) for 5 min at RT prior to loading onto the MSND dispenser. For each sample, 3 dispenses were made on one 5,184-well chip resulting in 348 and 459 wells containing single cells after selection in CellSelect software (Takara Bio). Selected wells were processed for RT and in-chip preamplification, and cDNA was pooled for final library preparation using standard Nextera protocol (Illumina). The library was checked in Labchip Gx touch 24 (Perkin Elmer) and sequenced on Nextseq500 (Illumina) using V2 chemistry and paired-end setup (11 bp Read1 for cell barcode and 80 bp Read2 for cDNA template sequencing). A total of 537 M reads were obtained.
Single-cell RNA-seq of tdTom+ cells isolated from Gli1Cre−ERT2; tdTomatoflox lungs was carried out using the 10x Genomics Single-Cell 3′ v3.1 RNA-seq kit. Gene expression libraries were prepared according to the manufacturer’s protocol. MULTI-seq barcode libraries were retrieved from the samples, and libraries were prepared independently.
Single-cell data analysis
ICELL8 platform: cDNA reads were trimmed for poly(A) tails using BBMAP (unpublished, https://jgi.doe.gov/data-and-tools/bbtools/). Reads of length under 50 bases were discarded. The 456 M remaining reads were then aligned using STAR (release v2.4.0a) against Mus musculus build mm10 following ENCODE RNA-seq mapping recommendations. STAR indices were generated using Ensembl GTF file (release 83). For counting based on read counts, we used the Dropseq Core Computational Protocol version 1.12 (dropseq.jar) [19]. Gene-to-cell count table matrix was then used for statistical analysis performed within the Seurat package (http://satijalab.org/seurat/) [20]. The pipeline goes through cell and gene filtering, data normalization, then finding the most variable genes to perform principal component analysis (PCA) and t-distributed stochastic neighbor embedding (t-SNE) dimension reduction for visualization purpose. We first created a Seurat object including all cells and all genes present in at least 5 different cells. We then filtered out cells with more than 95% dropouts or less than 50,000 read counts. We also discarded cells with more than 20% of counts from mitochondrial genes or 10% counts from ribosomal genes. We then normalized the count values for the 305 remaining cells to a scaling factor corresponding to the median read counts per cell. We next identified most variable genes by plotting genes into bins based on X-axis (average expression) and Y-axis (dispersion) using cutoffs X = 3 and Y = 0.1, identifying 1,212 genes. We then performed a PCA, after scaling and centering of the data across these genes. This ensured robust identification of the primary structures in the data. We identified the first 8 principal components as statistically significant and used it for t-SNE calculation (perplexity = 30). The t-SNE procedure returns a two-dimensional (2D) embedding of single cells utilized for dataset visualization. Cells with similar expression signatures of genes within the variable set, and therefore, similar PC loadings, are most likely to localize near each other in the embedding, and hence, distinct cell types form 2D point clouds across the t-SNE map. Identification of the clusters of cells was determined using the FindClusters Seurat function (resolution = 1). The Seurat FindAllMarkers function was used to identify differentially expressed genes across each cluster (min.pct = 0.25, thresh.use = 0.25).
Chromium 10x platform: Single-cell suspensions were processed using the Chromium Next GEM Single-Cell 3ʹ Reagent Kits v3.1 (10x Genomics). Each sample was run separately on a lane in Chromium controller, and gene expression libraries were prepared according to the manufacturer’s protocol. Library sample index sequences were incorporated as the sample index read and sequenced together on a Nextseq2000 sequencer. Raw reads were aligned against the mouse genome (mm10, ensemble assembly 104) and mapped and counted by StarSolo [21]. Secondary analyses were carried out following the Seurat workflow. We removed 285 cells that did not express more than 1,000 genes, with less than 20,000 detected UMIs or had a mitochondrial content above 15%. Raw counts were normalized and scaled using the SCTransform function. The dimension of the count matrix was reduced by PCA. We used the first 40 principal components to compute the UMAP embedding and construct the nearest neighbor graph. Clusters were identified using a resolution of 0.3 and annotated according to the expression of markers described in the literature [22]. Gene expression exploration and differential analyses were carried out on log-normalized data.
Bronchiolosphere assay
Sorted mesenchymal cells (tdTom+) from Gli1Cre−ERT2; tdTomatoflox or Gli1Cre−ERT2; tdTomatoflox; Fgf10flox/flox lungs and epithelial cells (tdTom+) from Scgb1a1CreERT; tdTomatoflox lungs or epithelial progenitors from wild-type mice (defined as EpCAMhigh CD24low) were centrifuged, resuspended in cell culture medium (Dulbecco’s Modified Eagle Medium, Life Technologies) and subjected to organoid cultures as we previously described [14, 15, 23]. Cells were incubated under air–liquid conditions at 37 °C with 5% CO2 for 2 weeks, and the culture medium was changed 3 times per week.
Statistical analysis and figure assembly
Quantitative data were assembled and analyzed using GraphPad Prism software (GraphPad Software). These data are presented as average values ± standard error of mean (sem). Student’s t-test was used to compare 2 groups, one-way ANOVA was used to compare 3 or more groups in the presence of 1 variable (e.g., treatment, time or genotype), and two-way ANOVA was used to compare multiple groups in the presence of 2 variables (e.g., treatment and genotype, time and genotype). Differences were considered significant if P < 0.05. The number of biological replicates is indicated in the corresponding figure legends. Figures were assembled using Adobe Illustrator (Adobe).
Results
Mesenchymal cells transiently expressing Acta2 emerge during epithelial injury and repair
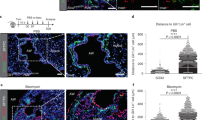
In order to analyze the spatial distribution of the ACTA2+ lineage around the injured airways, including our recently reported RSMC population [14], the Acta2-Cre-ERT2; tdTomatoflox lineage-tracing model was employed. Animals were randomly allocated into 3 groups receiving either tamoxifen food prior to corn oil injection (Tam-Oil), tamoxifen food prior to NA injection (Tam-NA) or NA injection followed by tamoxifen food (NA-Tam) (Fig. 1A). Animals were euthanized at day 14 (d14) after NA or Oil injections. Such approach allowed labeling (and visualizing) steady-state ACTA2+ cells, including those around the airways (ASMCs) (Tam-Oil), pre-existing ACTA2+ cells (including ASMCs) during repair after injury (Tam-NA) and total ACTA2+ cells during repair after injury (bona fide SMCs and newly recruited ACTA2+ cells; RSMCs) (NA-Tam). While the lineage label, tdTom, colocalized with the ACTA2 stain in the Tam-Oil (Fig. 1B, E, H, K) and Tam-NA groups (Fig. 1C, F, I, L), imaging of NA-Tam lungs revealed, in addition to ACTA2+ tdTom+ cells in the ASMC layer, the presence of ACTA2− tdTom+ cells between the ASMC layer and the airway epithelium, corresponding to RSMCs (Fig. 1D, G, J, M). These cells were clearly distinct from ASMCs and were mostly observed around the proximal lobular airways. They were more abundant around the mainstem bronchus and at the branch points. Quantification of the distance between the ASMC layer and airway epithelium showed significant thickening of the peribronchial region in Tam-NA, similarly to NA-Tam, compared to the Tam-Oil condition (Fig. 1N), confirming that the additional layer of RSMCs also forms in the Tam-NA condition but cannot be visualized due to the timing of tamoxifen administration (before injury).
A population of mesenchymal cells transiently expressing Acta2 is recruited after naphthalene injury. A Experimental setup and timeline of tamoxifen and naphthalene/corn oil treatment. Mice were fed tamoxifen-containing food. B–D Tile scans of left lung lobes from control oil-treated mice (Tam-Oil), naphthalene-treated mice where pre-existing ACTA2+ cells were labeled (Tam-NA) and naphthalene-treated mice where pre-existing and newly formed ACTA2+ cells were labeled (NA-Tam). Lung sections were stained with anti-ACTA2 antibodies (green), and the endogenous tdTomato signal (red) was detected. E–G Zoomed-in images of the conducting airway. H–J Zoomed-in images of the boxes in E–G. K–M Zoomed-in images of the boxes in H–J. Note the appearance of an additional population of ACTA2− tdTom+ cells (RSMC) in the NA-Tam group (J, M). N Quantification of the distance between ASMC and the epithelium. The double arrows in L and M illustrate the measured distance. O Gating strategy and quantification of tdTom+ cells in Tam-Oil, Tam-NA and NA-Tam lungs at d14. P Acta2, Pdgfra, Fgf10 and Gli1 expression levels analyzed by qPCR using sorted tdTom+ cells from Tam-Oil, Tam-NA and NA-Tam lungs at d14. ASMC airway smooth muscle cells, Epi epithelium, FSC forward scatter, NA naphthalene, Par parenchyma, RSMC repair-supportive mesenchymal cells, Tam tamoxifen. Scale bars: (B-D) 1400 μm. (N) n = 3 for Tam-Oil, n = 4 for Tam-NA and n = 4 for NA-Tam; (O) n = 4 for Tam-Oil, n = 5 for Tam-NA and n = 9 for NATam; (P) n = 3-4 for Tam-Oil, n = 4 for Tam-NA and n = 5 for NA-Tam. *P < 0.05, **P < 0.01, ***P < 0.001
The number of tdTom+ cells in these three experimental conditions was quantified by flow cytometry (Fig. 1O). Compared to the number of tdTom+ cells in Tam-Oil, our results demonstrate an increase in NA-Tam compared to Tam-Oil and Tam-NA, confirming that additional cells acquire ACTA2 expression after NA injury. We also investigated Acta2, Pdgfra, Fgf10 and Gli1 expression in sorted tdTom+ cells isolated from Tam-Oil, Tam-NA and NA-Tam lungs. Our results indicate that Pdgfra and Fgf10 expression are indeed upregulated in NA-Tam tdTom+ cells (RSMC-enriched) compared to Tam-Oil or Tam-NA conditions (both SMC-enriched), while Acta2 expression showed the opposite trend (Fig. 1P). Gli1 expression showed a trend for upregulation in NA-Tam compared to the other groups (Fig. 1P).
The emergence of tdTom+ RSMCs in NA-Tam, but not Tam-NA, lungs indicated that those cells were descendants of a mesenchymal population that transiently acquired Acta2 expression after NA injury. To study the dynamics of tdTom+ cells that are labeled after injury, NA-Tam lungs were analyzed by flow cytometry at d3, d7 and d14 (Fig. S1A, B). The results revealed a significant increase in the abundance of tdTom+ cells from d3 to d7 after NA injury (Fig. S1C). Interestingly, such difference in tdTom+ cells was no longer seen between d7 and d14 (Fig. S1C), indicating that RSMCs peaked in terms of abundance at d7.
Single-cell transcriptomic analysis reveals a mesenchymal niche that is associated with epithelial regeneration
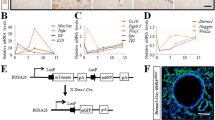
We next performed scRNA-seq on tdTom+ cells derived from Acta2-Cre-ERT2; tdTomatoflox lungs to delineate the complexity and heterogeneity of these cells, and to identify the RSMC cluster. FACS-sorted tdTom+ cells from Tam-NA (n = 3 animals) and NA-Tam (n = 3 animals) were used to dispense single cells onto an ICELL8 chip (Fig. 2). Following library preparation and RNA-seq, quality control yielded 122 cells from the Tam-NA group and 183 cells from the NA-Tam group (Fig. 2A–C). Bioinformatic analysis revealed the presence of five cellular clusters corresponding to SMCs, endothelial cells, macrophages and 2 additional clusters that showed matrix fibroblast signatures, one expressing Col13a1 and the other one Col14a1 (Figs. 2A–D, S2A). Similar matrix fibroblast clusters have been previously described [22, 24]. Importantly, the analysis showed a significant emergence of the COL13A1+ matrix fibroblast-like population in NA-Tam lungs compared to Tam-NA lungs (Fig. 2B–D). We have previously reported that deletion of Ctnnb1 and Fgf10 in NA-Tam mice led to repair impairment and that Pdgfra marks the RSMC population in the NA-Tam condition [14]. In the current study, Pdgfra indeed marked the emerging COL13A1+ population corresponding to RSMCs (Figs. 2G and S2A). This cluster expressed typical alveolar fibroblast/lipofibroblast markers such as Tcf21, Limch1 and Pdgfra (Figs. 2G and S2). Comparison between NA-Tam RSMCs and Tam-NA SMC confirmed that Pdgfra was among the top genes discriminating between RSMCs and pre-existing SMCs in the context of NA injury (Fig. S2B and Supplementary Table 1).
Single-cell RNA sequencing of tdTom+ cells isolated from Tam-NA and NA-Tam Acta2-Cre-ERT2; tdTomatoflox lungs identifies PDGFRα+ RSMCs. A–C t-SNE plots showing the distribution of tdTom+ cells among five cellular clusters: RSMC, SMC, COL14A1+ fibroblasts, endothelial cells and macrophages. Note the enrichment of the RSMC cluster in C. D–F Frequency of each cluster in Tam-NA and NA-Tam lungs. G Heatmap showing the top highly enriched genes in the different clusters. Groups of selected genes are shown in the corresponding boxes. Endo endothelial cells, Mac macrophages, NA naphthalene, RSMC repair-supportive mesenchymal cells, SMC smooth muscle cells, Tam tamoxifen. n = 3 (pooled 3 independent animals) for both Tam-NA and NA-Tam experiments
Differentially expressed genes in COL14A1+ matrix fibroblasts included Dcn, Col1a1 and Col14a1 (Figs. 2G and S2A). Cells within the SMC cluster expressed typical SMC markers such as Acta2, Tagln and Myh11 (Figs. 2G and S2A). Differentially expressed genes in the endothelial cluster included Ptprb, Sox17 and Cdh5 (Figs. 2G and S2A) while in macrophages, genes such as Ccl9, Cd14 and Cd86 were differentially expressed (Figs. 2G and S2A).
The GLI1+ lineage is a source of RSMCs in the lung
The peribronchial and perivascular regions of the lung are populated by hedgehog responsive GLI1+ mesenchymal cells (Fig. S1F). These cells have been shown to contribute to bleomycin-associated myofibroblasts in lung fibrosis [25] and were also shown to be amplified around the airways following NA injury [26]. A proportion of GLI1+ cells is intermingled with ASMCs in the peribronchial space (Fig. S1F). We previously used the Acta2-Cre-ERT2; tdTomatoflox mice to FACS-isolate the RSMC-enriched population (PDGFRα+ tdTom+) and the other mesenchymal populations (SMC-enriched PDGFRα− tdTom+) from NA-Tam animals at d3, d7 and d14 [14] (Fig, S1D). Gene expression analysis showed that Gli1 is highly expressed in the RSMC-enriched fraction, and its expression peaks at d7 (Fig. S1E). Therefore, we suspected that GLI1+ cells might serve as a source of RSMCs following NA injury.
To test the hypothesis that pre-existing GLI1+ cells are a source of RSMCs, Gli1Cre−ERT2; tdTomatoflox mice were fed tamoxifen-containing pellets for 2 weeks followed by a 2-week washout period and then treated with NA or Oil (Fig. 3A). This approach allowed labeling of pre-existing GLI1+ cells and studying their fate during epithelial repair after injury. A similar experimental setup was carried out using Gli1Cre−ERT2; tdTomatoflox; Fgf10flox/flox mice to investigate whether FGF10 specifically derived from these cells is required for the repair process (Fig. 3A). Genetic deletion of both Fgf10 alleles in pre-existing GLI1+ cells significantly impaired club cell replenishment (Fig. 3B, C). Immunofluorescence for KI67 showed a significant decrease in club cell proliferation in experimental (Gli1Cre−ERT2; tdTomatoflox; Fgf10flox/flox) vs. control (Gli1Cre−ERT2; tdTomatoflox) lungs following NA treatment (Fig. S3). Quantitative real-time PCR using lung homogenates showed significant downregulation of Scgb1a1 at d14 after NA treatment in experimental mice compared to controls (Fig. 3D). Interestingly, we did not observe a significant drop in Fgf10 expression in non-injured (Oil condition) experimental vs. control lungs, suggesting that at the resting state, GLI1+ cells are not the main source of Fgf10 expression in the lung (Fig. 3D). However, following injury, a clear drop in Fgf10 expression is observed in experimental vs. control lungs fitting with the proposed increase in Fgf10 expression in the lung during the repair process (Fig. 3D). Of note, we could not detect significant proliferation in the surrounding mesenchyme in the NA vs. Oil-treated control lungs at d14 (Fig. S3), which is in line with the results shown in Fig. S1C, where no change in tdTom+ cells was seen between d7 and d14, thus supporting the notion that RSMCs were no longer expanding after d7.
Genetic deletion of Fgf10 in GLI1+ cells leads to impaired repair in the bronchial epithelium. A Experimental setup and timeline of tamoxifen and naphthalene/corn oil treatment. Mice were fed tamoxifen-containing food. B Immunofluorescence for SCGB1A1 using control and experimental lungs treated with corn oil or NA and analyzed at d14. C Quantification of the immunofluorescence shown in B demonstrating decreased abundance of SCGB1A1+ cells in the lungs of experimental mice, indicating impairment of epithelial regeneration. D Quantitative real-time PCR using lung homogenates and showing decreased levels of Scgb1a1 and Fgf10 gene expression in experimental mice compared to controls. IF immunofluorescence, NA naphthalene, Tam tamoxifen. Scale bars: 100 μm. n = 3 per group. *P < 0.05, **P < 0.01
Examination of Oil-treated Gli1Cre−ERT2; tdTomatoflox mice revealed significant abundance of tdTom+ cells around the airways (Fig. 4A, B). The number of tdTom+ cells was significantly increased in this region upon NA treatment (Figs. 4B, C, S3), as previously described [26]. Such increase was attenuated in experimental lungs vs. controls (Fig. S3). Strikingly, descendants of the GLI1+ lineage were also observed in similar regions as Acta2-Cre-ERT2-traced RSMCs (compare Fig. 4B with Fig. 1J, M).
GLI1+ cells give rise to RSMCs and support club cell growth. A Experimental setup and timeline of tamoxifen and naphthalene/corn oil treatment. Mice were fed tamoxifen-containing food. B Immunofluorescence for ACTA2 using d14 NA and d14 Oil Gli1Cre−ERT2; tdTomatoflox lungs. C Quantification of the number of tdTom+ cells around the airways. D Quantification of tdTom+ cells by flow cytometry. E Expression levels of Gli1, Acta2 and Fgf10 in tdTom+ cells isolated from Oil, d3 NA and d14 NA lungs. F Gating strategy and quantification of cells based on tdTom and ACTA2 expression. G Experimental setup for isolating tdTom+ club cells or epithelial progenitors from wild-type lungs, and co-culturing them with tdTom+ cells isolated from oil- or naphthalene-treated Gli1Cre−ERT2; tdTomatoflox lungs. (H, I) Representative images of large bronchiolospheres grown from tdTom+ mesenchymal and tdTom+ epithelial cells. J, K Quantification of organoid number and diameter. IF Immunofluorescence, NA Naphthalene, Tam Tamoxifen, ns Not significant. Scale bar: (B, H, I) 100 μm. n = 4 for d3 and d = 3 for d14; (C) n = 3; (D) n = 5 for Oil and n = 7 for NA, (E) n = 5 for Oil, (F) n = 5 for d3 Oil and n = 4 for d3 NA ; (J, K) n = 5 for Oil and n = 3 for NA. In (K), data from individual organoids are shown. *P < 0.05, **P < 0.01
To verify if the increase in Fgf10 expression (Fig. 3D) is linked to Fgf10 upregulation in GLI1+ cells, we analyzed Fgf10 expression in sorted GLI1+ cells. Our results indicate no significant difference in Fgf10 expression between the d14 group and the Oil group (Fig. 4E). Therefore, the global increase in Fgf10 expression is likely linked to a quantitative increase in GLI1+ cells. Furthermore, qPCR on tdTom+ cells sorted from Oil- and NA-treated Gli1Cre−ERT2; tdTomatoflox lungs revealed a transient downregulation of Gli1 at d3 as previously described [26], in parallel to a significant, transient upregulation of Acta2 at d3 (Fig. 4E). Flow cytometry-based quantification confirmed the acquisition of Acta2 expression in the GLI1+ lineage upon NA injury (Fig. 4F). Altogether, these data suggest that pre-existing GLI1+ cells 1) transiently acquire Acta2 expression and give rise to RSMCs during epithelial regeneration (and are therefore captured when ACTA2+ cells are labeled after injury (Fig. 1)), 2) are a source of FGF10 during both steady state and after injury and 3) are massively amplified and recruited to the airways in response to NA injury.
GLI1+ cells support club cell growth in an in vitro co-culture organoid system
We then opted to test whether GLI1+ cells derived from d14 Oil- and NA-treated lungs possess an intrinsic capacity to support the growth of club cells in a co-culture organoid system. Club cells sorted using uninjured Scgb1a1CreERT; tm14 tdTomatoflox mice or epithelial progenitors sorted from wild-type mice were co-cultured with tdTom+ cells derived from Tam-Oil or Tam-NA Gli1Cre−ERT2; tdTomatoflox mice (Fig. 4G). Representative images of large bronchiolospheres are shown in Fig. 4H, I. Further analysis of these bronchiolospheres indicated a slight trend for increased organoid number and size in the NA vs. Oil condition (Fig. 4J, K), suggesting that the beneficial effect of GLI1+ cells appearing after injury is likely linked to their quantity/recruitment rather than a qualitative change. Additionally, loss of function of Fgf10 in GLI1+ cells led to decreased organoid size compared to controls (Fig. S4).
Single-cell RNA-seq of GLI1+ cells suggests that alveolar fibroblasts, co-expressing Gli1 and Fgf10, converge on the RSMC lineage
The organoid assay suggested that GLI1+ cells have an intrinsic club cell-supportive potential even in the absence of injury (Fig. 4G-K). We therefore explored the heterogeneity of GLI1+ cells in non-injured lungs using scRNA-seq (Fig. 5). UMAP analysis showed the presence of seven clusters (Fig. 5A). Cluster 1 corresponds to alveolar fibroblasts, while cluster 6 represents a subcluster of alveolar fibroblasts expressing high levels of Mmp3 (Fig. 5A, B). Clusters 0 and 2 contain two adventitial fibroblast subsets. Clusters 3 and 4 include peribronchial fibroblasts, while cluster 5 corresponds to SMCs. Examination of the RSMC transcriptomic signature defined in Fig. 2 shows that most of the signature genes are contained mostly in alveolar fibroblasts (clusters 1 and 6), and to a less extent in adventitial fibroblasts (Fig. 5B lower panel). Interestingly, cluster 5 (SMCs) did not display significant RSMC signature, thus supporting the notion that RSMCs are distinct from SMCs. Given the important role played by Fgf10 expressed by GLI1+ cells in the repair process and the fact that Fgf10 is not significantly upregulated in these cells after NA injury, we reasoned that GLI1+ FGF10+ cells exist in the non-injured lung. This is rather counter-intuitive as Gli1, a downstream target of sonic hedgehog signaling, and Fgf10 expression are thought to be mutually exclusive especially during lung development [27]. Confirming our hypothesis, these cells were found in alveolar fibroblasts and adventitial fibroblasts but completely absent in the SMCs (Fig. 5C). Quantification of the expression of Gli1, Fgf10 and Acta2 confirmed that alveolar fibroblasts represent the main cluster displaying high levels of Fgf10 as well as moderate levels of Gli1 and Acta2 (Fig. 5D). Using our novel Fgf10Cre−ERT2 mice recently reported [28], we also examined the localization of FGF10+ cells in non-injured lungs. In addition to being located in the alveolar region [28], FGF10+ cells were also observed around the bronchi and distinct from ASMCs (Fig. 5E), similarly to what was observed for GLI1+ cells (Fig. S1F).
Discussion
In this study, we demonstrated that GLI1+ cells are a source of RSMCs during airway epithelial regeneration. Lineage tracing of ACTA2+ cells (using the Acta2-Cre-ERT2 driver line) labeled either before (Tam-NA) or after (NA-Tam) injury indicates that these cells appear mostly following injury. Nevertheless, lineage-tracing experiments using the Gli1Cre−ERT2 driver line indicate that RSMCs also arise from pre-existing GLI1+ cells that acquire Acta2 expression and accumulate in the peribronchial region. Deletion of Fgf10 in GLI1+ cells leads to impaired repair. Using bronchiolosphere assays, we show that GLI1+ cells isolated from either non-injured or NA-treated lungs similarly support club cell growth in vitro. Overall, our data indicate that GLI1+ cell-derived RSMCs produce FGF10 that is required for the repair process after NA injury. As such, RSMCs represent a privileged mesenchymal niche for club cell progenitors.
The lineage map of the lung mesenchyme is still poorly understood
Despite extensive research, the lineage map of the lung mesenchyme is still poorly understood, with the best-characterized cell types being ASMCs/VSMCs and pericytes, and, to a less extent, alveolar fibroblasts (mostly during neonatal stages) and myofibroblasts. The perivascular region of the lung is populated by GLI1+ mesenchymal cells that have been proposed to constitute a subpopulation of PDGFRβ+ pericytes [25]. These cells have been shown to possess mesenchymal stem cell (MSC)-like characteristics such as trilineage differentiation potential in vitro (giving rise to adipocytes, osteoblasts and chondrocytes) and also represent a repertoire of myofibroblast progenitors in fibrotic disease [25, 29]. On the other hand, the peribronchial domain of the postnatal mouse lung consists of cartilaginous rings (around main-stem bronchi), ASMCs (around conducting airways) and other poorly characterized mesenchymal cells, some of which also express Gli1.
RSMCs are a component of the niche required for activating club cell progenitors
In this study, we demonstrate the complexity of the mesenchymal niche in the mature lung and identify GLI1+ cell-derived RSMCs as a component of the niche required for activating club cell progenitors. GLI1+ cells are amplified, transiently acquire Acta2 expression and contribute to the repair process by producing FGF10 (Fig. 6). We previously published side-by-side comparison of PDGFRα+ tdTom+ cells (RSMC-enriched population) and PDGFRα− tdTom+ cells (SMC-enriched population), indicating that RSMCs display higher WNT signaling activation (using Axin2 mRNA levels as readout), higher Fgf10 expression and greater potential in supporting club cell growth in an in vitro organoid system [14]. However, one limitation was that the PDGFRα− tdTom+ population contains a mixture of bona fide ASMCs/VSMCs and other subsets such as matrix fibroblasts rather than a pure population of ASMCs, and this might reduce the colony-forming efficiency and consequently bronchiolosphere number and size. Moreover, we could not exclude the possibility that parenchymal lineage-labeled cells (such as alveolar fibroblasts) also contribute to repair in the conducting airway epithelium. Our data strongly indicate that GLI1+ cells, which are normally found in the peribronchial, perivascular and alveolar regions, serve as a source of FGF10+ RSMCs in the lung.
scRNA-seq analysis of GLI1+ cells confirms the presence of an RSMC cluster. (A) UMAP clustering of GLI1+ cells using uninjured Gli1Cre−ERT2; tdTomatoflox lungs. (B) Upper panel: Heatmap showing the top highly enriched genes in the different clusters. Lower panel: Heatmap showing the top enriched RSMC signature genes in different clusters. (C) Analysis of the co-expression of Gli1 and Fgf10 in different clusters. (D) Gli1, Fgf10 and Acta2 expression throughout the different clusters. (E) Immunofluorescence images of Fgf10Cre−ERT; tdTomatoflox lungs showing FGF10+ cells around the bronchi and distinct from ASMCs. ASMC airway smooth muscle cells, br bronchus, IF immunofluorescence
Model summarizing the role of RSMCs as a niche that facilitates epithelial regeneration after naphthalene injury. GLI1+ cells are distinct from ASMCs. After naphthalene injury, GLI1+ cells transiently acquire Acta2 expression and get amplified, giving rise to RSMCs and contributing to epithelial regeneration by producing FGF10. Pre-existing ASMCs have also been shown to mediate the repair process via a WNT-FGF10-mediated mechanism [12, 13]. CGRP calcitonin gene-related peptide, CYP2F2 cytochrome P450 2F2, NE neuroendocrine, TUB tubulin
How different are RMSCs from the resident mesenchymal cell (rMC) niche?
Resident mesenchymal cells supporting the growth of alveolar stem cells were further characterized [30]. Using flow cytometry to isolate cells as well as the alveolosphere model, CD31− CD45− EpCAM− cells (defined as rMC) could be subdivided into SCA1+ and SCA1−. Only SCA1+ rMCs displayed niche activity for AT2 stem cells. Using reporter lines for Fgf10 expression, these cells were further subdivided into SCA1+ FGF10+ rMCs and SCA1+ FGF10− rMCs, with the FGF10+ subset displaying the niche activity. These cells are also positive for PDGFRα.
We also recently used scRNA-seq to further characterize the different subpopulations present in SCA1+ rMCs [31]. We identified eighteen different clusters. Among them, five different clusters matched different subsets of lipofibroblasts and specifically expressed Fgf10, Fgf7 and Pdgfra. These clusters could also be differentiated via the level of Col13a1, Col1a1 and Col14a1 expression. One of these five clusters expresses high levels of Col13a1, Fgf10, Limch1, inmt, Npnt and Pdgfra, and could therefore be very similar to RSMCs. Interestingly, this cluster expresses lower levels of Sca1 compared to the other clusters. This puzzling similarity between RSMCs around the bronchi and rMC niche cells in the alveolar region raises the possibility that rMC niche cells could be recruited from the adjacent alveolar region surrounding the bronchi. Further investigation using high-resolution imaging and more specific lineage tracing using the Dre/Cre technology should allow answering this important question.
Data availability
Primary data can be made available upon reasonable request. Single-cell data have been deposited on gene expression omnibus (GEO) GSE215094.
References
Clara M (1937) Zur histobiologie des bronchalepithels. Z mikrosk anat Forsch 41:321–347
Winkelmann A, Noack T (2010) The clara cell: a “third reich eponym”? Eur Respir J 36:722–727. https://doi.org/10.1183/09031936.00146609
Rawlins EL, Okubo T, Xue Y et al (2009) The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell 4:525–534. https://doi.org/10.1016/j.stem.2009.04.002
Stripp BR, Maxson K, Mera R, Singh G (1995) Plasticity of airway cell proliferation and gene expression after acute naphthalene injury. Am J Physiol 269:L791–L799
Boyd MR (1977) Evidence for the Clara cell as a site of cytochrome P450-dependent mixed-function oxidase activity in lung. Nature 269:713–715
Fanucchi MV, Murphy ME, Buckpitt AR et al (1997) Pulmonary cytochrome P450 monooxygenase and Clara cell differentiation in mice. Am J Respir Cell Mol Biol 17:302–314. https://doi.org/10.1165/ajrcmb.17.3.2774
Mahvi D, Bank H, Harley R (1977) Morphology of a naphthalene-induced bronchiolar lesion. Am J Pathol 86:558–572
Giangreco A, Reynolds SD, Stripp BR (2002) Terminal bronchioles harbor a unique airway stem cell population that localizes to the Bronchoalveolar duct junction. Am J Pathol 161:173–182. https://doi.org/10.1016/S0002-9440(10)64169-7
Hong KU, Reynolds SD, Giangreco A et al (2001) Clara cell secretory protein-expressing cells of the airway neuroepithelial body microenvironment include a label-retaining subset and are critical for epithelial renewal after progenitor cell depletion. Am J Respir Cell Mol Biol 24:671–681. https://doi.org/10.1165/ajrcmb.24.6.4498
Reynolds SD, Giangreco A, Power JHT, Stripp BR (2000) Neuroepithelial bodies of pulmonary airways serve as a reservoir of progenitor cells capable of epithelial regeneration. Am J Pathol 156:269–278. https://doi.org/10.1016/S0002-9440(10)64727-X
Jones MR, Lingampally A, Dilai S et al (2019) Characterization of Tg(Etv4-GFP) and Etv5 RFP reporter lines in the context of fibroblast growth factor 10 signaling during mouse embryonic lung development. Front Genet 10:178. https://doi.org/10.3389/fgene.2019.00178
Volckaert T, Dill E, Campbell A et al (2011) Parabronchial smooth muscle constitutes an airway epithelial stem cell niche in the mouse lung after injury. J Clin Invest 121:4409–4419. https://doi.org/10.1172/JCI58097
Lee JH, Tammela T, Hofree M et al (2017) Anatomically and functionally distinct lung mesenchymal populations marked by Lgr5 and Lgr6. Cell 170:1149-1163.e12. https://doi.org/10.1016/j.cell.2017.07.028
Moiseenko A, Vazquez-Armendariz AI, Kheirollahi V et al (2020) Identification of a repair-supportive mesenchymal cell population during airway epithelial regeneration. Cell Rep 33:108549. https://doi.org/10.1016/j.celrep.2020.108549
Vazquez-Armendariz AI, Seeger W, Herold S, El Agha E (2021) Protocol for the generation of murine bronchiolospheres. STAR Protoc 2:100594. https://doi.org/10.1016/j.xpro.2021.100594
Wendling O, Bornert J-M, Chambon P, Metzger D (2009) Efficient temporally-controlled targeted mutagenesis in smooth muscle cells of the adult mouse. Genesis 47:14–18. https://doi.org/10.1002/dvg.20448
Abler LL, Mansour SL, Sun X (2009) Conditional gene inactivation reveals roles for Fgf10 and Fgfr2 in establishing a normal pattern of epithelial branching in the mouse lung. Dev Dyn 238:1999–2013. https://doi.org/10.1002/dvdy.22032
Urness LD, Paxton CN, Wang X et al (2010) FGF signaling regulates otic placode induction and refinement by controlling both ectodermal target genes and hindbrain Wnt8a. Dev Biol 340:595–604. https://doi.org/10.1016/j.ydbio.2010.02.016
Macosko EZ, Basu A, Satija R et al (2015) Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161:1202–1214. https://doi.org/10.1016/j.cell.2015.05.002
Butler A, Hoffman P, Smibert P et al (2018) Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36:411–420. https://doi.org/10.1038/nbt.4096
Dobin A, Davis CA, Schlesinger F et al (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. https://doi.org/10.1093/bioinformatics/bts635
Tsukui T, Sun K-H, Wetter JB et al (2020) Collagen-producing lung cell atlas identifies multiple subsets with distinct localization and relevance to fibrosis. Nat Commun 11:1920. https://doi.org/10.1038/s41467-020-15647-5
Vazquez-Armendariz AI, Heiner M, El Agha E et al (2020) Multilineage murine stem cells generate complex organoids to model distal lung development and disease. EMBO J 39:1476. https://doi.org/10.15252/embj.2019103476
Xie T, Wang Y, Deng N et al (2018) Single-cell deconvolution of fibroblast heterogeneity in mouse pulmonary fibrosis article single-cell deconvolution of fibroblast heterogeneity in mouse pulmonary fibrosis. Cell Rep 22:3625–3640. https://doi.org/10.1016/j.celrep.2018.03.010
Kramann R, Schneider RK, DiRocco DP et al (2015) Perivascular Gli1+ progenitors are key contributors to injury-induced organ fibrosis. Cell Stem Cell 16:51–66. https://doi.org/10.1016/j.stem.2014.11.004
Peng T, Frank DB, Kadzik RS et al (2015) Hedgehog actively maintains adult lung quiescence and regulates repair and regeneration. Nature 526:578–582. https://doi.org/10.1038/nature14984
Li C, Hu L, Xiao J et al (2005) Wnt5a regulates Shh and Fgf10 signaling during lung development. Dev Biol 287:86–97. https://doi.org/10.1016/j.ydbio.2005.08.035
Chu X, Taghizadeh S, Vazquez-Armendariz AI et al (2021) Validation of a novel Fgf10 Cre-ERT2 knock-in mouse line targeting FGF10Pos cells postnatally. Front Cell Dev Biol 9:671841. https://doi.org/10.3389/fcell.2021.671841
El Agha E, Kramann R, Schneider RK et al (2017) Mesenchymal stem cells in fibrotic disease. Cell Stem Cell 21:166–177. https://doi.org/10.1016/j.stem.2017.07.011
Taghizadeh S, Heiner M, Vazquez-Armendariz AI et al (2021) Characterization in mice of the resident mesenchymal niche maintaining AT2 stem cell proliferation in homeostasis and disease. Stem Cells 39:1382–1394. https://doi.org/10.1002/stem.3423
Taghizadeh S, Chao C-M, Guenther S et al (2022) FGF10 triggers de novo alveologenesis in a BPD model: impact on the resident mesenchymal niche cells. Stem Cells. https://doi.org/10.1093/stmcls/sxac025
Funding
Open Access funding enabled and organized by Projekt DEAL. MT and BM were supported by CNRS and the French Government (National Research Agency, ANR) through ANR-PRCI-18-CE92-0009-01 FIBROMIR. SB was supported by the Cardio-Pulmonary Institute (CPI; EXC 2026, Project ID: 390649896), the German Center for Lung Research (DZL) and the German Research Foundation (DFG; BE4443/1-1, BE4443/4-1, BE4443/6-1, KFO309 P7 and SFB1213 268555672 projects A02 and A04). EEA acknowledges the support of the Institute for Lung Health (ILH), the DFG (EL 931/4-1, KFO309 P7 and SFB CRC1213 268555672 project A04), the CPI and the DZL.
Author information
Authors and Affiliations
Contributions
SB and EEA contributed to the study conception and design. Material preparation, data collection and analysis were performed by XC, AL, AM, VK, AIVA, JK, AK, GK, MSF, MZ, IS, IA, SG, KL, MT, SB and EEA. AG, TB, BM, CS, XL, WS, SH, JSZ, SB and EEA contributed to platforms and methodology. The first draft of the manuscript was written by SB and EEA, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
Animal experiments were conducted at JLU Giessen after approval by the Regierungspräsidium Giessen (protocol # G 2/2016) and at Wenzhou Medical University (protocol # 2021–0017).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
18_2022_4599_MOESM1_ESM.csv
Supplementary file1 Supplementary Table 1. Top differentially expressed genes in tdTom+ cells between NA-Tam RSMCs and Tam-NA SMCs. (CSV 108 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chu, X., Lingampally, A., Moiseenko, A. et al. GLI1+ cells are a source of repair-supportive mesenchymal cells (RSMCs) during airway epithelial regeneration. Cell. Mol. Life Sci. 79, 581 (2022). https://doi.org/10.1007/s00018-022-04599-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04599-2