Abstract
The transcription factor SOX9 is essential for the development of multiple organs including bone, testis, heart, lung, pancreas, intestine and nervous system. Mutations in the human SOX9 gene led to campomelic dysplasia, a haploinsufficiency disorder with several skeletal malformations frequently accompanied by 46, XY sex reversal. The mechanisms underlying the diverse SOX9 functions during organ development including its post-translational modifications, the availability of binding partners, and tissue-specific accessibility to target gene chromatin. Here we summarize the expression, activities, and downstream target genes of SOX9 in molecular genetic pathways essential for organ development, maintenance, and function. We also provide an insight into understanding the mechanisms that regulate the versatile roles of SOX9 in different organs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mammalian embryogenesis is a complicated process; a cross-talking network of instructions for specialized developmental processes. Many transcription factors are essential for multiple developmental pathways, or to direct a large variety of processes. One such transcription factor is sex-determining region Y (SRY)-box 9 (SOX9)—a member of the SOX (SRY-type HMG box) family of transcription factors. These all harbor a high mobility group (HMG) box DNA-binding domain, first identified in SRY. Twenty SOX proteins have since been identified in mice and humans, and are grouped into nine subfamilies (A, B1, B2, C–H) depending on the structural homology outside of the HMG box [1]. Heterozygous mutations in SOX9 lead to the human disorder campomelic dysplasia (CMPD, OMIM# 114, 290) characterized by skeletal dysplasia and variable 46, XY sex reversal [2, 3]. Studies of Sox9-deficient mice, and SOX9 function in other species, have demonstrated its diverse roles during the development in multiple tissues and organs. These include cartilage [4], testis [5], nervous system [6], retina [7], lung [8], heart valve [9], pancreas [10], bile duct [11], intestine [12], prostate [13], and hair follicle [14]. In this review, we describe common and unique functions of SOX9 among tissues and organs, and the varied mechanisms through which SOX9 acts. Overall, this review comprehensively highlights the essential role that SOX9 plays in mammalian embryogenesis and organogenesis, thereby providing insights into the role this transcription factor plays to both initiate and maintain regulatory processes.
SOXE, SOX9, and their functional domains
The SOX9/Sox9 gene lies on human chromosome 17q and mouse chromosome 11q. SOX9 is located in a gene desert containing long-range spatiotemporal specific enhancers. The human SOX9 protein comprises 509 amino acids and consists of a HMG box, a dimerization domain (DIM), two transactivation domains located in the middle (TAM) and the C-terminus (TAC) of the protein, and a proline/glutamine/alanine (PQA)-rich domain (Fig. 1).
Schematic structure of the human SOX9 protein. The dimerization domain (DIM) precedes the HMG box. Two transactivation domains are located in the middle (TAM) and at the C-terminus (TAC). The proline, glutamine and alanine (PQA)-rich domain is required for transactivation. Phosphorylation of serine (S) residues, acetylation and sumoylation of lysine (K) residues are highlighted
SOX9, SOX8 and SOX10 form the SOXE subgroup, sharing homology in the HMG, DIM, and TAM and TAC domains, with two nuclear localization signals (NLS) and one nuclear export signal (NES). The HMG domain facilitates sequence-specific DNA binding and binds to the minor groove of DNA, inducing DNA bending by forming an L-shaped complex. The consensus DNA-binding motif of SOX9 is AGAACAATGG, with AACAAT being the core-binding element, and flanking 5′ AG and 3′ GG nucleotides specific to SOX9 [15].
SOX9 is capable of homodimerization through the DIM domain; this is required for DNA binding and transactivation of cartilage-specific genes [16, 17]. SOXE proteins heterodimerize via the DIM domain of one protein and the HMG box of the other protein [18]. In contrast, SOX9 functions as a monomer in testicular Sertoli cells [16]. Active SOX9-binding dimer motifs in regulatory regions of target genes are cell-type specific: SOX9 dimerizes on palindromic composite DNA motifs separated by 3–5 nucleotides in melanoma cells [18] and chondrocytes [19], whereas no enrichment of palindromic sequences is observed in hair follicle stem cells [20].
SOX9 transactivation domains interact with transcriptional co-activators or basal transcriptional machinery components. The transactivation domain at the C-terminus (TAC) physically interacts with MED12 (mediator complex subunit 12), CBP/p300 (CREB binding protein/E1A binding protein p300), TIP60 (Tat interactive protein-60), and WWP2 (WW domain containing E3 ubiquitin protein ligase 2), enhancing transcriptional activity of SOX9 [21,22,23,24]. The TAC domain is required for inhibition of β-catenin during chondrocyte differentiation [25]. The transactivation domain in the middle (TAM) synergizes with the TAC domain to activate cartilage-specific genes in vitro [26].
SOX9 also contains a unique PQA-rich domain that enhances transactivation in vitro [27] but lacks autonomous transactivation capability [26]. Deletion of the PQA-rich domain reduced SOX9’s capacity to transactivate a reporter plasmid with tandemly repeated SOX9 binding sites [27]. These domains thus allow SOX9 to enact different roles within the complex molecular pathways in which it plays such a crucial part.
SOX9 in organ and tissue development
SOX9 plays an important role in organogenesis during mammalian embryo development. The expression profile, function, and target genes that SOX9 directly binds to and activates during organ development will be discussed. A diagrammatic summary of cell-type specific expression of SOX9 during the development of cartilage, testis, nervous system, retina, lung, heart valve, pancreas, bile duct, intestine, prostate, and hair follicle is shown in Supplementary Fig. 1. The functions and target genes activated by SOX9 are summarized in Table 1.
Cartilage
Chondrogenesis and SOX9 expression
Endochondral ossification creates fetal bone tissue from a cartilage template and later forms bones of the axial and appendicular skeleton (Fig. S1A). At the beginning, mesenchymal cells condense and migrate toward the sites of future bone. Bipotential osteochondroprogenitors commit to the chondrocytic fate, differentiate into prechondrocytes and deposit cartilage matrix. The early chondrocytes in the cartilage anlagen undergo proliferation, maturation, cell cycle exit, and hypertrophy. Between the epiphyses and bone shaft, the cartilage growth plate is composed of four zones of activity, allowing longitudinal bone growth.
The resting or reserve zone contains small round chondrocytes, serving as progenitor-like cells for subsequent proliferation and differentiation. The proliferating zone contains proliferating chondrocytes organized into longitudinal columns. The prehypertrophic zone comprises maturing chondrocytes which undergo cell cycle arrest and initiate expression of the hypertrophic markers Indian hedgehog (Ihh) [28] and collagen type 10 gene (Col10a1) [29]. The hypertrophic zone is composed of massively expanded chondrocytes [30]. These hypertrophic chondrocytes undergo terminal differentiation and remodeling, mineralize the cartilage matrix, and may undergo apoptosis or transdifferentiate into osteoblasts [31, 32].
During chondrogenesis, SOX9 is expressed in condensed mesenchymal cells and differentiated chondrocytes of the cartilage growth plate, except in hypertrophic chondrocytes in which its expression is shut off [33]. In the developing limbs, SOX9 is abundant in mesenchymal condensation at embryonic day E11.5–E12.5 [4]. Most cartilage is well formed by E15.5, and SOX9 is expressed in the resting, proliferating and prehypertrophic chondrocytes within the growth plates of the long bones. SOX9 is a master regulator in chondrogenesis and is required for successive steps of chondrocyte differentiation which are described in detail below.
Chondrogenic mesenchymal condensation and survival of mesenchymal cells
During chondrogenesis, SOX9 is required for chondrogenic mesenchymal condensations. In mouse embryo chimeras, Sox9−/− cells are excluded from mesenchymal condensations and fail to express chondrocyte-specific markers Col2a1, Col9a2, Col11a2, and Acan (aggrecan) [34]. Accordingly, inactivation of Sox9 in all mesenchymal cells of the limb buds results in complete loss of cartilage and bone in limbs [33, 34]. At E13.5, the mesenchymal cells in Sox9−/− limb buds display extensive apoptosis, suggesting that SOX9 is necessary for their survival [33]. SOX9 likely controls prechondrocyte condensation by upregulating several genes that are crucial for actin cytoskeleton assembly, homotypic cell–cell adhesion and heterotypic cell–cell repulsion [35]. These genes include Fam101a (filamin-interacting protein), Myh14 (myosin heavy chain 14), Sema3c (semaphorin 3c) and Sema3d, which are directly transactivated by SOX9 via binding to their enhancers.
Chondrocyte differentiation and proliferation
Condensed prechondrocytes differentiate into early-stage chondrocytes, which start to produce cartilage extra-cellular matrix (ECM). If Sox9 is inactivated in mice from E13.5, after mesenchymal condensations, severe chondrodysplasia is observed as most condensed mesenchymal cells fail to differentiate into chondrocytes [33]. The expression of Sox5, Sox6 and extracellular matrix genes Col2a1, Acan, and Comp (cartilage oligomeric matrix protein) are markedly down-regulated in metacarpals of these mice at E15.5. Moreover, SOX9 is necessary for chondrocyte proliferation as the size of proliferating zones in these mice is dramatically reduced in the absence of Sox9 [33].
During early chondrogenesis, SOX9 cooperates with SOX5/SOX6 to activate expression of target genes, such as Col2a1 and Acan by binding to their enhancers [36]. The three SOX proteins associate with insulin-like growth factor-I (IGF-I), transcription factors Sp1/Sp3, and euchromatin-associated factors CBP/p300 and TIP60 to increase COL2A1 transcription by interacting with its enhancer region [37]. In addition, the SOX trio physically interacts with the Runt domain transcription factor (RUNX1) to enhance activation of chondrocyte-specific genes thereby increasing cartilage matrix production [38]. Moreover, in non-hypertrophic chondrocytes, GLI2 and GLI3 functionally interact with SOX9 by binding to a conserved GLI-binding element near the SOX9 binding site to repress the expression of Col10a1 which is a hypertrophic chondrocyte marker [39].
Chondrocyte hypertrophy and survival
SOX9 is also required for chondrocyte hypertrophy. Inactivation of Sox9 in flat chondrocytes near the prehypertrophic chondrocytes results in absence of hypertrophy, enhanced apoptosis and absence of Col10a1 expression [40]. Additionally, SOX9 prevents premature activation of prehypertrophy and subsequent acquisition of an osteoblastic fate. This is achieved in part through inhibition of Runx2 expression and β-catenin signaling [30]. SOX9 is also necessary to maintain proper homeostasis of cartilaginous tissues in adult mice [41].
Although Sox9 RNA is turned off abruptly when chondrocytes undergo hypertrophy, SOX9 protein outlives its mRNA and is necessary for chondrocyte hypertrophy [30, 40]. SOX9 interacts with AP-1 (activator protein 1) family members such as JUN and FOSL2 and they directly co-bind at target motifs to promote transcription of hypertrophic genes in the early hypertrophic chondrocytes [42]. Upon hypertrophy and as SOX9 levels decline, FOXA (forkhead box A) competes off SOX9 to accesses the regulatory elements of target genes such as Col10a1, to promote terminal chondrocyte maturation together with RUNX2/3, AP1 and MEF2 (myocyte enhancer factor 2) [43].
SOX9 targets classification in chondrocytes
ChIP-Seq analysis shows that SOX9 targets in rat proliferating/early prehypertrophic growth plate chondrocytes comprise all major cartilage ECM genes and many cartilage-specific regulatory genes, including the SOX trio target genes [44]. Two distinct categories of SOX9 targets have been identified in mouse chondrocytes [45]. Class I targets are non-chondrocyte-specific genes such as Kelch-like family member 21 (Klhl21), solute carrier family 29 member 1 (Slc29a1) and RNA binding motif protein 25 (Rbm25), which control basic cellular functions, such as translation, non-coding RNA metabolic process and protein folding. SOX9 associates with basal transcriptional components around the transcriptional start sites (TSS) of these genes. Class II targets are chondrocyte-specific genes in which a SOX9 homodimer directly binds to evolutionarily conserved active enhancers and super-enhancers. Moreover, the abundance of binding sites for AP-1, NFAT (nuclear factor of activated T-cells), FOX, RUNX, and HOX (homeobox) transcription factor families near the SOX trio motifs implies their interactions are required to regulate enhancer activities in chondrocytes [45].
Testis
Testis organogenesis and SOX9 expression
Fetal testicular development in mice is depicted in Fig. S1B. In mice, Sox9 expression is detected in both XY and XX gonads at very low levels at E10.5. Once the Y-linked sex-determining gene Sry is expressed in the developing XY gonad, Sox9 is strongly up-regulated by E11.5 in the bipotential supporting cells which differentiate into pre-Sertoli cells. In contrast, in the developing XX gonad Sox9 is down-regulated and the supporting cells differentiate into pre-granulosa cells [46]. Sox9 then continues to be expressed in Sertoli cells throughout testis development and initiates a cascade of gene interactions and cellular events that drive testis differentiation [47]. Sertoli cells start to proliferate and migrate to surround the germ cells and release signals that are required for the differentiation of fetal Leydig cells (FLCs) and peritubular myoid cells (PMCs) [48]. By E13.5, the male gonad is composed of well-defined testis cords and interstitial space. The testis cords comprise germ cells surrounded by Sertoli cells, PMCs and ECM to provide structural support. The interstitium contains the FLCs which will secret testosterone for virilization. Between E13.5 and E14.5, germ cells in the XY gonad undergo mitotic arrest, whereas germ cells in the XX gonad enter meiosis triggered by retinoid acid [49, 50].
Role of SOX9 in fetal testis development
Heterozygous mutations in human SOX9 lead to Campomelic dysplasia (CMPD), with associated partial or complete XY sex reversal in around 75% of cases [2, 3, 51]. Thus, SOX9 is an important factor during human testis determination. In mice, conditional homozygous inactivation of Sox9 in the XY gonads results in complete male-to-female sex reversal. Sox9 knockout males show lack of Sertoli cells and testis cords, and Leydig cells. Also, Sox9 mutant XY gonads show up-regulation of the ovarian markers Wnt4 (Wnt family member 4) and Foxl2 (forkhead box protein L2), and the presence of meiotic germ cells [5, 52]. These data demonstrate that SOX9 is essential for Sertoli cell differentiation and testis determination in mice [5, 52]. SOX9 directly regulates the expression of many target genes in Sertoli cells that have important functions during sex determination and differentiation such as Ptgds (prostaglandin D2 synthase), Fgf9 (fibroblast growth factor 9), Amh (anti-Müllerian hormone), Dhh (desert hedgehog), Cyp26b1 (cytochrome P450 family 26 subfamily B member 1), Sox8, Sox9, and Foxl2. These and other genes are discussed below, classified into groups according to their cellular function.
Repression of the ovarian pathway
The ectopic expression of the ovarian fate markers Wnt4 and Foxl2 [53] in Sox9 knockout gonads suggests that SOX9 prevents granulosa cell differentiation. Sertoli cell differentiation is promoted by repressing these genes. SOX9 inhibits Wnt4 via FGF9. In XY gonads, SOX9 upregulates Fgf9 transcription [54], and it does so directly [55]. In turn, FGF9 represses Wnt4 transcription, as shown by ectopic addition of FGF9 to XX mouse gonadal cultures [54]. In contrast, ChIP-Seq experiments show that SOX9 binds to the Foxl2 promoter in the fetal mouse testis, and SOX9 can reduce the basal activity of the Foxl2 promoter in a mouse Sertoli cell line MSC1 cells [56]. SOX9 also binds to the FOXL2 locus in the bovine testis, supporting a direct repression of Foxl2 by SOX9 [55].
Activation of the SoxE genes Sox8 and Sox10
Sox8 and Sox10 are potential target genes of SOX9 as they are down-regulated in Sox9 knockout mice [52, 55]. Sox8 and Sox9 double-mutant mice show more severe phenotypes than Sox8 or Sox9 single knockouts during testis differentiation. This implies that they act redundantly to trigger and maintain the expression of downstream genes in the testis [57]. Similarly, SOX10 can activate transcriptional targets of SOX9, and ectopic expression of Sox10 in the developing XX gonads can cause sex reversal. This explains its ability to direct male development [58]. ChIP-Seq demonstrates that SOX9 binds to the Sox8 promoter in the fetal mouse testis, and SOX9 can up-regulate endogenous Sox8 expression and transactivate the Sox8 promoter in MSC1 cells [56]. SOX9 also binds to the SOX8 locus in the bovine testis [55], indicating a direct regulation of Sox8 by SOX9. ChIP-Seq also shows SOX9 binding at the Sox10/SOX10 loci in the fetal mouse and bovine testis [55, 56].
SOX9 expression maintenance
Continuous and sufficient SOX9 expression is required for Sertoli cell differentiation and testis development. There are at least three ways to maintain SOX9 expression in Sertoli cells. The cell-autonomous Sox9 auto-regulation via SOX9 and steroidogenic factor 1 (SF1), the FGF9/FGFR2-mediated pathway, and the PTGDS/Prostaglandin D2 (PGD2)-mediated pathway. SOX9 maintains its own expression by directly regulating the expression of Ptgds, Sox9 (see “Regulation of the SOX9/Sox9 gene”) and possibly of Fgf9. In XY Sox9 knockout mice, Ptgds expression is almost absent [59]. Conversely, XY Ptgds knockout gonads show reduced Sox9 expression levels and a delay in testicular differentiation [59]. In vitro studies show that SOX9 can transactivate the Ptgds promoter through binding to a paired SOX-site within the promoter of Ptgds as a homodimer [60]. ChIP assays confirm that SOX9 can also bind to the paired SOX binding sites in the Ptgds promoter in vivo. These findings imply that Sox9 and Ptgds form a positive feed-forward loop which maintains Sox9 expression in the XY gonads. SOX9 is essential for Fgf9 expression. In XY Sox9−/− gonads at E11.5, Fgf9 expression is reduced or absent [54]. In turn, FGF9 is necessary for maintaining Sox9 expression in Sertoli precursor cells at E12.5. Thus, Sox9 and Fgf9 generate a positive feed-forward loop in XY gonads and both genes are required for somatic cell proliferation. Support for Fgf9 being a direct target of SOX9 comes from the SOX9 ChIP-Seq studies in mouse and bovine testes, which show that SOX9 binds to the Fgf9/FGFG9 locus in both species [55].
Sertoli cell driven germ cell and Leydig cell differentiation
In XY Sox9 knockout testes, germ cells are undergoing meiosis, and Leydig cells do not form, like in XX gonads [5, 52]. SOX9 might directly control Cyp26b1 and Dhh gene expression in Sertoli cells to regulate the fate of germ cells and Leydig cell differentiation, respectively. CYP26B1 prevents premature entry of XY germ cells into meiosis through degradation of retinoic acid [50]. In Sox9 or Sf1 conditional knockout XY gonads, Cyp26b1 expression is significantly decreased at E13.5, indicating that SOX9 and SF1 are involved in positive regulation of Cyp26b1 expression [61]. Indeed, SOX9 and SF1 can synergistically up-regulate endogenous Cyp26b1 expression in the mouse Leydig cell line TM3 in a dose-dependent manner. In addition, SOX9 can potently activate the Cyp26b1 promoter in MSC1 cells and binds to the Cyp26b1 promoter in vivo in the fetal mouse testis [56]. SOX9 also binds to the CYP26B1 locus in the bovine testis. These data support a direct role for SOX9 in the regulation of the Cyp26b1 gene.
In humans, mutations in DHH gene cause 46, XY gonadal dysgenesis [62]. DHH promotes fetal Leydig cell differentiation and regulates peritubular myoid cell differentiation and male fertility in the mouse [63]. SOX9 together with SF1 can strongly activate the Dhh promoter in MSC1 cells, and SOX9 binds to the Dhh promoter in the fetal mouse testis [56]. ChIP-Seq also shows that SOX9 binds to the DHH locus in the bovine testis [55], supporting a direct regulation of Dhh by SOX9.
Cerebellin 4 precursor (Clbn4) is also likely to be target gene of SOX9 in the fetal testis, but its testicular function is yet to be elucidated. CBLN4 may direct the differentiation of other cell types in the developing testis as a paracrine factor produced by Sertoli cells [64]. Cbln4 expression is reduced in XY Sox9-deficient mice at E13.5, whereas overexpression of Sox9 in XX mice upregulates Cbln4 expression. ChIP analysis shows that SRY and SOX9 can bind to the Cbln4 enhancer fragment both in vitro and in XY gonads at E11.5 in vivo, indicating a joint function for the regulation of Cbln4 male-specific expression [64].
AMH and Müllerian duct regression
The Müllerian ducts develop into the fallopian tubes, the uterus, and part of the vagina in females. AMH is secreted by Sertoli cells in the male gonads, causing the regression of the Müllerian ducts. There is strong evidence that SOX9 directly regulates the Amh gene to regress the Müllerian ducts in males. In XY Sox9 knockout mice, the Müllerian ducts show no signs of degeneration at E15.5, consistent with the absence of AMH protein expression at E13.5 [5]. Arango et al. [65] have shown that when the SOX9-binding site within the mouse Amh promoter is homozygously mutated, Amh transcription is not initiated, resulting in males with both male and female internal genitalia. In addition, ChIP-Seq demonstrates that SOX9 binds to the Amh/AMH locus in mouse and bovine testes, supporting a direct regulation of Amh by SOX9 [55].
Adult Sertoli cell function and spermatogenesis
To illustrate the role of SOX9 after the sex determination stage, AMH-Cre;Sox9flox/flox mice were generated with conditional inactivation of Sox9 in Sertoli cells at E13.5 [57]. AMH-Cre;Sox9flox/flox show normal fetal testis development but progressive seminiferous tubule and spermatogenesis defects, and infertility at about 5 months. Two likely SOX9 target genes with important roles mainly during spermatogenesis are ETV5 (Ets variant gene 5) and GDNF (glial cell-line derived neurotrophic factor). ETV5 is a transcription factor that plays an important role in maintaining fertility. Mice lacking Etv5 lose maintenance of spermatogonial stem cell self-renewal, leading to progressive germ cell loss and a Sertoli-cell-only syndrome [66]. In XY Ck19-Cre;Sox9flox/flox knockout mice, Etv5 expression is decreased. Furthermore, ChIP assays show that SOX9 binds directly to a conserved SOX-binding site within the regulatory region of ETV5 [67]. GDNF is a growth factor produced by Sertoli cells which regulates the cell fate decision of spermatogonia and spermatogonial self-renewal [68]. Gdnf expression is reduced in XY Sox9 knockout gonads [57] and ChIP-Seq on fetal mouse testes identified that both SRY and SOX9 bind to the Gdnf promoter [55, 56].
Nervous system
CNS development and SOX9 expression
The central nervous system (CNS) develops from a neural plate that folds to form a neural groove and eventually the neural tube (Fig. S1C). Neural stem cells (NSCs) are a subtype of progenitor cells in the nervous system that can differentiate into both neurons and glial cells. Oligodendrocytes and astrocytes are the two main types of glial cells in the CNS. Different types of neurons and glial cells are generated in different domains of the ventricular zone. pMN and p2 domains first generate motoneurons and V2 interneurons, respectively, and later there is a switch to the generation of oligodendrocytes and astrocytes, respectively.
In mice, Sox9 is weakly expressed in cells of the ventral neural tube at E9.5 [69]. At E10.5, SOX9 expression is detectable in a few SOX2-positive neuroepithelia throughout the ventricular zone and increases in many cells over the next 24 h [69]. Concurrent with this, multipotent NSCs appear in large quantity after E10.5 [6]. At E12.5, SOX9-positive oligodendrocyte progenitors appear at the border of the ventricular zone, indicating a switch from neurogenesis to gliogenesis [69]. The number of SOX9-expressing cells increases in the surrounding parenchyma while decreases in the ventricular zone, accordant with the reduction of the ventricular zone and neuroepithelial cells. Sox9 expression is also detected in the migrating, actively proliferating oligodendrocyte progenitors, but downregulated in oligodendrocytes at the onset of terminal differentiation [69].
Induction and maintenance of neural stem cells
SOX9 is required for induction and maintenance of neural stem cells [6]. Loss of Sox9 in the CNS from E11.5 can significantly reduce neurosphere formation which is a widely used in vitro culture system to quantitate the frequency of NSCs. In contrast, overexpression of Sox9 can induce precocious neurosphere formation from embryonic CNS. In addition, the removal of SOX9 leads to NSCs loss of multipotency.
Glial specification
SOX9 is required for the switch from neurogenesis to gliogenesis [69]. Specific deletion of Sox9 from E10.5 in the CNS in Nestin-Cre;Sox9flox/flox mice causes a reduction in oligodendrocyte precursors at E12.5, whereas the number of motoneurons is increased. In addition, at E14.5 and E16.5, neural tissue in Sox9 null mice contains virtually no astrocytes, but more V2 interneurons are present compared with controls [6]. These data indicate that the generation of oligodendrocyte precursors from the pMN domain and astrocyte precursors from other domains of the ventricular zone including the p2 domain is disrupted in the absence of Sox9, and that stem cells in these domains continue to produce neurons.
Astrocyte differentiation
After glial specification, Sox9 expression is maintained in the astrocyte lineage [69]. SOX9 induces the crucial transcription factor NFIA (nuclear factor-I A) by binding to its enhancer element and then cooperates with this factor to mediate the initiation of gliogenesis [70]. After glial initiation, SOX9 and NFIA form a complex and cooperatively activate a set of astrocytic genes. These include Apcdd1 (APC down-regulated 1), Mmd2 (monocyte to macrophage differentiation associated 2) and Zcchc24 (zinc finger CCHC-type containing 24), by binding to their promoter regions. Additionally, SOX9 positively regulates the expression of Nfe2l1 (nuclear factor erythroid-2-like 1), a transcription factor gene that promotes glial maturation, by binding to its promoter in vitro [71]. Thus, SOX9 not only plays a role in astrocyte specification, but also in its differentiation and maturation.
Oligodendrocyte differentiation
During oligodendro-gliogenesis, SOX9 and NFIA are required for oligodendroglial specification in the pMN domain of the ventricular zone [70]. After specification, Sox9 is down-regulated in the oligodendrocyte precursors emigrating from the pMN domain. SOX9 is important for oligodendroglial specification and SOX10 is essential for oligodendrocyte terminal differentiation [72]. Intriguingly, there is a time between specification and terminal differentiation when these two SOXE proteins can work together to regulate Pdgfra (platelet-derived growth factor receptor A) for cell survival, proliferation and migration [73].
Retina
Retina development and SOX9 expression
Mouse retina development begins at around E9.5 when the early retina is populated by a pool of multipotent retinal progenitor cells (RPCs) (Fig. S1D) [74]. Six types of neurons and one type of glial cell within the retina arise from the RPCs. During retinogenesis, ganglion cells are first to differentiate with Müller glial (MG) cells the last.
Sox9 is expressed in the proliferating, multipotent progenitors throughout retinogenesis and then exclusively in Müller glial cells and retinal pigment epithelia (RPE) until adulthood [7, 75]. At E11.5, Sox9 is expressed in the progenitors at a low level in the center of the retina, while highly in the RPE [74, 75]. At E13.5, Sox9 expression is clearly observed within the neuroblastic layer of RPCs, and gradually increased in the proliferating RPCs until the postnatal day 1 (P1). From P3, some RPCs exit cell cycle and begin to differentiate, and Sox9 is down-regulated in the differentiating populations but remains in Müller glial cells and RPE [7, 75].
SOX9 functions
In mice with retinal-specific inactivation of Sox9 starting from E10.5 (Chx10-Cre;Sox9flox/flox), MG cells are absent in Sox9-deficient retinal areas, as evidenced by loss of MG markers p27Kip1 and glutamine synthetase (GS) [7]. In addition, knockdown of Sox9 in retina organ cultures prepared from E17.5 mouse embryos reduces the number of MG cells and increases the relative proportion of rod photoreceptors, indicating that SOX9 is required for MG cell specification [75].
Considering continuous Sox9 expression in the RPE, SOX9 is important for RPE differentiation and maturation [76, 77]. RNA-Seq analyses of mouse RPE reveal that genes related to visual perception and transporter activity are up-regulated while genes related to cell proliferation are down-regulated at P5 compared to E15.5 [76]. When Sox9 is inactivated in the developing RPE in Dct-Cre;Sox9flox/flox mice from E11.5, RPE maturation related genes are down-regulated, indicating that SOX9 plays a role in regulating the late stages of RPE differentiation [76, 78]. Furthermore, Sox9 is expressed in the nuclei of mature RPE cells and regulates the visual cycle genes during postnatal stages [77].
Although Sox9 expression is not detected in choroid, SOX9 is essential for choroid vasculature development via regulating angiogenesis genes such as vascular endothelial growth factor A (Vegfa) and angiopoietin-like 4 (Angptl4) in RPE [76]. During choroid development, Vegfa, Vegfb and Angptl4 expression is progressively increased in the mouse RPE and also in RPE generated from human embryonic stem cells [76]. In contrast, angiogenesis genes are down-regulated in the developing RPE when Sox9 is conditionally deleted, leading to poor vasculature and little pigmentation [76, 78].
SOX9 targets
SOX9 regulates genes of the visual cycle, as specific deletion of Sox9 in mouse RPE leads to decreased expression of several visual cycle genes, especially Rpe65 (retinal pigment epithelium 65) and Rgr (retinal G protein-coupled receptor) [77]. Promoter-luciferase assays reveal that SOX9 and OTX2 (orthodenticle homeobox 2) synergistically activate the human RPE65 and RLBP1 (retinaldehyde binding protein 1) promoters. Moreover, SOX9 and OTX2 bind to the promoter regions of RPE65, RLBP1, and RGR in bovine RPE and human fetal RPE cells. SOX9 ChIP-Seq analysis in differentiated hES-RPE revealed that several SOX9 target genes are involved in the early stages of age-related macular degeneration (AMD) [76]. Seven out of these eight genes were also downregulated in the Dct-Cre;Sox9flox/flox mouse. ChIP-PCR assays elucidated that SOX9 and PAX6 (paired box protein 6) cooperatively bind to the first exon of ANGPTL4 in hES-RPE [76].
Lung
Lung development and SOX9 expression
The lungs and trachea arise from the anterior foregut endoderm (Fig. S1E). Morphogenesis of the respiratory system begins around E9.0 when the transcription factor Nkx-2.1 is expressed in a subset of the ventral foregut endoderm cells in mice. By E9.5, the Nkx-2.1-positive epithelial cells evaginate the surrounding mesenchyme to form two primary lung buds and the trachea. Between E9.5 and E12.5, the primary lung buds continue to proliferate and invade the mesenchyme. The trachea gradually separates from the esophagus. During the pseudoglandular stage of fetal lung development (E12.5–E16.5), the primary lung buds undergo branching morphogenesis to generate a tree-like network of airways. Two distinct progenitor cell lineages develop along the proximal–distal axis with SOX2 marking the proximal progenitors, while SOX9 and ID2 (inhibitor of DNA binding 2) marking the distal progenitors. The proximal progenitors give rise to airway neuroendocrine cells, secretory cells, ciliated cells and mucosal cells. The distal progenitors give rise to type 1 and type 2 alveolar epithelial cells (AEC1 and AEC2). During the canalicular (E16.5–E17.5) and saccular (E18.5–P5) stages, branching morphogenesis ceases and the terminal branches expand to form epithelial sacs which later develop into alveoli.
Sox9 expression can be detected in the distal epithelial progenitors during branching morphogenesis (E11.5–E16.5) in mice. After E16.5, Sox9 is down-regulated, coincident with the initiation of AEC1 and AEC2 differentiation [8]. Sox9 is also expressed in a population of mesenchymal cells, which surround the trachea, bronchi, and bronchioles, and that will develop into the future cartilage rings [79].
SOX9 functions
SOX9 is essential for branching morphogenesis and alveolar epithelial differentiation [8, 80]. SOX9 functions downstream of FGF10/FGFR2b to promote branching and suppress premature initiation of alveolar differentiation. Specific deletion of Sox9 using Shh-Cre in the lung epithelium before E12 leads to smaller lungs and fewer and dilated airway branches [80]. Conversely, Sox9 overexpression leads to large and cyst-like structures at the distal epithelial branch tips [8]. Microarray analysis of control and Sox9 mutant lungs identifies several genes that are down-regulated upon Sox9 deletion, such as Clusterin (Clu) and Melanoma inhibitory activity (Mia) which are related to lung development. Fgf10 expression is up-regulated in the Sox9-deficient lung, implying that SOX9 controls epithelial feed-back regulation of Fgf10 in the mesenchyme. Moreover, SOX9 prevents premature differentiation of the lung epithelium. Inactivation of Sox9 causes precocious differentiation of AEC2, whereas Sox9 overexpression inhibits terminal differentiation of distal lung progenitors [8]. Genes expressed by differentiated alveolar cells, such as Sftpb (surfactant protein B), Sftpc (surfactant protein C), Lamp3 (lysosomal associated membrane protein 3), Napsa (napsin A aspartic peptidase), and Ctsh (cathepsin H), are up-regulated in Sox9-deficient lungs [80]. These results suggest that SOX9 functions to maintain the undifferentiated status of distal lung progenitors.
SOX9 also plays multiple roles in regulating proliferation, differentiation, ECM organization, cell polarity and migration during lung development. Deletion of Sox9 using Shh-Cre results in cytoskeletal disorganization, ECM defects, and abnormal epithelial movement [8]. Microvilli are common extensions of the apical surface of epithelial cells and are uniform in size and shape. Sox9-mutant epithelial cells show reduced or absent microvilli and a rounded apical surface [8]. The control epithelial cells have a flat and uniform basal surface, while the Sox9-mutant epithelial cells have a rounded and irregular basal surface. Strikingly, E-cadherin immunofluorescence shows that SOX9 does not affect apical-basal cell polarity [8, 80]. SOX9 also regulates ECM genes in the lung epithelium. Two ECM proteins, COL2A1 and Laminin, are disrupted when Sox9 expression is altered. SOX9 directly regulates Col2a1 by binding to its consensus binding site, and loss of Sox9 leads to reduced expression of Col2a1/COL2A1 mRNA and protein in lungs [8]. Sox9 knockout and SOX9-overexpressing lungs show mis-localized and reduced Laminin staining at the basement membrane, respectively. However, these defects may be indirect, as no Laminin mRNA levels change is found by qRT-PCR. In addition, Sox9-deficient epithelial cells show reduced cytoskeleton marker acetylated tubulin along the basal surface. The ECM and microtubule dynamics are strongly associated with cell migration. In vitro assays of E12.5 isolated lung epithelial buds demonstrate that loss of Sox9 indeed inhibits cell migration [8]. Taken together, SOX9 plays multiple roles in regulating proliferation, differentiation, ECM organization, cell polarity and migration during lung development.
SOX9 targets
Based on the SOX9 functions mentioned above, the potential SOX9 targets in the lung could be grouped into branching-related genes and alveolar epithelial differentiation genes [80]. However, these potential SOX9 targets are only inferred from the microarray expression analyses of the control and Sox9 knockout fetal lungs. Whether SOX9 binds to the regulatory regions of these potential targets has yet to be demonstrated. Another group of SOX9 targets in the lung are ECM genes such as Col2a1 and Laminin [8], which are considered SOX9 targets in various tissues.
Heart valve
Heart valve development and SOX9 expression
Heart valves arise from endocardial cushions in the atrioventricular canal (AVC) and outflow tract (Fig. S1F). Endocardial cushion formation is initiated with endothelial-to-mesenchymal transformation (EMT) of endocardial cells and expansion of the ECM called cardiac jelly. The newly transformed mesenchymal cells continue to proliferate, differentiate and remodel to eventually form the mature heart valve leaflets and supporting chordae tendineae. Sox9 is directly activated in endocardial cells by Notch signaling which induces EMT at E9.5 in mice during early valve development [80]. By E12.5, SOX9 is highly expressed in the newly transformed mesenchymal cells throughout the developing valves [9].
SOX9 functions
SOX9 is essential for endocardial cushion formation and maturation. Mice with germline inactivation of Sox9 show hypoplastic endocardial cushions whose mesenchymal cells fail to complete EMT, and mice die at E11.5–E12.5. This indicates that SOX9 plays a role in the early stage of endocardial cushion formation [9]. This is further validated in Tie2-Cre;Sox9flox/flox mice, with conditional inactivation of Sox9 in endocardial mesenchymal cells. The Sox9 knockout mice die between E11.5 and E14.5, and manifest with hypoplastic endocardial cushions, reduced proliferation of endocardial mesenchymal cells and altered ECM deposition [81]. In addition, Col2a1-Cre;Sox9flox/flox mice in which Sox9 is inactivated in a subpopulation of valve cells during later stages of remodeling die at birth. Mutant mice show thickened valves, reduced expression of cartilage-associated proteins and abnormal ECM patterning [81]. Together, these studies support the idea that SOX9 is required early in valve development for expansion of the heart valve progenitor cell pool after initiation of EMT, and later for normal expression and organization of the ECM.
SOX9 targets
SOX9 binds to the promoters or potential regulatory regions of proliferation genes including Cops5 (COP9 signalosome subunit 5), Junb, Fosl1, Fosl2, Fos, Srpk2 (SRSF protein kinase 2), Akt2, Eed (embryonic ectoderm development), Hdac1 (histone deacetylase 1), and Hdac2 in AVC and limb at E12.5, as identified by ChIP-Seq [82]. This illustrates the common function of SOX9 in regulating cell proliferation across different cell types and many developing tissues. In mice, SOX9 is also needed for expression of ErbB3 (erb-b2 receptor tyrosine kinase 3), as shown by the lack of ErbB3 expression in Sox9-deficient endocardial mesenchymal cells [9]. ERBB3 is a member of the epidermal growth factor receptor tyrosine kinase family and is required for endocardial cushion mesenchymal cell proliferation and myocardium differentiation [83]. Loss of Sox9 led to altered ECM deposition and organization. SOX9 controls the expression of Hapln1 (hyaluron and proteoglycan link protein 1), Acan, Col2a1, Eln (elastin), Postn (periostin) and other collagens in the developing valve [82]. ChIP-Seq and RNA-Seq analyses of control and Tie2-Cre;Sox9flox/flox mice identified several transcription factors as SOX9 targets that participate in heart valve development [82], including Mecom (MDS1 and EVI1 complex locus protein) [84], Lef1 (lymphoid enhancer binding factor 1) [85], Pitx2 (pituitary homeobox 2) [86], Hand2 (hand and neural crest derivatives expressed 2) [87], Twist1 (twist-related protein 1) [88] and Sox4 [89]. Among these, SOX9 directly regulates the expression of Mecom by binding to its enhancer [82]. The ChIP-Seq study also determined other SOX9 AVC-specific targets, which are categorized into genes involved in DNA binding, cardiac neural crest cell development, and ascending aorta morphogenesis.
SOX9 can also act as a transcriptional repressor during valvulogenesis. Using ChIP-Seq and RNA-Seq, 58 genes were identified as being both bound and repressed by SOX9 in the E12.5 heart valve, including Junb, Fos, Bhlhe40 (basic helix-loop-helix family member e40) and Ddit3 (DNA-damage inducible transcript 3). The authors further demonstrated that SOX9 inhibited the activity of the Junb promoter in vitro. Another study showed that SOX9 binds and represses transactivation of the osteogenic glycoprotein Spp1 (secreted phosphoprotein 1) in maturing heart valves and chondrocytes to prevent pathologic matrix mineralization [90].
Pancreas
Pancreas development and SOX9 expression
Mouse pancreas development begins when two dorsal and ventral buds of the foregut endoderm emerge at the boundary between the stomach and duodenum at around E9 (Fig. S1G). During the primary transition (E9.5-E12.5), pancreatic progenitors rapidly proliferate and increase the size of the two buds, where tubulogenesis and patterning occur concomitantly. The two pancreatic buds fuse into a unified organ at the end of the primary transition. The pancreatic epithelium is segregated into two domains. The distal tip domain gives rise to endocrine, ductal and acinar cells while the proximal trunk domain forms ductal and endocrine cells. During the secondary transition (E13.5-birth), the pancreatic epithelium undergoes branching morphogenesis. Simultaneously, the differentiation of endocrine and exocrine cells take place. By the end of the secondary transition, the pancreas has acquired the topological organization of the mature organ while functional maturation continues postnatally.
SOX9 is first detected at E9 and colocalizes with the pancreatic progenitor marker PDX1 (pancreatic and duodenal homeobox 1, also known as MODY4) within the foregut endoderm [10]. From E10.5 to E12.5, Sox9 remains strongly expressed in all PDX1 + pancreatic epithelial cells, showing that Sox9 is expressed in pluripotent pancreatic progenitors. At E14, Sox9 expression continues in a subpopulation of PDX1 + epithelial cells located in the central epithelial cords. At E15.5, Sox9 expression is restricted to a subset of ductal epithelial cells while absent from committed endocrine progenitors. From E16.5, Sox9 expression is significantly downregulated and remains exclusive to a small subset of the ductal cells and centroacinar cells from E18.5 to adulthood. SOX9 expression is also detected in human pancreas during embryogenesis [91]. SOX9 is strongly expressed in the pancreatic primordium at 32 days post-conception (dpc) and remains high at later embryonic stages (52 dpc). Following embryogenesis, SOX9 expression declines in the fetal pancreas and particularly in the islets at 10 weeks gestation. From 14 weeks gestation on, SOX9 is substantially down-regulated and absent from the developing islets. This implies that mouse and human Sox9/SOX9 show a similar expression pattern during pancreatic endocrine differentiation.
SOX9 functions
SOX9 maintains pluripotent pancreatic progenitors by stimulating their proliferation and survival and is also required for the generation of endocrine progenitors [92]. Endocrine and exocrine progenitors are specified within the SOX9-positive epithelial cords. In Pdx1-Cre;Sox9flox/flox mice with Sox9 inactivated in pancreatic progenitors, the pancreas displays a reduced PDX1 + progenitor cell pool attributable to reduced cell proliferation and increased apoptosis at E11.5 [10]. In the absence of SOX9, pancreatic cells show reduced Hes1 (hairy and enhancer of split-1) expression at E11.5, an important marker of multipotent, Notch-responsive and exocrine-restricted progenitors [93]. When Pdx1-Cre mediates deletion of a single Sox9flox allele, mice show about 50% reduction in islet mass, while the islet architecture and the exocrine compartment is normal at E18.5 [92]. This shows that SOX9 is required for the specification of endocrine progenitors, whereas it is dispensable for the generation of exocrine progenitors. Another study shows that Sox9 haploinsufficiency causes 50% reduced β-cell mass at birth and glucose intolerance during adulthood [94]. This is consistent with a previous study of CMPD patients with heterozygous SOX9 mutations who show abnormal islet cell morphology and reduced expression of hormone and β cell markers [91]. These findings indicate that SOX9 plays a similar role during pancreas development in humans and mice.
SOX9 is essential for repressing hepatic and intestinal genes in the pancreatic domain. Microarray analysis on E12.5 Pdx1-Cre;Sox9flox/flox and control pancreata shows up-regulation of α-fetoprotein (AFP) and Albumin, markers of liver cells, verified by qRT-PCR analysis. This indicates that Sox9 ablation results in a pancreas-to-liver fate conversion [95]. Conditional deletion of Sox9 at E11.5 [96] also induced a pancreatic-to-hepatic fate switch, suggesting that the competence window for pancreatic-to-hepatic fate switch in Sox9-deleted progenitors closes by E12.5 [95].
Primary cilia are cellular organelles extending from the apical membrane of epithelial cells. In the pancreas, cilia are found in duct and islet cells and are essential for pancreatic organization and function. Loss or disruption of cilia causes ductal abnormalities, polarity defects and dysregulated β-cell function [97]. Pancreatic primary cilia are markedly reduced in Sox9-deficient embryos and adults, showing that SOX9 is essential for primary cilia formation in the pancreas [98].
SOX9 targets
SOX9 maintains both the pancreatic progenitor identity and expansion through a FGF10/FGFR2b/SOX9 feed-forward loop [95]. Mesenchymal FGF10 binds to FGFR2b of pancreatic progenitors and activates Sox9 expression, which in turn, activates expression of Fgfr2b, thereby maintaining FGF10 receptivity of pancreatic progenitors.
SOX9 activates the pro-endocrine gene Neurog3 (neurogenin 3) cell-autonomously in bipotent progenitors which can develop either into endocrine cells or ductal cells [98]. NEUROG3 in turn represses Sox9, committing the bipotent progenitors to the endocrine program. ChIP assays confirmed that SOX9 binds to the Neurog3 promoter in both E15.5 mouse pancreata [92] and the mouse pancreatic duct cell line mPAC L20 [99].
SOX9 and PDX1 work together to activate a transcription factor network for pancreatic development and repress an intestinal program by co-occupying the regulatory sequences of particular transcription factor genes [100]. ChIP-Seq analysis of pancreatic progenitors derived from human embryonic stem cells (hESCs) showed that SOX9 and PDX1 are both recruited to regulatory regions of pancreatic transcription factor genes. These include PTF1A (pancreas associated transcription factor 1a), PAX6, and NEUROG3 and several intestinal transcription factor genes, such as CDX2 (caudal type homeobox 2), ONECUT-2 (one cut homeobox 2) and NKX6-3. Transcriptional profiling of pancreatic progenitors from compound Pdx1;Sox9 heterozygous mutant mice shows up-regulation of Cdx2, Onecut-2 and Nkx6.3, suggesting direct repression of intestinal cell fate regulators by PDX1 and SOX9 [100].
Following Sox9 deletion in mouse pancreatic progenitors by E10.5, PDX1 protein expression appears normal at E10.5 [10] but is dramatically down-regulated at E12.5 [94]. This suggests that SOX9 is required for the maintenance of Pdx1 expression. Conversely, there was no apparent change in SOX9 protein expression in the E10.5 Pdx1-deficient pancreas [95]. Therefore, neither PDX1 nor SOX9 is required for the initiation of each other’s expression, but there is evidence that they mutually reinforce expression of each other directly as PDX1 occupies SOX9 regulatory sequences and vice versa [100]. Thus, positive autoregulation of Pdx1 [101] and Sox9 [99] and the positive cross-regulatory loop between Pdx1 and Sox9 [94, 95] strengthen pancreatic development.
Bile duct
Bile duct development and SOX9 expression
Hepatocytes and biliary epithelial cells (cholangiocytes) arise from the same progenitors, hepatoblasts, during liver development (Fig. S1H). SOX9 expression is first detected in the hepatoblasts near the portal vein mesenchyme at E11.5 in mice [11]. These SOX9-positive hepatoblasts differentiate into cholangiocyte precursors and form the ductal plate which is composed of a single layer of SOX9-expressing cells encompassing the branches of the portal vein. SOX9 expression is then restricted to cholangiocytes. Therefore, SOX9 is the earliest marker of the biliary cell lineage. At E15.5, the ductal plate gradually forms the primitive ductal structure (PDS) which comprises the portal layer of SOX9-positive cholangiocytes and the parenchymal layer of SOX9-negative hepatoblasts [11]. The PDS matures to bile ducts which are surrounded by ECM and mesenchyme, and all cells lining the ducts become typical cholangiocytes.
SOX9 functions
SOX9 controls the timing of bile duct morphogenesis during development. Homozygous inactivation of floxed Sox9 alleles driven by Alfp1-Cre (Alfp1-Cre;Sox9flox/flox) in the mouse liver starting at E11.5 leads to delayed maturation of the PDS into bile ducts [11]. The expression of Laminin α5, which is secreted by cholangiocytes and needed for bile duct maturation, is absent in the Sox9-deficient liver [102]. Since Sox9 inactivation only causes mild and transient biliary defects, other members of the SOX family may compensate for the loss of SOX9. Combined ablation of Sox4 and Sox9 in the developing mouse liver shows significantly perturbed biliary differentiation both prenatally and postnatally [102]. In contrast, the loss of either Sox4 or Sox9 only induced delayed biliary differentiation, and cholangiocyte differentiation returned to normal after birth. Additionally, both SOX4 and SOX9 are required for establishing polarity of cholangiocytes. Inactivation of Sox4 causes loss of apico-basal polarity both at prenatal and postnatal stages, whereas inactivation of Sox9 results in the absence of polarization only at the fetal stage, and polarization is progressively restored after birth. Notably, Sox4;Sox9 double knockout mice show major polarity defects and irregular cell shape. The apical pole of cholangiocytes has a primary cilium. This functions to detect stimuli from bile and transmit signals into cells regulating several signaling pathways that are required for biliary development such as Notch, TGF-β and Hippo pathways. Sox4;Sox9 double knockouts show perturbed cilia formation leading to a reduced number of cilia after birth. In contrast, deletion of either Sox4 or Sox9 display normal cilia formation [102].
SOX4 and SOX9 cooperate to control the signaling pathways TGFβ and NOTCH which are involved in biliary development. Transforming growth factor β receptor II (TβRII) transmits mesenchymal TGF-β signals to hepatoblasts which differentiate into the cholangiocyte lineage, and ΤβRII then becomes repressed in cholangiocytes during duct maturation [11]. At E15.5, TβRII is repressed on the portal side of PDS in wild type but still detectable in all PDS cells in the absence of SOX4 and/or SOX9. At E18.5, TβRII is absent in wild-type cholangiocytes but still detectable in a subset of duct cells in the absence of Sox4 and in Sox4;Sox9 double knockout mice [102]. The NOTCH target Hes1 is not affected at E15.5 but is down-regulated in cholangiocytes at E18.5 in the absence of SOX4 and/or SOX9.
SOX9 targets
In Sox9 conditional knockout mice studies potential SOX9 targets may include genes associated with cholangiocyte differentiation and maturation [102]. RT-qPCR results of the control and Sox9 knockout liver show that the expression of the apical markers Osteopontin (Spp1) and Mucin-1 (Muc) is controlled by SOX9 and SOX4, respectively. In addition, SOX9 and SOX4 conjointly affect several signaling pathways controlling biliary development.
Intestine
Intestine development and SOX9 expression
The intestine develops from a flattened, tubular structure, and by E9.5 the gut tube is fully formed and becomes pseudostratified (Fig. S1I). Between E9.5 and E13.5, the intestinal epithelia undergo rapid proliferation, resulting in elongation and increase in circumference and lumen size. From E14.5, the villus morphogenesis begins to form villi and crypt structures. This is followed by epithelial cytodifferentiation into absorptive enterocytes, mucus-producing goblet cells and hormone-producing enteroendocrine cells at around E16.5 [103]. In the small intestine, the antimicrobial agents-secreting Paneth cells settle at the bottom of the crypt as terminally differentiated cells [12]. The mature intestinal epithelium can be divided into a proliferative compartment comprising the crypt of Lieberkühn, and a differentiated compartment comprising the villus in the small intestine and the luminal surface in the colon.
During intestinal development in the mouse, Sox9 is expressed in all epithelial cells at E13.5 [12]. At E15.5, Sox9 expression is detected in the rapidly proliferating progenitors, and gradually becomes restricted to intestinal stem cells, transit amplifying progenitor cells, Paneth cells, and enteroendocrine cells by adulthood.
SOX9 functions
Differential SOX9 levels mark two distinct populations of Sox9HI and Sox9LO cell types in the crypts [104]. Sox9HI cells represent mature enteroendocrine cells whereas, Sox9LO cells represent proliferative progenitor cells. Paneth cells also express a high level of SOX9 [105]. Overexpression of Sox9 in an intestinal progenitor cell line blocks proliferation and induces a transition from an epithelial cell morphology to a neuroendocrine morphology [104]. Moreover, in Sox9-deficient mice the proliferative compartment expands to occupy the whole crypt base, leading to crypt hyperplasia throughout the small intestine [106]. These data indicate that a low level of SOX9 promotes progenitor proliferation, whereas a high SOX9 level promotes cell maturation.
SOX9 is essential for the differentiation of Paneth cells and goblet cells [12, 106]. Specific deletion of Sox9 in mice using Villin-Cre (Vil-Cre;Sox9flox/flox) from E10.5 leads to a decreased number of both the Paneth and goblet cells, whereas the enterocyte or enteroendocrine cell populations are unaffected. In addition, overexpression of human SOX9 upregulates several Paneth cell markers such as LYZ (lysozyme), MM7 and ANGPT4 in vitro.
SOX9 targets
SOX9 maintains intestinal progenitor cells likely through direct repression of the differentiation genes CDX2 and MUC2 [107]. CDX2 is the master regulator of intestine development. ChIP-Seq analysis of hESC-derived pancreatic progenitors showed that SOX9 and PDX1 directly co-bind and repress intestinal cell fate regulators such as CDX2, ONECUT-2 and NKX6-3 to inhibit intestinal fate in pancreas [100]. Microarray analysis of E12.5 mouse pancreatic epithelia also identified that SOX9 and PDX1 co-regulate Cdx2, Onecut-2 and Nkx6.3 [100]. MUC2 is the main intestinal mucin covering the intestinal surface and an in vitro assay showed that SOX9 transcriptionally represses the MUC2 gene via an intermediate repressor protein in human intestinal adeno-carcinoma cells [107].
Prostate
Prostate development and SOX9 expression
The mammalian prostate arises from the urogenital sinus (UGS), which consists of an inner layer of UGS epithelium and an outer layer of UGS mesenchyme (Fig. S1J) [108]. Prostate development relies on androgen, produced by testes from E13-E14 in mice. Prostate morphogenesis starts as the epithelia buds first appear and penetrate the surrounding mesenchyme at E17.5. Then prostate organogenesis continues through birth and prepubertal stages until reaching maturation during puberty. The budding stage is followed by epithelial outgrowth and branching morphogenesis. During the canalization and differentiation stages, the epithelial cords form the ductal lumen and differentiate into luminal secretory cells and basal cells. The surrounding mesenchymal cells differentiate into smooth muscle and fibroblasts. The mature mouse prostate can be divided into four distinct lobes: anterior, dorsal, lateral and ventral. SOX9 is first detected throughout the UGS epithelium and in the mesenchyme near the epithelium at E15 [109]. When the prostatic buds appear at E17.5, Sox9 is strongly expressed in the prostatic epithelial cells [13]. During bud elongation, SOX9 expression is mainly found in the epithelium with strongest expression at the tips of the growing bud.
SOX9 functions
SOX9 plays an important role in the proliferation and branching activity of the prostate. Prostate specific deletion of Sox9 in Nkx3.1-Cre;Sox9flox/flox mice leads to a lack of ventral prostate development and abnormal anterior prostate differentiation [13]. Cell proliferation is significantly decreased in the ventral prostate but not in other lobes in the Sox9 mutant animals compared with controls at E18.5. During later stages of prostate development, SOX9 appears to be less critical. The anterior, dorsal and lateral prostatic lobes show seemingly normal histological structures and cytodifferentiation. The lack of an abnormal phenotype in the dorsal-lateral buds could be due to the inefficient early Sox9 inactivation in Nkx3.1-Cre;Sox9flox/flox mice [13].
To delete Sox9 throughout prostate development, a tamoxifen (TAM)-inducible ER-Cre;Sox9flox/flox conditional knockout system was utilized [110]. When Sox9 is ablated at E14.5, the UGS fails to initiate prostate development. When Sox9 is abrogated at E16.5, only a few prostate buds form. Thus, SOX9 is required for prostate organogenesis before the onset of androgen exposure at E16.5. However, SOX9 is dispensable for adult prostate maintenance.
Shh-Cre;Sox9flox/flox mice where Sox9 is specifically deleted from the UGS epithelia by E16.5 immediate prior to bud initiation, shows that SOX9 is important for bud elongation [109]. The number of prostatic buds is not changed while the length of buds is shortened in Sox9-deficient mice. Microarray analysis of the control and Sox9 mutant mice identified that cell migration is most severely impaired in the Sox9 knockouts. These results suggest that SOX9 is not required for prostatic bud initiation but is essential for bud elongation via promoting cell migration.
SOX9 targets
SOX9 regulates cell proliferation via several genes in the ventral prostate lobe. The expression of Nkx3.1, Shh, Fgfr2, and Spry2 (sprouty RTK signaling antagonist 2) genes are severely reduced in the Sox9 deficient mice at early stages [13]. Microarray analysis from E16.75 control and Sox9 mutant UGS epithelia identified a subset of SOX9-dependent genes such as Cdc42 (cell division control protein 42), Barx2 (BARX homeobox 2), Cxcr4 (C-X-C chemokine receptor type 4) and Col14a1 that mediate cell migration during prostatic bud elongation [109].
Hair follicle
Hair follicle development and SOX9 expression
In mice, hair follicle morphogenesis begins with hair placode formation at E14 in response to signals from neighboring epidermal cells and underlying mesenchymal cells (Fig. S1K). The epithelial cells proliferate, invaginate downward into the dermis and induce the formation of the dermal papilla (DP). The epithelial cells in close contact with DP constitute highly proliferative matrix cells at the base. Matrix cells continue to proliferate and, while moving upwards, differentiate into cells to form the hair shaft (HS) and the inner root sheath (IRS). The IRS is surrounded by the outer root sheath (ORS), which is contiguous with the epidermis. In the upper ORS and just below the sebaceous gland (SG), there is a region called bulge which contains hair follicle stem cells (HF-SCs) [111].
In adult mice, hair follicles cycle through phases of growth. In anagen, matrix cells undergo proliferation and differentiation leading to downgrowth of the hair follicle into the dermis and upward growth of the hair shaft. During catagen, the lower part of the hair follicle below the bulge undergoes apoptosis. This causes the retraction of the hair follicle and the dermal papilla to rest beneath the bulge. Finally, during telogen, the hair follicle becomes inactive, and the bulge anchors the old hair (club) before starting a new hair cycle. Hair follicle regeneration involves the stimulation of stem cells residing in the bulge [112]. Some HF-SCs exit the bulge and migrate toward the matrix where they become early progenitor cells and subsequently undergo proliferation and differentiation into a new hair follicle [113]. In mice, Sox9 expression occurs in hair placodes at the time of induction at E14.5 but later it can only be detected in the ORS and the bulge of the hair follicle [14]. In adult mice, Sox9 is mainly expressed in the quiescent HF-SCs residing in the bulge [20].
SOX9 functions
SOX9 is required for matrix cell proliferation and the maintenance of HF-SCs and ORS. Specific ablation of SOX9 in the epithelial layer of the developing mouse skin, starting at E14.5 and complete by P2, led to dramatically impaired proliferative capacity in the matrix cells [14]. Moreover, Sox9 mutant skin shows loss of the HF-SC marker CD34, loss of external hair, and acquirement of epidermal characteristics in ORS. To demonstrate whether SOX9 is required for hair placode induction, the upper lips of germline-specific E11.5 Sox9 knockout embryos were trans-planted under the kidney capsules of nude mice. Histological analysis of the transplants after 10 days showed that hair follicles developed, indicating that Sox9 is dispensable for early hair follicle formation [14].
SOX9 is critical for initial stem cell specification which occurs during the earliest stages of HF morphogenesis [114]. Furthermore, SOX9-expressing stem cells can contribute to all skin epithelial lineages and sebaceous glands. In the absence of SOX9, the early stem cell population never forms, and HF differentiation, sebaceous gland formation and epidermal wound-healing are severely impaired. Thus, SOX9 is required for the maintenance of ORS identity, the formation of the HF-SCs, the proliferation of matrix cells, and epidermal wound-healing, although SOX9 seems to be dispensable for initial hair induction.
When Sox9 is conditionally ablated in established adult mouse HF-SCs, they lose stemness. This leads to arrested HF downgrowth as hair cycle proceeds. Moreover, the Sox9-deficient HF-SCs showed signs of epidermal differentiation. These results suggest that SOX9 is required for maintaining adult HF-SCs identity [20]. Transcriptional profiling of wild-type and Sox9-deficient HF-SCs revealed that Sox9-deficient bulge cells lose the expression of HF-SC signature genes while they acquire the expression of some interfollicular epidermis signature genes. Together, these findings suggest that SOX9 is essential for adult HF-SC maintenance and suppression of epidermal differentiation in the bulge niche [20].
SOX9 targets
SOX9 plays a role in maintaining HF progenitor/stem cells and cell proliferation. However, little is known about SOX9 targets associated with these functions during fetal HF development. In the adult HF-SCs, SOX9 ChIP-Seq together with RNA-Seq data identified a short list of both SOX9-bound and activated genes. These include TF genes such as Lhx2 (LIM homeobox 2), Nfatc1 and Tcf7l2 (transcription factor 7-like 2), ECM genes such as Timp2 (TIMP metallopeptidase inhibitor 2), Wfdc3 (WAP four-disulfide core domain 3) and Col8a2, and genes encoding TGF-β/Activin signaling factors such as Sulf2 (sulfatase 2), Inhbb (inhibin subunit beta B) and Wwp2 [20].
Mechanisms underlying SOX9 common and unique roles
SOX9 acts commonly in roles like cell-fate determination, progenitor maintenance, and cell proliferation, yet also has unique roles within specific tissue. We propose that the mechanisms underlying the SOX9 common functions include control of broadly expressed genes, Sox9 autoregulation and common signaling pathways like FGF and Notch signaling.
In this section, we focus on the mechanisms by which the unique roles of SOX9 are achieved among different organs. The specificity of SOX9 roles and functions may be explained by the transcriptional regulation of the SOX9/Sox9 genes, the posttranslational modifications of the SOX9 protein, SOX9 binding partners, and enhancers of target genes.
Regulation of the SOX9/Sox9 gene
Enhancers
The large gene desert region upstream and downstream of SOX9/Sox9 contains regulatory sequences controlling SOX9/Sox9 tissue-specific expression patterns. Comparison of human and pufferfish genomic sequences highlighted eight highly conserved sequence elements (E1–E8) between 290 kb 5′ and 450 kb 3′ to Sox9. These were tested for enhancer activity in transgenic mice using a lacZ reporter gene [115]. The E3 enhancer, located 251 kb upstream of SOX9, drove lacZ expression specifically within cranial neural crest cells and the inner ear. The E1 enhancer, located 28 kb upstream of SOX9, controls lacZ expression in the node, notochord, gut, bronchial epithelium and pancreas. The E7 enhancer, located 95 kb downstream of SOX9, regulates lacZ expression in the telencephalon and midbrain [115].
Yao et al. [116] could show that the E4 sequence element (termed E250), located 250 kb 5′ to mouse Sox9, shows enhancer activity, driving lacZ reporter gene expression in condensed prechondrocytes but not later in the developing cartilage. They also identified two additional chondrocyte lineage enhancers. E195, located 195 kb 5′ to Sox9, drives lacZ reporter gene expression in proliferating chondrocytes. E84, located 84 kb 5′ to Sox9, shows reporter activity in differentiated chondrocytes but not in the non Sox9-expressing hypertrophic chondrocytes. These data suggests that during cartilage development endogenous Sox9 expression is regulated at least in part by the sequential activities of enhancers E250, E195 and E84. Moreover, a cartilage-specific enhancer (rib cage-specific enhancer, RCSE) was identified 1 Mb upstream of the Sox9 TSS, and CRISPR/Cas9-mediated deletion of RCSE leads to smaller rib cages in mice [117].
Microdeletions and translocations define a locus around 1.2–1.5 Mb upstream of SOX9 associated with Pierre-Robin Syndrome (PRS), a rare congenital condition characterized by a hypoplastic mandible. Several enhancers have been identified in this region which drive reporter gene expression in the fetal mouse mandibles, suggesting that SOX9-related PRS is caused by the removal of these enhancers [118, 119].
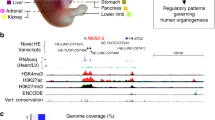
Gonad-specific enhancers of SOX9/Sox9 were identified by several studies. The 3.2 kb TES enhancer (Testis-specific Enhancer of Sox9), located − 13 to − 10 kb upstream of the Sox9 TSS, can fully recapitulate the gonadal expression pattern of Sox9 in the mouse [120]. Both SRY and SF1 bind to TES and drive Sox9 expression in Sertoli cells. Subsequently, SOX9 replaces SRY and interacts with the core element within TES (TESCO), forming a feed-forward regulatory loop to maintain its transcription level. In addition, four novel gonadal enhancers were identified in mouse, with Enh13 and Enh14 showing strong testicular expression [121]. Enh13 is located 565 kb upstream of Sox9 and, like TESCO, it is bound by SRY and SOX9 in vivo, suggesting that both TESCO and Enh13 are regulated by SRY and SOX9. CRISPR-mediated deletion of either TESCO, Enh13 or Enh14 in mice showed that Enh13 is a critical enhancer for testicular Sox9 expression and thus testis development. XY gonads lacking Enh13 only expressed 21% of wildtype Sox9 levels, leading to complete sex reversal. In contrast, XY gonads lacking TESCO or Enh14 still expressed 45% of or normal wildtype Sox9 levels, respectively, and developed as normal testes. Enh13 might also be important for human testicular SOX9 expression since it is located within a 32.5 kb locus, whose removal is associated with XY sex reversal [121]. By analyzing the genome data of DSD patients, three testis-specific enhancers eALDI, eSR-A and eSR-B 5′ of SOX9 were identified in humans which showed synergistic action to drive SOX9 expression in the testis [122].
While SOX9 autoregulation in the testis is mediated via TESCO and possibly also Enh13, in most other Sox9-expressing tissues SOX9 is directly autoregulated via the enhancer SOM, which is located 70 kb 5′ to Sox9. CRISPR-mediated deletion of SOM leads to an average reduction of 18–37% in Sox9 expression levels in the gut, pancreas, liver, lung, kidney and salivary gland [123].
Promoters
In addition to enhancers, other regulatory regions may include promoters, silencers, and 3D chromatin organization. Regulatory elements of the SOX9/Sox9 gene have been expertly reviewed elsewhere [124,125,126,127]. In brief, in vitro studies of chondrocytic and mesenchymal cell lines show that within the SOX9/Sox9 proximal promoter, functional binding sites occur for transcription factors hypoxia-inducible factor-1α (HIF1α) [128], Notch signaling mediator RBPJ [129], NF-κB subunit RELA [130], CREB and SP1 [131], bone morphogenetic protein 2 (BMP-2) [132], signal transducer and activator of transcription (STAT3) [133], and Yes-associated protein 1 (YAP1)/TEA domain transcription factor (TEAD) [134]. During testicular development, the Sox9 proximal promoter located − 193 bp to − 73 bp shows higher activity in the mouse testis than in the ovary and liver at E13.5 [135], implying the action of testis-specific factors. However, this Sox9 proximal promoter region shows only neural tube activity in vivo [115]. This suggests that important tissue-specific factors act on genomic regions outside the promoter, such as SOX9 and SF1 interacting with enhancers TESCO, eALDI, eSR-A and eSR-B [120, 122].
Topologically associating domains (TADs)
Topologically associating domains (TADs) are megabase-sized chromatin domains in which regulatory elements such as enhancers and promoters interact with one another more frequently than outside TADs [127]. TADs are separated by boundaries which harbor binding sites for zinc finger transcription factor CTCF and the cohesin protein complexes. The human and mouse SOX9/Sox9 loci contain two major TADs. One harbors the SOX9/Sox9 gene and the large gene desert that contains many enhancers deleted or duplicated in human disease [136]. The other TAD encompasses the genes KCNJ2/Kcnj2 and KCNJ16/Kcnj16 which is separated from the SOX9/Sox9 TAD by a boundary in the gene desert. By modelling Cooks syndrome, a congenital limb malformation, genomic duplications upstream of SOX9/Sox9 creates a new TAD (neo-TAD) containing both the Kcnj2 gene and the Sox9 regulatory region leading to limb malformation due to ectopic Kcnj2 expression [136]. In silico analysis of chromatin conformation of the SOX9 locus shows that SOX9 chromatin folding domains associate with real and putative regulatory elements of SOX9. Also, there is conservation of chromatin domain boundaries across multiple cell lines [137]. Detailed studies of SOX9/Sox9 chromatin organization await further chromosome conformation capture methods such as capture Hi-C and 4C-seq.
Epigenetic regulation of SOX9/Sox9
Expression of the SOX9/Sox9 gene may be controlled by epigenetic regulatory mechanisms, such as DNA methylation, histone acetylation and methylation. However, most epigenetic studies of SOX9/Sox9 have been in cancer tissues, rather than during organogenesis. For example, in pancreatic cancer, the NF-κB pathway is highly activated. Demethylation of an adjacent CpG island may enhance binding of NF-κB to the SOX9 promoter thereby increasing SOX9 expression [138]. Conversely, in bladder cancer [139] and melanoma [140], hypermethylation of the SOX9 promoter leads to SOX9 silencing.
In cartilage, DNA methylation regulates Sox9 expression [141]. During articular cartilage development, Wnt signals recruit DNA methyl transferase DNMT3A to the Sox9 promoter, leading to hypermethylation, and thus SOX9 downregulation in the limb bud mesenchymal cells [142]. In contrast, FGF signals block DNMT3A recruitment to the Sox9 promoter by promoting phosphorylation of DNMT3A. BMP-2 stimulates Sox9 expression during chondrogenesis in primary mouse embryo fibroblasts [143]. It increases the association of the transcription factor NF-Y with histone acetyltransferase p300, resulting in a complex binding to the Sox9 promoter. In addition, BMP-2 induces histone hyperacetylation and hypomethylation at the Sox9 proximal promoter, suggesting that BMP-2 affects Sox9 gene transcription through histone modification and chromatin remodeling.
Posttranscriptional regulation of SOX9
microRNA
SOX9 regulation by microRNAs, which act as repressors of gene expression, has been widely studied in different types of cancer, but less so during organ development. miR-124-mediated repression of SOX9 protein is important for neuronal differentiation in the adult mouse brain [144]. Moreover, miR-124 can repress both SOX9 translation and transcription in mouse ovarian cells [145]. Over-expression of miR-145 decreases expression of SOX9 protein and miR-145 inhibition significantly elevates SOX9 protein levels [146]. Thus, miR-145 is likely a key negative regulator of SOX9 during early chondrogenic differentiation. IL-1β-induced miR-101 binds to the 3’UTR of Sox9 in rat chondrocytes, leading to decreased expression of Sox9 mRNA and SOX9 protein and degeneration of cartilage ECM [147]. miR-1247 targets a highly conserved sequence in the SOX9 coding region to repress SOX9 protein expression [148]. miR-495 also directly targets the 3’UTR of SOX9 in human mesenchymal stem cells, leading to reduced SOX9 protein level and inhibition of chondrogenic differentiation [149].
Long non-coding RNA
Long non-coding RNAs (lncRNAs) often modulate gene expression in a tissue-specific manner. lncRNAs exceed 200 nucleotides but are not translated. Interactions between lncRNAs and SOX9 have been explored in the context of cancers [150]. The regulatory role of lncRNAs in organogenesis are less understood. LOC102723505 (also termed ROCR, regulator of chondrogenesis RNA) may act as an upstream regulator of SOX9 in chondrocytes [151]. ROCR is upregulated during chondrogenic differentiation, and upon depletion of ROCR via RNAi, SOX9 mRNA and SOX9 protein levels are significantly reduced, cartilage-specific genes are downregulated, and ECM formation is perturbed. Microarray analysis of mouse neonatal and adult testes shows that over 3,000 lncRNAs are differentially expressed, with many lncRNAs overlapping or adjacent to important transcription factors in testicular development, such as Sox9, Lhx1 and Wt1 [152].
Posttranslational modifications of SOX9
Posttranslational modifications (PTMs) affect the stability, intracellular localization, activity, and partner protein interactions of SOX9. PTMs of SOX9 in chondrogenesis is explored in detail by Lefebvre and Dvir-Ginzberg [125], and evolutionary conservation of PTMs by Vining et al. [153].
Serine phosphorylation
SOX9 is phosphorylated at three serines (S64, S181 and S211) by cyclic AMP-dependent protein kinase A (PKA) in chondrocytes [154, 155], and also at S181 by cGKII (cGMP-dependent protein kinase type II) [156]. Phosphorylation of SOX9 enhances its DNA-binding affinity and transcriptional activity on a Col2a1 chondrocyte-specific enhancer. In addition, the parathyroid hormone-related peptide (PTHrP) signal can increase SOX9 phosphorylation and transcriptional activity in the prehypertrophic zone of the growth plate to inhibit chondrocyte maturation. The same phosphorylated sites of SOX9 have been found in Sertoli cells, in which phosphorylation induces SOX9 nuclear translocation and increases its DNA binding affinity [157]. Notably, phosphorylation of SOX9 on S64 and S181 is required for Sox9-Snail2 interaction to trigger neural crest delamination in the developing chick neural tube [158]. Phosphorylation of S64 and S181 also facilitates SUMO-ylation of SOX9 [158] (see below).
Arginine methylation
Coactivator associated arginine methyltransferase 1 (CARM1), a member of the arginine-specific protein methyltransferases (PRTMs), methylates SOX9 at multiple arginine residues within the HMG domain in vitro and in vivo [159]. Methylated SOX9 then dissociates from beta-catenin which increases Cyclin D1 mRNA expression and thus chondrocyte proliferation. CARM1−/− mouse embryos die immediately after birth, have significantly delayed endochondral bone formation, and inhibited chondrocyte proliferation. These data indicate that SOX9 methylation plays an important role in chondrocyte proliferation.
Lysine acetylation and deacetylation
Acetylation and deacetylation of SOX9 affect cartilage-specific gene expression. TIP60 acetylates SOX9 at lysine 61, 253 and 398 residues. This leads to more diffuse nuclear localization of both proteins and subnuclear colocalization [23]. The transcriptional activity of SOX9 on Col2a1 is enhanced by TIP60. However, this effect is independent of enhanced acetylation of SOX9 by TIP60 as the SOX9 mutants K61/253/398A show enhanced transcriptional activity by TIP60 as well. On the contrary, sirtuin 1 (SIRT1) deacetylates SOX9 and increases its transcriptional activity on COL2A1 in human articular chondrocytes [160]. A further study reveals that deacetylation of SOX9 by SIRT1 promotes SOX9 nuclear translocation and enhances its binding to a -10 kb ACAN enhancer in adult human chondrocytes [161].
Lysine SUMO-ylation
Small ubiquitin-like modifier (SUMO)-ylation affects SOX9 transcriptional activity in a context-dependent manner. SUMO-ylation of both SF1 and SOX9 represses their synergistic transcription of the mouse Amh reporter gene in vitro but does not affect their DNA binding activity or interaction with each other [162]. The acetylated lysine residues of SOX9 (K61, K253 and K398) are also SUMO-ylation sites [23, 163]. Protein inhibitor of activated STAT (PIAS) proteins, which act as SUMO ligases, increase cellular SOX9 protein levels and SOX9-dependent transcription of a Col2a1 promoter-enhancer reporter [163]. Conversely, PIAS1 enhances the sumoylation of the mouse SOX9 protein but represses SOX9-dependent transcription of the Col11a2 enhancer reporter gene [164]. Intriguingly, in Xenopus, SUMO-ylation of SOX9 and SOX10 directs inner ear development, whereas non-SUMO-ylation promotes neural crest development [165].
Ubiquitination and UFMylation
SOX9 is also subject to ubiquitination, which results in degradation of SOX9 via the ubiquitin-proteosome pathway. E6-AP ubiquitinates SOX9 in vitro and in vivo, and SOX9 may be ubiquitinated by E6-AP in hypertrophic chondrocytes given the opposing distribution of SOX9 and E6-AP: with E6-AP high in hypertrophic chondrocytes, but low in prehypertrophic chondrocytes, and the converse for SOX9 levels [166]. Further, E6-AP-deficient mice have an accumulation of SOX9 in chondrocytes and the brain [166].
On the contrary, UFMylation, a posttranslational modification where a ubiquitin-like fold modifier 1 (UFM1) covalently binds to its targets, prevents SOX9 proteasomal degradation [167]. SOX9 UFMylation is likely regulated by the DDRGK domain-containing protein 1 (DDRGK1). In vitro assay shows that DDRGK1 directly binds to SOX9 and inhibits its ubiquitination and proteasomal degradation. In addition, Ddrgk1 deficient mice show decreased SOX9 protein levels and chondrogenic mesenchymal condensation. Furthermore, a loss-of-function mutation in the DDRGK1 gene has been identified in Shohat-type spondyloepimetaphyseal dysplasia (SEMD), a chondrodysplasia characterized by vertebral, epiphyseal, and metaphyseal abnormalities.
SOX9 and its transcriptional partners
SOX9 exhibits diverse functions in a vast number of tissues in part through forming complexes with functional partners to cooperatively activate and/or repress target genes (Fig. 2A). Target genes of SOX9 often harbor binding motifs for SOX9 partner proteins adjacent to the SOX binding sites to facilitate cooperative transcriptional activity. For example, in cartilage, SOX9 homodimer binds to the homodimer-binding sequences on the enhancer regions of chondrogenic genes such as Col2a1 and Col11a2, while SOX5 and SOX6 bind to pairs of SOX motifs close to SOX9 and potentiate SOX9 transcriptional activity [16, 168]. ChIP-Seq analysis has identified abundant binding sites for AP-1, NFAT, RUNX, HOX and FOX transcription factor families near the SOX trio motifs, indicating multiple transcription factors cooperate to regulate cartilage-specific genes [45]. In the testis, SOX9 and SF1 bind to their respective binding elements and synergistically activate AMH/Amh expression [65, 169]. Based on ChIP-Seq and RNA-Seq analyses of murine and bovine fetal testes, it was found that SOX9-bound genomic regions are enriched in binding motifs for three Sertoli cell reprogramming factors SOX9, DMRT1 (doublesex and mab-3 related transcription factor 1) and GATA4 (GATA binding protein 4) [55]. This SOX9, DMRT1 and GATA4 DNA-binding motif enrichment is specific to fetal Sertoli cells and conserved among mammals and thus called Sertoli Cell Signature (SCS). In the pancreas, SOX9 works with PDX1 to activate pancreatic genes by co-binding the regulatory sequences with other pancreatic transcription factors [100].
Regulation of target genes by SOX9 and partners. A SOX9 homodimer binds to inverted SOX binding sites in multiple enhancers of chondrocyte-specific genes. It physically interacts with SOX5/SOX6 and other partners which bind to DNA sequences close to SOX9 binding sites. SRY and SF1 function together to regulate Amh and Sox9 itself in Sertoli cells. B Apart from transactivating targets, SOX9 forms complex with GLI factors to repress genes such as Col10a1 and Vegfa. C SOX9 and partner proteins regulate stepwise progression of tissue development. In Sertoli cells, SRY and SF1 induce Sox9 expression and then SOX9 forms complex with SF1 to promote subsequent gene expression. During astrocyte differentiation, SOX9 induces Nfia expression and then interacts with NFIA protein to promote astrocytic gene expression. D Multiple TFs bind to super-enhancers to regulate cartilage-specific gene expression in chondrocytes, while SOX9 indirectly engages the genome via interacting with basal transcriptional components to regulate widely expressed gene expression in chondrocytes
SOX9 predominantly acts as a transcriptional activator but can also function as a repressor. The transcriptional activation or repression effects appear to be in part modulated by protein partners and co-activators or co-repressors. For instance, SOX9 and SOX5/6 function together to activate ECM genes in early stage and proliferating chondrocytes, whereas SOX9 interacts with GLI factors to repress the transcription of Col10a1, a marker for chondrocyte maturation in hypertrophic chondrocytes (Fig. 2B) [39]. Furthermore, SOX9 induces expression of Notch coactivator Mastermind-like transcriptional activator 2 (MAML2), which, independently of Notch signaling, can promote beta-catenin turnover [170]—indicating that SOX9 can indirectly work as a repressor via other factors. SOX9 can also bind directly to β-catenin, preventing TCF/LEF-mediated signaling and promoting β-catenin degradation [25].
SOX9 interacts with AP-1 family members such as JUN and FOSL2, and they directly co-bind at target motifs to promote hypertrophic gene expression [42]. In addition, peroxisome proliferator-activated receptor γ co-activator 1α (PGC-1α) directly interacts with SOX9 as a coactivator to directly regulate Col2a1 transcription by binding to its enhancer during chondrogenesis [171]. SOX9 can regulate expression of Runx2 via several mechanisms: indirectly via activation of the transcriptional repressor vertebrate homologue of Drosophila bagpipe (Bapx1), which negatively regulates expression of Runx2 [172]; and also directly, via modulating the ubiquitination status of RUNX2, decreasing ubiquitination and redirecting it toward lysosomal degradation [173].
Furthermore, SOX9 enables a stepwise progression of development by forming complexes with different partners or co-factors (Fig. 2C). In the male gonad, SRY and SF1 directly bind to TESCO to induce Sox9 expression. SOX9 then substitutes SRY, and physically interacts with SF1 to maintain its own expression and promote subsequent developmental processes [120]. SOX9 is required for glial specification and subsequent astrocyte differentiation in the CNS [69]. SOX9 activates the transcription of Nfia by binding to its enhancer element and then cooperates with NFIA to promote gliogenesis by inducing a set of astrocytic genes [70].
These data thus highlight the diversity and complexity of the roles of SOX9, directly and indirectly activating or repressing the expression of many target genes to modulate tissue-specific expression profiles.
Cell-specific enhancers and super-enhancers of SOX9 target genes
Master transcription factors regulate cell identity genes by co-occupying clusters of enhancers, i.e., super-enhancers, which are different from typical enhancers in size, transcription factor binding motif density and content, transcriptional activity, and sensitivity to perturbation [174]. In contrast, transcription factors interact with basal transcriptional components to indirectly bind to promoters of broadly expressed genes. In non-hypertrophic chondrocytes, ChIP-Seq analysis shows that SOX9 and SOX5/6 bind to enhancers and super-enhancers of cartilage-specific genes (Fig. 2D) [44, 45]. These super-enhancers also feature binding sites for multiple transcription factor families close to the SOX trio binding sequences, supporting the notion that cell-specific genes are controlled by several key transcription factors. In mouse HF-SCs, SOX9 was identified as the pioneer factor that promotes and maintains cell fate [175]. Also, SOX9 binds to super-enhancers with multiple transcription factors to regulate cell-specific genes. SOX9 is sensitive to local microenvironment changes and responds to changes by remodeling super-enhancers.
Conclusions and future directions
The transcription factor SOX9 is critical for mammalian embryonic development. Sox9 ablation affects the development of multiple organs, such as bone, testis, heart, retina, pancreas, and lung. Investigations into the regulation of the SOX9/Sox9 gene, posttranslational modifications and partner factors have helped to understand the mechanisms underlying the diverse SOX9 functions in different tissues and at different stages in mammals. In view of the complexity and size of the regulatory regions of SOX9/Sox9 and numerous target genes, further studies are needed to identify novel regulatory sequences such as enhancers, transcriptional partners and the mechanisms by which SOX9 regulates target genes across different tissues.
Availability of data and material
Not applicable.
References
Schepers GE, Teasdale RD, Koopman P (2002) Twenty pairs of sox: extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev Cell 3(2):167–170. https://doi.org/10.1016/s1534-5807(02)00223-x
Wagner T, Wirth J, Meyer J et al (1994) Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 79(6):1111–1120. https://doi.org/10.1016/0092-8674(94)90041-8
Foster JW, Dominguez-Steglich MA, Guioli S et al (1994) Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature 372(6506):525–530. https://doi.org/10.1038/372525a0
Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B (1999) Sox9 is required for cartilage formation. Nat Genet 22(1):85–89. https://doi.org/10.1038/8792
Barrionuevo F, Bagheri-Fam S, Klattig J et al (2006) Homozygous inactivation of Sox9 causes complete XY sex reversal in mice. Biol Reprod 74(1):195–201. https://doi.org/10.1095/biolreprod.105.045930
Scott CE, Wynn SL, Sesay A et al (2010) SOX9 induces and maintains neural stem cells. Nat Neurosci 13(10):1181–1189. https://doi.org/10.1038/nn.2646
Poché RA, Furuta Y, Chaboissier MC, Schedl A, Behringer RR (2008) Sox9 is expressed in mouse multipotent retinal progenitor cells and functions in Müller glial cell development. J Comp Neurol 510(3):237–250. https://doi.org/10.1002/cne.21746
Rockich BE, Hrycaj SM, Shih HP et al (2013) Sox9 plays multiple roles in the lung epithelium during branching morphogenesis. Proc Natl Acad Sci U S A 110(47):E4456-4464. https://doi.org/10.1073/pnas.1311847110
Akiyama H, Chaboissier MC, Behringer RR et al (2004) Essential role of Sox9 in the pathway that controls formation of cardiac valves and septa. Proc Natl Acad Sci U S A 101(17):6502–6507. https://doi.org/10.1073/pnas.0401711101
Seymour PA, Freude KK, Tran MN et al (2007) SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc Natl Acad Sci U S A 104(6):1865–1870. https://doi.org/10.1073/pnas.0609217104
Antoniou A, Raynaud P, Cordi S et al (2009) Intrahepatic bile ducts develop according to a new mode of tubulogenesis regulated by the transcription factor SOX9. Gastroenterology 136(7):2325–2333. https://doi.org/10.1053/j.gastro.2009.02.051
Mori-Akiyama Y, van den Born M, van Es JH et al (2007) SOX9 is required for the differentiation of paneth cells in the intestinal epithelium. Gastroenterology 133(2):539–546. https://doi.org/10.1053/j.gastro.2007.05.020
Thomsen MK, Butler CM, Shen MM, Swain A (2008) Sox9 is required for prostate development. Dev Biol 316(2):302–311. https://doi.org/10.1016/j.ydbio.2008.01.030
Vidal VP, Chaboissier MC, Lützkendorf S et al (2005) Sox9 is essential for outer root sheath differentiation and the formation of the hair stem cell compartment. Curr Biol 15(15):1340–1351. https://doi.org/10.1016/j.cub.2005.06.064
Mertin S, McDowall SG, Harley VR (1999) The DNA-binding specificity of SOX9 and other SOX proteins. Nucleic Acids Res 27(5):1359–1364. https://doi.org/10.1093/nar/27.5.1359
Bernard P, Tang P, Liu S et al (2003) Dimerization of SOX9 is required for chondrogenesis, but not for sex determination. Hum Mol Genet 12(14):1755–1765. https://doi.org/10.1093/hmg/ddg182
Coustry F, Oh CD, Hattori T et al (2010) The dimerization domain of SOX9 is required for transcription activation of a chondrocyte-specific chromatin DNA template. Nucleic Acids Res 38(18):6018–6028. https://doi.org/10.1093/nar/gkq417
Huang YH, Jankowski A, Cheah KS, Prabhakar S, Jauch R (2015) SOXE transcription factors form selective dimers on non-compact DNA motifs through multifaceted interactions between dimerization and high-mobility group domains. Sci Rep 5:10398. https://doi.org/10.1038/srep10398
Bridgewater LC, Walker MD, Miller GC et al (2003) Adjacent DNA sequences modulate Sox9 transcriptional activation at paired Sox sites in three chondrocyte-specific enhancer elements. Nucleic Acids Res 31(5):1541–1553. https://doi.org/10.1093/nar/gkg230
Kadaja M, Keyes BE, Lin M et al (2014) SOX9: a stem cell transcriptional regulator of secreted niche signaling factors. Genes Dev 28(4):328–341. https://doi.org/10.1101/gad.233247.113
Zhou R, Bonneaud N, Yuan CX et al (2002) SOX9 interacts with a component of the human thyroid hormone receptor-associated protein complex. Nucleic Acids Res 30(14):3245–3252. https://doi.org/10.1093/nar/gkf443
Tsuda M, Takahashi S, Takahashi Y, Asahara H (2003) Transcriptional co-activators CREB-binding protein and p300 regulate chondrocyte-specific gene expression via association with Sox9. J Biol Chem 278(29):27224–27229. https://doi.org/10.1074/jbc.M303471200
Hattori T, Coustry F, Stephens S et al (2008) Transcriptional regulation of chondrogenesis by coactivator Tip60 via chromatin association with Sox9 and Sox5. Nucleic Acids Res 36(9):3011–3024. https://doi.org/10.1093/nar/gkn150
Nakamura Y, Yamamoto K, He X et al (2011) Wwp2 is essential for palatogenesis mediated by the interaction between Sox9 and mediator subunit 25. Nat Commun 2:251. https://doi.org/10.1038/ncomms1242
Akiyama H, Lyons JP, Mori-Akiyama Y et al (2004) Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Dev 18(9):1072–1087. https://doi.org/10.1101/gad.1171104
Haseeb A, Lefebvre V (2019) The SOXE transcription factors-SOX8, SOX9 and SOX10-share a bi-partite transactivation mechanism. Nucleic Acids Res 47(13):6917–6931. https://doi.org/10.1093/nar/gkz523
McDowall S, Argentaro A, Ranganathan S et al (1999) Functional and structural studies of wild type SOX9 and mutations causing campomelic dysplasia. J Biol Chem 274(34):24023–24030. https://doi.org/10.1074/jbc.274.34.24023
Kronenberg HM (2003) Developmental regulation of the growth plate. Nature 423(6937):332–336. https://doi.org/10.1038/nature01657
Kwan KM, Pang MK, Zhou S et al (1997) Abnormal compartmentalization of cartilage matrix components in mice lacking collagen X: implications for function. J Cell Biol 136(2):459–471. https://doi.org/10.1083/jcb.136.2.459
Dy P, Wang W, Bhattaram P et al (2012) Sox9 directs hypertrophic maturation and blocks osteoblast differentiation of growth plate chondrocytes. Dev Cell 22(3):597–609. https://doi.org/10.1016/j.devcel.2011.12.024
Lui JC, Yue S, Lee A et al (2019) Persistent Sox9 expression in hypertrophic chondrocytes suppresses transdifferentiation into osteoblasts. Bone 125:169–177. https://doi.org/10.1016/j.bone.2019.05.027
Zhou X, von der Mark K, Henry S et al (2014) Chondrocytes transdifferentiate into osteoblasts in endochondral bone during development, postnatal growth and fracture healing in mice. PLoS Genet 10(12):e1004820. https://doi.org/10.1371/journal.pgen.1004820
Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B (2002) The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev 16(21):2813–2828. https://doi.org/10.1101/gad.1017802
Bi W, Huang W, Whitworth DJ et al (2001) Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc Natl Acad Sci U S A 98(12):6698–6703. https://doi.org/10.1073/pnas.111092198
Liu CF, Angelozzi M, Haseeb A, Lefebvre V (2018) SOX9 is dispensable for the initiation of epigenetic remodeling and the activation of marker genes at the onset of chondrogenesis. Development. https://doi.org/10.1242/dev.164459
Han Y, Lefebvre V (2008) L-Sox5 and Sox6 drive expression of the aggrecan gene in cartilage by securing binding of Sox9 to a far-upstream enhancer. Mol Cell Biol 28(16):4999–5013. https://doi.org/10.1128/mcb.00695-08
Renard E, Porée B, Chadjichristos C et al (2012) Sox9/Sox6 and Sp1 are involved in the insulin-like growth factor-I-mediated upregulation of human type II collagen gene expression in articular chondrocytes. J Mol Med (Berl) 90(6):649–666. https://doi.org/10.1007/s00109-011-0842-3
Yano F, Ohba S, Murahashi Y et al (2019) Runx1 contributes to articular cartilage maintenance by enhancement of cartilage matrix production and suppression of hypertrophic differentiation. Sci Rep 9(1):7666. https://doi.org/10.1038/s41598-019-43948-3
Leung VY, Gao B, Leung KK et al (2011) SOX9 governs differentiation stage-specific gene expression in growth plate chondrocytes via direct concomitant transactivation and repression. PLoS Genet 7(11):e1002356. https://doi.org/10.1371/journal.pgen.1002356
Ikegami D, Akiyama H, Suzuki A et al (2011) Sox9 sustains chondrocyte survival and hypertrophy in part through Pik3ca-Akt pathways. Development 138(8):1507–1519. https://doi.org/10.1242/dev.057802
Henry SP, Liang S, Akdemir KC, de Crombrugghe B (2012) The postnatal role of Sox9 in cartilage. J Bone Miner Res 27(12):2511–2525. https://doi.org/10.1002/jbmr.1696
He X, Ohba S, Hojo H, McMahon AP (2016) AP-1 family members act with Sox9 to promote chondrocyte hypertrophy. Development 143(16):3012–3023. https://doi.org/10.1242/dev.134502
Tan Z, Niu B, Tsang KY et al (2018) Synergistic co-regulation and competition by a SOX9-GLI-FOXA phasic transcriptional network coordinate chondrocyte differentiation transitions. PLoS Genet 14(4):e1007346. https://doi.org/10.1371/journal.pgen.1007346
Liu CF, Lefebvre V (2015) The transcription factors SOX9 and SOX5/SOX6 cooperate genome-wide through super-enhancers to drive chondrogenesis. Nucleic Acids Res 43(17):8183–8203. https://doi.org/10.1093/nar/gkv688
Ohba S, He X, Hojo H, McMahon AP (2015) Distinct transcriptional programs underlie Sox9 regulation of the mammalian chondrocyte. Cell Rep 12(2):229–243. https://doi.org/10.1016/j.celrep.2015.06.013
Sekido R, Bar I, Narváez V, Penny G, Lovell-Badge R (2004) SOX9 is up-regulated by the transient expression of SRY specifically in Sertoli cell precursors. Dev Biol 274(2):271–279. https://doi.org/10.1016/j.ydbio.2004.07.011
Kent J, Wheatley SC, Andrews JE, Sinclair AH, Koopman P (1996) A male-specific role for SOX9 in vertebrate sex determination. Development 122(9):2813–2822
Svingen T, Koopman P (2013) Building the mammalian testis: origins, differentiation, and assembly of the component cell populations. Genes Dev 27(22):2409–2426. https://doi.org/10.1101/gad.228080.113
Durcova-Hills G, Capel B (2008) Development of germ cells in the mouse. Curr Top Dev Biol 83:185–212. https://doi.org/10.1016/s0070-2153(08)00406-7
Bowles J, Knight D, Smith C et al (2006) Retinoid signaling determines germ cell fate in mice. Science 312(5773):596–600. https://doi.org/10.1126/science.1125691
Mansour S, Hall CM, Pembrey ME, Young ID (1995) A clinical and genetic study of campomelic dysplasia. J Med Genet 32(6):415–420. https://doi.org/10.1136/jmg.32.6.415
Chaboissier MC, Kobayashi A, Vidal VI et al (2004) Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development 131(9):1891–1901. https://doi.org/10.1242/dev.01087
Ottolenghi C, Pelosi E, Tran J et al (2007) Loss of Wnt4 and Foxl2 leads to female-to-male sex reversal extending to germ cells. Hum Mol Genet 16(23):2795–2804. https://doi.org/10.1093/hmg/ddm235
Kim Y, Kobayashi A, Sekido R et al (2006) Fgf9 and Wnt4 act as antagonistic signals to regulate mammalian sex determination. PLoS Biol 4(6):e187. https://doi.org/10.1371/journal.pbio.0040187
Rahmoun M, Lavery R, Laurent-Chaballier S et al (2017) In mammalian foetal testes, SOX9 regulates expression of its target genes by binding to genomic regions with conserved signatures. Nucleic Acids Res 45(12):7191–7211. https://doi.org/10.1093/nar/gkx328
Li Y, Zheng M, Lau YF (2014) The sex-determining factors SRY and SOX9 regulate similar target genes and promote testis cord formation during testicular differentiation. Cell Rep 8(3):723–733. https://doi.org/10.1016/j.celrep.2014.06.055
Barrionuevo F, Georg I, Scherthan H et al (2009) Testis cord differentiation after the sex determination stage is independent of Sox9 but fails in the combined absence of Sox9 and Sox8. Dev Biol 327(2):301–312. https://doi.org/10.1016/j.ydbio.2008.12.011
Polanco JC, Wilhelm D, Davidson TL, Knight D, Koopman P (2010) Sox10 gain-of-function causes XX sex reversal in mice: implications for human 22q-linked disorders of sex development. Hum Mol Genet 19(3):506–516. https://doi.org/10.1093/hmg/ddp520
Moniot B, Declosmenil F, Barrionuevo F et al (2009) The PGD2 pathway, independently of FGF9, amplifies SOX9 activity in Sertoli cells during male sexual differentiation. Development 136(11):1813–1821. https://doi.org/10.1242/dev.032631
Wilhelm D, Hiramatsu R, Mizusaki H et al (2007) SOX9 regulates prostaglandin D synthase gene transcription in vivo to ensure testis development. J Biol Chem 282(14):10553–10560. https://doi.org/10.1074/jbc.M609578200
Kashimada K, Svingen T, Feng CW et al (2011) Antagonistic regulation of Cyp26b1 by transcription factors SOX9/SF1 and FOXL2 during gonadal development in mice. Faseb J 25(10):3561–3569. https://doi.org/10.1096/fj.11-184333
Umehara F, Tate G, Itoh K et al (2000) A novel mutation of desert hedgehog in a patient with 46, XY partial gonadal dysgenesis accompanied by minifascicular neuropathy. Am J Hum Genet 67(5):1302–1305. https://doi.org/10.1016/s0002-9297(07)62958-9
Clark AM, Garland KK, Russell LD (2000) Desert hedgehog (Dhh) gene is required in the mouse testis for formation of adult-type Leydig cells and normal development of peritubular cells and seminiferous tubules. Biol Reprod 63(6):1825–1838. https://doi.org/10.1095/biolreprod63.6.1825
Bradford ST, Hiramatsu R, Maddugoda MP et al (2009) The cerebellin 4 precursor gene is a direct target of SRY and SOX9 in mice. Biol Reprod 80(6):1178–1188. https://doi.org/10.1095/biolreprod.108.071480
Arango NA, Lovell-Badge R, Behringer RR (1999) Targeted mutagenesis of the endogenous mouse Mis gene promoter: in vivo definition of genetic pathways of vertebrate sexual development. Cell 99(4):409–419. https://doi.org/10.1016/s0092-8674(00)81527-5
Chen C, Ouyang W, Grigura V et al (2005) ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature 436(7053):1030–1034. https://doi.org/10.1038/nature03894
Alankarage D, Lavery R, Svingen T et al (2016) SOX9 regulates expression of the male fertility gene Ets variant factor 5 (ETV5) during mammalian sex development. Int J Biochem Cell Biol 79:41–51. https://doi.org/10.1016/j.biocel.2016.08.005
Meng X, Lindahl M, Hyvönen ME et al (2000) Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 287(5457):1489–1493. https://doi.org/10.1126/science.287.5457.1489
Stolt CC, Lommes P, Sock E et al (2003) The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev 17(13):1677–1689. https://doi.org/10.1101/gad.259003
Kang P, Lee HK, Glasgow SM et al (2012) Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron 74(1):79–94. https://doi.org/10.1016/j.neuron.2012.01.024
Molofsky AV, Glasgow SM, Chaboub LS et al (2013) Expression profiling of Aldh1l1-precursors in the developing spinal cord reveals glial lineage-specific genes and direct Sox9-Nfe2l1 interactions. Glia 61(9):1518–1532. https://doi.org/10.1002/glia.22538
Stolt CC, Rehberg S, Ader M et al (2002) Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev 16(2):165–170. https://doi.org/10.1101/gad.215802
Finzsch M, Stolt CC, Lommes P, Wegner M (2008) Sox9 and Sox10 influence survival and migration of oligodendrocyte precursors in the spinal cord by regulating PDGF receptor alpha expression. Development 135(4):637–646. https://doi.org/10.1242/dev.010454
Zhu MY, Gasperowicz M, Chow RL (2013) The expression of NOTCH2, HES1 and SOX9 during mouse retinal development. Gene Expr Patterns 13(3–4):78–83. https://doi.org/10.1016/j.gep.2012.12.001
Muto A, Iida A, Satoh S, Watanabe S (2009) The group E Sox genes Sox8 and Sox9 are regulated by Notch signaling and are required for Müller glial cell development in mouse retina. Exp Eye Res 89(4):549–558. https://doi.org/10.1016/j.exer.2009.05.006
Cohen-Tayar Y, Cohen H, Mitiagin Y et al (2018) Pax6 regulation of Sox9 in the mouse retinal pigmented epithelium controls its timely differentiation and choroid vasculature development. Development. https://doi.org/10.1242/dev.163691
Masuda T, Wahlin K, Wan J et al (2014) Transcription factor SOX9 plays a key role in the regulation of visual cycle gene expression in the retinal pigment epithelium. J Biol Chem 289(18):12908–12921. https://doi.org/10.1074/jbc.M114.556738
Davis N, Yoffe C, Raviv S et al (2009) Pax6 dosage requirements in iris and ciliary body differentiation. Dev Biol 333(1):132–142. https://doi.org/10.1016/j.ydbio.2009.06.023
Turcatel G, Rubin N, Menke DB et al (2013) Lung mesenchymal expression of Sox9 plays a critical role in tracheal development. BMC Biol 11:117. https://doi.org/10.1186/1741-7007-11-117
Chang DR, Martinez Alanis D, Miller RK et al (2013) Lung epithelial branching program antagonizes alveolar differentiation. Proc Natl Acad Sci U S A 110(45):18042–18051. https://doi.org/10.1073/pnas.1311760110
Lincoln J, Kist R, Scherer G, Yutzey KE (2007) Sox9 is required for precursor cell expansion and extracellular matrix organization during mouse heart valve development. Dev Biol 305(1):120–132. https://doi.org/10.1016/j.ydbio.2007.02.002
Garside VC, Cullum R, Alder O et al (2015) SOX9 modulates the expression of key transcription factors required for heart valve development. Development 142(24):4340–4350. https://doi.org/10.1242/dev.125252
Erickson SL, O’Shea KS, Ghaboosi N et al (1997) ErbB3 is required for normal cerebellar and cardiac development: a comparison with ErbB2-and heregulin-deficient mice. Development 124(24):4999–5011
Bard-Chapeau EA, Szumska D, Jacob B et al (2014) Mice carrying a hypomorphic Evi1 allele are embryonic viable but exhibit severe congenital heart defects. PLoS ONE 9(2):e89397. https://doi.org/10.1371/journal.pone.0089397
Cai X, Zhang W, Hu J et al (2013) Tbx20 acts upstream of Wnt signaling to regulate endocardial cushion formation and valve remodeling during mouse cardiogenesis. Development 140(15):3176–3187. https://doi.org/10.1242/dev.092502
Hill MC, Kadow ZA, Li L et al (2019) A cellular atlas of Pitx2-dependent cardiac development. Development. https://doi.org/10.1242/dev.180398
Laurent F, Girdziusaite A, Gamart J et al (2017) HAND2 target gene regulatory networks control atrioventricular canal and cardiac valve development. Cell Rep 19(8):1602–1613. https://doi.org/10.1016/j.celrep.2017.05.004
Chakraborty S, Wirrig EE, Hinton RB et al (2010) Twist1 promotes heart valve cell proliferation and extracellular matrix gene expression during development in vivo and is expressed in human diseased aortic valves. Dev Biol 347(1):167–179. https://doi.org/10.1016/j.ydbio.2010.08.021
Schilham MW, Oosterwegel MA, Moerer P et al (1996) Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature 380(6576):711–714. https://doi.org/10.1038/380711a0
Peacock JD, Huk DJ, Ediriweera HN, Lincoln J (2011) Sox9 transcriptionally represses Spp1 to prevent matrix mineralization in maturing heart valves and chondrocytes. PLoS ONE 6(10):e26769. https://doi.org/10.1371/journal.pone.0026769
Piper K, Ball SG, Keeling JW et al (2002) Novel SOX9 expression during human pancreas development correlates to abnormalities in Campomelic dysplasia. Mech Dev 116(1–2):223–226. https://doi.org/10.1016/s0925-4773(02)00145-4
Seymour PA, Freude KK, Dubois CL et al (2008) A dosage-dependent requirement for Sox9 in pancreatic endocrine cell formation. Dev Biol 323(1):19–30. https://doi.org/10.1016/j.ydbio.2008.07.034
Kopinke D, Brailsford M, Shea JE et al (2011) Lineage tracing reveals the dynamic contribution of Hes1+ cells to the developing and adult pancreas. Development 138(3):431–441. https://doi.org/10.1242/dev.053843
Dubois CL, Shih HP, Seymour PA et al (2011) Sox9-haploinsufficiency causes glucose intolerance in mice. PLoS ONE 6(8):e23131. https://doi.org/10.1371/journal.pone.0023131
Seymour PA, Shih HP, Patel NA et al (2012) A Sox9/Fgf feed-forward loop maintains pancreatic organ identity. Development 139(18):3363–3372. https://doi.org/10.1242/dev.078733
Kawaguchi Y, Cooper B, Gannon M et al (2002) The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet 32(1):128–134. https://doi.org/10.1038/ng959
Hughes JW, Cho JH, Conway HE et al (2020) Primary cilia control glucose homeostasis via islet paracrine interactions. Proc Natl Acad Sci U S A 117(16):8912–8923. https://doi.org/10.1073/pnas.2001936117
Shih HP, Kopp JL, Sandhu M et al (2012) A Notch-dependent molecular circuitry initiates pancreatic endocrine and ductal cell differentiation. Development 139(14):2488–2499. https://doi.org/10.1242/dev.078634
Lynn FC, Smith SB, Wilson ME et al (2007) Sox9 coordinates a transcriptional network in pancreatic progenitor cells. Proc Natl Acad Sci U S A 104(25):10500–10505. https://doi.org/10.1073/pnas.0704054104
Shih HP, Seymour PA, Patel NA et al (2015) A gene regulatory network cooperatively controlled by Pdx1 and Sox9 governs lineage allocation of foregut progenitor cells. Cell Rep 13(2):326–336. https://doi.org/10.1016/j.celrep.2015.08.082
Marshak S, Benshushan E, Shoshkes M et al (2000) Functional conservation of regulatory elements in the pdx-1 gene: PDX-1 and hepatocyte nuclear factor 3beta transcription factors mediate beta-cell-specific expression. Mol Cell Biol 20(20):7583–7590. https://doi.org/10.1128/mcb.20.20.7583-7590.2000
Poncy A, Antoniou A, Cordi S et al (2015) Transcription factors SOX4 and SOX9 cooperatively control development of bile ducts. Dev Biol 404(2):136–148. https://doi.org/10.1016/j.ydbio.2015.05.012
Chin AM, Hill DR, Aurora M, Spence JR (2017) Morphogenesis and maturation of the embryonic and postnatal intestine. Semin Cell Dev Biol 66:81–93. https://doi.org/10.1016/j.semcdb.2017.01.011
Formeister EJ, Sionas AL, Lorance DK et al (2009) Distinct SOX9 levels differentially mark stem/progenitor populations and enteroendocrine cells of the small intestine epithelium. Am J Physiol Gastrointest Liver Physiol 296(5):G1108-1118. https://doi.org/10.1152/ajpgi.00004.2009
Sato T, van Es JH, Snippert HJ et al (2011) Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469(7330):415–418. https://doi.org/10.1038/nature09637
Bastide P, Darido C, Pannequin J et al (2007) Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J Cell Biol 178(4):635–648. https://doi.org/10.1083/jcb.200704152
Blache P, van de Wetering M, Duluc I et al (2004) SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J Cell Biol 166(1):37–47. https://doi.org/10.1083/jcb.200311021
Cunha GR, Donjacour AA, Cooke PS et al (1987) The endocrinology and developmental biology of the prostate. Endocr Rev 8(3):338–362. https://doi.org/10.1210/edrv-8-3-338
Schneider AJ, Gawdzik J, Vezina CM, Baker TR, Peterson RE (2019) Sox9 in mouse urogenital sinus epithelium mediates elongation of prostatic buds and expression of genes involved in epithelial cell migration. Gene Expr Patterns 34:119075. https://doi.org/10.1016/j.gep.2019.119075
Huang Z, Hurley PJ, Simons BW et al (2012) Sox9 is required for prostate development and prostate cancer initiation. Oncotarget 3(6):651–663. https://doi.org/10.18632/oncotarget.531
Fuchs E (2007) Scratching the surface of skin development. Nature 445(7130):834–842. https://doi.org/10.1038/nature05659
Morris RJ, Liu Y, Marles L et al (2004) Capturing and profiling adult hair follicle stem cells. Nat Biotechnol 22(4):411–417. https://doi.org/10.1038/nbt950
Oshima H, Rochat A, Kedzia C, Kobayashi K, Barrandon Y (2001) Morphogenesis and renewal of hair follicles from adult multipotent stem cells. Cell 104(2):233–245. https://doi.org/10.1016/s0092-8674(01)00208-2
Nowak JA, Polak L, Pasolli HA, Fuchs E (2008) Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell 3(1):33–43. https://doi.org/10.1016/j.stem.2008.05.009
Bagheri-Fam S, Barrionuevo F, Dohrmann U et al (2006) Long-range upstream and downstream enhancers control distinct subsets of the complex spatiotemporal Sox9 expression pattern. Dev Biol 291(2):382–397. https://doi.org/10.1016/j.ydbio.2005.11.013
Yao B, Wang Q, Liu CF et al (2015) The SOX9 upstream region prone to chromosomal aberrations causing campomelic dysplasia contains multiple cartilage enhancers. Nucleic Acids Res 43(11):5394–5408. https://doi.org/10.1093/nar/gkv426
Mochizuki Y, Chiba T, Kataoka K et al (2018) Combinatorial CRISPR/Cas9 approach to elucidate a far-upstream enhancer complex for tissue-specific Sox9 expression. Dev Cell 46(6):794-806.e796. https://doi.org/10.1016/j.devcel.2018.07.024
Gordon CT, Attanasio C, Bhatia S et al (2014) Identification of novel craniofacial regulatory domains located far upstream of SOX9 and disrupted in Pierre Robin sequence. Hum Mutat 35(8):1011–1020. https://doi.org/10.1002/humu.22606
Benko S, Fantes JA, Amiel J et al (2009) Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat Genet 41(3):359–364. https://doi.org/10.1038/ng.329
Sekido R, Lovell-Badge R (2008) Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature 453(7197):930–934. https://doi.org/10.1038/nature06944
Gonen N, Futtner CR, Wood S et al (2018) Sex reversal following deletion of a single distal enhancer of Sox9. Science 360(6396):1469–1473. https://doi.org/10.1126/science.aas9408
Croft B, Ohnesorg T, Hewitt J et al (2018) Human sex reversal is caused by duplication or deletion of core enhancers upstream of SOX9. Nat Commun 9(1):5319. https://doi.org/10.1038/s41467-018-07784-9
Mead TJ, Wang Q, Bhattaram P et al (2013) A far-upstream (− 70 kb) enhancer mediates Sox9 auto-regulation in somatic tissues during development and adult regeneration. Nucleic Acids Res 41(8):4459–4469. https://doi.org/10.1093/nar/gkt140
Lefebvre V, Angelozzi M, Haseeb A (2019) SOX9 in cartilage development and disease. Curr Opin Cell Biol 61:39–47. https://doi.org/10.1016/j.ceb.2019.07.008
Lefebvre V, Dvir-Ginzberg M (2017) SOX9 and the many facets of its regulation in the chondrocyte lineage. Connect Tissue Res 58(1):2–14. https://doi.org/10.1080/03008207.2016.1183667
Migale R, Neumann M, Lovell-Badge R (2021) Long-range regulation of key sex determination genes. Sex Dev 15(5–6):360–380. https://doi.org/10.1159/000519891
Ridnik M, Schoenfelder S, Gonen N (2021) Cis-regulatory control of mammalian sex determination. Sex Dev 15(5–6):317–334. https://doi.org/10.1159/000519244
Amarilio R, Viukov SV, Sharir A et al (2007) HIF1alpha regulation of Sox9 is necessary to maintain differentiation of hypoxic prechondrogenic cells during early skeletogenesis. Development 134(21):3917–3928. https://doi.org/10.1242/dev.008441
Chen S, Tao J, Bae Y et al (2013) Notch gain of function inhibits chondrocyte differentiation via Rbpj-dependent suppression of Sox9. J Bone Miner Res 28(3):649–659. https://doi.org/10.1002/jbmr.1770
Ushita M, Saito T, Ikeda T et al (2009) Transcriptional induction of SOX9 by NF-kappaB family member RelA in chondrogenic cells. Osteoarthr Cartil 17(8):1065–1075. https://doi.org/10.1016/j.joca.2009.02.003
Piera-Velazquez S, Hawkins DF, Whitecavage MK et al (2007) Regulation of the human SOX9 promoter by Sp1 and CREB. Exp Cell Res 313(6):1069–1079. https://doi.org/10.1016/j.yexcr.2007.01.001
Pan Q, Yu Y, Chen Q et al (2008) Sox9, a key transcription factor of bone morphogenetic protein-2-induced chondrogenesis, is activated through BMP pathway and a CCAAT box in the proximal promoter. J Cell Physiol 217(1):228–241. https://doi.org/10.1002/jcp.21496
Hall MD, Murray CA, Valdez MJ, Perantoni AO (2017) Mesoderm-specific Stat3 deletion affects expression of Sox9 yielding Sox9-dependent phenotypes. PLoS Genet 13(2):e1006610. https://doi.org/10.1371/journal.pgen.1006610
Goto H, Nishio M, To Y et al (2018) Loss of Mob1a/b in mice results in chondrodysplasia due to YAP1/TAZ-TEAD-dependent repression of SOX9. Development. https://doi.org/10.1242/dev.159244
Kanai Y, Koopman P (1999) Structural and functional characterization of the mouse Sox9 promoter: implications for campomelic dysplasia. Hum Mol Genet 8(4):691–696. https://doi.org/10.1093/hmg/8.4.691
Franke M, Ibrahim DM, Andrey G et al (2016) Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538(7624):265–269. https://doi.org/10.1038/nature19800
Smyk M, Akdemir KC, Stankiewicz P (2017) SOX9 chromatin folding domains correlate with its real and putative distant cis-regulatory elements. Nucleus 8(2):182–187. https://doi.org/10.1080/19491034.2017.1279776
Sun L, Mathews LA, Cabarcas SM et al (2013) Epigenetic regulation of SOX9 by the NF-κB signaling pathway in pancreatic cancer stem cells. Stem Cells 31(8):1454–1466. https://doi.org/10.1002/stem.1394
Aleman A, Adrien L, Lopez-Serra L et al (2008) Identification of DNA hypermethylation of SOX9 in association with bladder cancer progression using CpG microarrays. Br J Cancer 98(2):466–473. https://doi.org/10.1038/sj.bjc.6604143
Cheng PF, Shakhova O, Widmer DS et al (2015) Methylation-dependent SOX9 expression mediates invasion in human melanoma cells and is a negative prognostic factor in advanced melanoma. Genome Biol 16(1):42. https://doi.org/10.1186/s13059-015-0594-4
Duan L, Liang Y, Ma B et al (2017) DNA methylation profiling in chondrocyte dedifferentiation in vitro. J Cell Physiol 232(7):1708–1716. https://doi.org/10.1002/jcp.25486
Kumar D, Lassar AB (2014) Fibroblast growth factor maintains chondrogenic potential of limb bud mesenchymal cells by modulating DNMT3A recruitment. Cell Rep 8(5):1419–1431. https://doi.org/10.1016/j.celrep.2014.07.038
Pan Q, Wu Y, Lin T et al (2009) Bone morphogenetic protein-2 induces chromatin remodeling and modification at the proximal promoter of Sox9 gene. Biochem Biophys Res Commun 379(2):356–361. https://doi.org/10.1016/j.bbrc.2008.12.062
Cheng LC, Pastrana E, Tavazoie M, Doetsch F (2009) miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci 12(4):399–408. https://doi.org/10.1038/nn.2294
Real FM, Sekido R, Lupiáñez DG et al (2013) A microRNA (mmu-miR-124) prevents Sox9 expression in developing mouse ovarian cells. Biol Reprod 89(4):78. https://doi.org/10.1095/biolreprod.113.110957
Yang B, Guo H, Zhang Y et al (2011) MicroRNA-145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PLoS ONE 6(7):e21679. https://doi.org/10.1371/journal.pone.0021679
Dai L, Zhang X, Hu X, Zhou C, Ao Y (2012) Silencing of microRNA-101 prevents IL-1β-induced extracellular matrix degradation in chondrocytes. Arthritis Res Ther 14(6):R268. https://doi.org/10.1186/ar4114
Martinez-Sanchez A, Murphy CL (2013) miR-1247 functions by targeting cartilage transcription factor SOX9. J Biol Chem 288(43):30802–30814. https://doi.org/10.1074/jbc.M113.496729
Lee S, Yoon DS, Paik S et al (2014) microRNA-495 inhibits chondrogenic differentiation in human mesenchymal stem cells by targeting Sox9. Stem Cells Dev 23(15):1798–1808. https://doi.org/10.1089/scd.2013.0609
Ashrafizadeh M, Zarrabi A, Orouei S et al (2021) Interplay between SOX9 transcription factor and microRNAs in cancer. Int J Biol Macromol 183:681–694. https://doi.org/10.1016/j.ijbiomac.2021.04.185
Barter MJ, Gomez R, Hyatt S et al (2017) The long non-coding RNA ROCR contributes to SOX9 expression and chondrogenic differentiation of human mesenchymal stem cells. Development 144(24):4510–4521. https://doi.org/10.1242/dev.152504
Sun J, Lin Y, Wu J (2013) Long non-coding RNA expression profiling of mouse testis during postnatal development. PLoS ONE 8(10):e75750. https://doi.org/10.1371/journal.pone.0075750
Vining B, Ming Z, Bagheri-Fam S, Harley V (2021) Diverse regulation but conserved function: SOX9 in vertebrate sex determination. Genes (Basel). https://doi.org/10.3390/genes12040486
Huang W, Chung UI, Kronenberg HM, de Crombrugghe B (2001) The chondrogenic transcription factor Sox9 is a target of signaling by the parathyroid hormone-related peptide in the growth plate of endochondral bones. Proc Natl Acad Sci U S A 98(1):160–165. https://doi.org/10.1073/pnas.011393998
Huang W, Zhou X, Lefebvre V, de Crombrugghe B (2000) Phosphorylation of SOX9 by cyclic AMP-dependent protein kinase A enhances SOX9’s ability to transactivate a Col2a1 chondrocyte-specific enhancer. Mol Cell Biol 20(11):4149–4158. https://doi.org/10.1128/mcb.20.11.4149-4158.2000
Chikuda H, Kugimiya F, Hoshi K et al (2004) Cyclic GMP-dependent protein kinase II is a molecular switch from proliferation to hypertrophic differentiation of chondrocytes. Genes Dev 18(19):2418–2429. https://doi.org/10.1101/gad.1224204
Malki S, Nef S, Notarnicola C et al (2005) Prostaglandin D2 induces nuclear import of the sex-determining factor SOX9 via its cAMP-PKA phosphorylation. Embo j 24(10):1798–1809. https://doi.org/10.1038/sj.emboj.7600660
Liu JA, Wu MH, Yan CH et al (2013) Phosphorylation of Sox9 is required for neural crest delamination and is regulated downstream of BMP and canonical Wnt signaling. Proc Natl Acad Sci U S A 110(8):2882–2887. https://doi.org/10.1073/pnas.1211747110
Ito T, Yadav N, Lee J et al (2009) Arginine methyltransferase CARM1/PRMT4 regulates endochondral ossification. BMC Dev Biol 9:47. https://doi.org/10.1186/1471-213x-9-47
Dvir-Ginzberg M, Gagarina V, Lee EJ, Hall DJ (2008) Regulation of cartilage-specific gene expression in human chondrocytes by SirT1 and nicotinamide phosphoribosyltransferase. J Biol Chem 283(52):36300–36310. https://doi.org/10.1074/jbc.M803196200
Bar Oz M, Kumar A, Elayyan J et al (2016) Acetylation reduces SOX9 nuclear entry and ACAN gene transactivation in human chondrocytes. Aging Cell 15(3):499–508. https://doi.org/10.1111/acel.12456
Komatsu T, Mizusaki H, Mukai T et al (2004) Small ubiquitin-like modifier 1 (SUMO-1) modification of the synergy control motif of Ad4 binding protein/steroidogenic factor 1 (Ad4BP/SF-1) regulates synergistic transcription between Ad4BP/SF-1 and Sox9. Mol Endocrinol 18(10):2451–2462. https://doi.org/10.1210/me.2004-0173
Hattori T, Eberspaecher H, Lu J et al (2006) Interactions between PIAS proteins and SOX9 result in an increase in the cellular concentrations of SOX9. J Biol Chem 281(20):14417–14428. https://doi.org/10.1074/jbc.M511330200
Oh HJ, Kido T, Lau YF (2007) PIAS1 interacts with and represses SOX9 transactivation activity. Mol Reprod Dev 74(11):1446–1455. https://doi.org/10.1002/mrd.20737
Taylor KM, Labonne C (2005) SoxE factors function equivalently during neural crest and inner ear development and their activity is regulated by SUMOylation. Dev Cell 9(5):593–603. https://doi.org/10.1016/j.devcel.2005.09.016
Hattori T, Kishino T, Stephen S et al (2013) E6-AP/UBE3A protein acts as a ubiquitin ligase toward SOX9 protein. J Biol Chem 288(49):35138–35148. https://doi.org/10.1074/jbc.M113.486795
Egunsola AT, Bae Y, Jiang MM et al (2017) Loss of DDRGK1 modulates SOX9 ubiquitination in spondyloepimetaphyseal dysplasia. J Clin Invest 127(4):1475–1484. https://doi.org/10.1172/jci90193
Smits P, Li P, Mandel J et al (2001) The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev Cell 1(2):277–290. https://doi.org/10.1016/s1534-5807(01)00003-x
De Santa Barbara P, Bonneaud N, Boizet B et al (1998) Direct interaction of SRY-related protein SOX9 and steroidogenic factor 1 regulates transcription of the human anti-Müllerian hormone gene. Mol Cell Biol 18(11):6653–6665. https://doi.org/10.1128/mcb.18.11.6653
Sinha A, Fan VB, Ramakrishnan AB et al (2021) Repression of Wnt/β-catenin signaling by SOX9 and Mastermind-like transcriptional coactivator 2. Sci Adv. https://doi.org/10.1126/sciadv.abe0849
Kawakami Y, Tsuda M, Takahashi S et al (2005) Transcriptional coactivator PGC-1alpha regulates chondrogenesis via association with Sox9. Proc Natl Acad Sci U S A 102(7):2414–2419. https://doi.org/10.1073/pnas.0407510102
Yamashita S, Andoh M, Ueno-Kudoh H et al (2009) Sox9 directly promotes Bapx1 gene expression to repress Runx2 in chondrocytes. Exp Cell Res 315(13):2231–2240. https://doi.org/10.1016/j.yexcr.2009.03.008
Cheng A, Genever PG (2010) SOX9 determines RUNX2 transactivity by directing intracellular degradation. J Bone Miner Res 25(12):2680–2689. https://doi.org/10.1002/jbmr.174
Whyte WA, Orlando DA, Hnisz D et al (2013) Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153(2):307–319. https://doi.org/10.1016/j.cell.2013.03.035
Adam RC, Yang H, Rockowitz S et al (2015) Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature 521(7552):366–370. https://doi.org/10.1038/nature14289
Liu CJ, Zhang Y, Xu K et al (2007) Transcriptional activation of cartilage oligomeric matrix protein by Sox9, Sox5, and Sox6 transcription factors and CBP/p300 coactivators. Front Biosci 12:3899–3910. https://doi.org/10.2741/2359
Acknowledgements
Figures created with BioRender.com.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. This work was funded by National Health and Medical Research Council Program Grant 2002426 (to VH) and Fellowship APP1154870 (to VH). This work was supported by the China Scholarship Council (CSC) (to ZM) and the Australian Government Research Training Program Scholarship (to BV). This work was supported by the Victorian Government’s Operational Infrastructure Support Program.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Original draft preparation was conducted by ZM. Writing, reviewing and editing of the draft manuscript was completed by BV, SB-F and VH. Visualization and figure creation was completed by ZM and BV. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ming, Z., Vining, B., Bagheri-Fam, S. et al. SOX9 in organogenesis: shared and unique transcriptional functions. Cell. Mol. Life Sci. 79, 522 (2022). https://doi.org/10.1007/s00018-022-04543-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04543-4