Abstract
Global genome nucleotide excision repair (GG-NER) eliminates a broad spectrum of DNA lesions from genomic DNA. Genomic DNA is tightly wrapped around histones creating a barrier for DNA repair proteins to access DNA lesions buried in nucleosomal DNA. The DNA-damage sensors XPC and DDB2 recognize DNA lesions in nucleosomal DNA and initiate repair. The emerging view is that a tight interplay between XPC and DDB2 is regulated by post-translational modifications on the damage sensors themselves as well as on chromatin containing DNA lesions. The choreography between XPC and DDB2, their interconnection with post-translational modifications such as ubiquitylation, SUMOylation, methylation, poly(ADP-ribos)ylation, acetylation, and the functional links with chromatin remodelling activities regulate not only the initial recognition of DNA lesions in nucleosomes, but also the downstream recruitment and necessary displacement of GG-NER factors as repair progresses. In this review, we highlight how nucleotide excision repair leaves a mark on chromatin to enable DNA damage detection in nucleosomes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
DNA damage recognition in nucleotide excision repair
Cells are continually exposed to different sources of DNA damage including solar UV light, environmental chemicals, food-borne mutagens, and reactive metabolites that generate a wide variety of structurally diverse genomic DNA lesions [1, 2]. Dedicated DNA repair mechanisms recognize and remove genomic DNA lesions to maintain genome integrity and prevent disease [3]. Nucleotide excision repair (NER) is a versatile DNA repair pathway that eliminates a wide range of structurally diverse DNA lesions from genomic DNA, including UV-induced photolesions, such as 6–4 pyrimidine–pyrimidone photoproducts (6-4PPs) and cyclobutane pyrimidine dimers (CPDs) [4].
DNA lesions in transcribed strands are substrates of transcription-coupled repair (TC-NER) [5], while elimination of DNA lesions throughout the genome is carried out by global genome repair (GG-NER) [6, 7]. Recognition through both sub-pathways ultimately leads to a common mechanism of verification, excision and re-synthesis of the damaged DNA, involving the same set of core NER proteins, including the TFIIH complex, XPA, RPA and the endonucleases XPG and ERCC1-XPF (Fig. 1a) [4]. The mechanisms involved in TC-NER initiation have been reviewed recently [5, 8, 9]. In this review we focus on recent insight into how GG-NER is initiated and operates in a chromatin context (see Table 1,2,3).
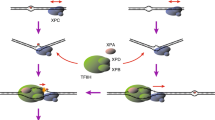
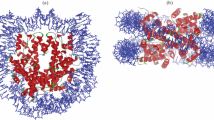
DNA damage-recognition factors initiate GG-NER. a Model of GG-NER initiation by the CRL4DDB2 complex (consisting of DDB2-DDB1-CUL4A-RBX1) and the XPC complex (consisting of XPC, RAD23B, CETN2), which is followed by the recruitment of the TFIIH complex, XPA, RPA and the endonucleases XPG and ERCC1-XPF. b Structure of yeast Rad4/XPC bound to a 6-4PP lesion. The lesion is displaced from the helix stack using the hairpins of the BHD2 and BHD3 domains. The BHD2/3 domains form a tight binding pocket for the dA residues in the non-damaged strand, but do not contact the lesion directly. The BHD1-TGD domains of Rad4/XPC bind in a damage and sequence non-specific manner and anchor the protein on DNA during the lesion search process. Figure generated using PDB 6CFI with PyMol. c Structure of UV-DDB (consisting of DDB2 and DDB1) bound to a 6-4PP in a nucleosome. The DDB2 protein binds to the nucleosome at a 60° angle and pushes the 6-4PP into a lesion-binding pocket using wedge residues (F334, Q335 and H336). Figure generated using PDB 6R8Y with PyMol
The recognition of DNA lesions during GG-NER is critically dependent on the DNA damage-recognition complex XPC-RAD23B, which utilizes an indirect recognition mechanism [10,11,12,13]. Structural studies of Rad4, the yeast homolog of XPC, have shown that the protein uses four domains for DNA and damage recognition [12, 14]. The BHD1 and TGD domains anchor the protein on DNA non-specifically to allow the BHD2 and BHD3 domains to probe for sites of thermodynamic destabilization induced by the lesion. BHD2-3 interacts with the lesion site through a binding pocket for two native bases on the undamaged strand and by inserting the tips of BHD3 into the duplex at the site of the lesion, displacing the lesion into an extrahelical position [14, 15] (Fig. 1b). XPC does not make any specific contacts with the lesion itself. This feature of XPC explains the broad substrate specificity of lesion binding by XPC and NER in general [15,16,17]. Furthermore, a “kinetic gating” mechanism for Rad4/XPC lesion binding has been proposed, which suggests that lesion recognition primarily depends on the local destabilization of the DNA duplex and the protein's retention time at the lesion site rather than the presence of a particular lesion. These observations explain why the XPC protein binds with high affinity to helix-destabilizing DNA lesions, such as 6-4PPs, while its affinity for the more abundant, but less helix-destabilizing UV-induced CPD photolesions is rather low [18].
For the recognition of CPDs, XPC needs the support of the CRL4DDB2 complex, consisting of DDB2, the damage-recognition protein, and DDB1, which serves as a link to a CUL4A-RBX1-based (CRL4) E3 ubiquitin ligase complex [19,20,21]. DDB2 directly associates with photolesions by extruding the lesion out of the helix into a hydrophobic pocket embedded in its WD40 domain using three residues that form a wedge to take the place of the lesion in the helix (Fig. 1c) [21, 22]. An overlay of the structures of XPC and DDB2 bound to 6-4PPs suggests that the two proteins cannot coexist on a lesion. Instead, DDB2 makes the lesion more accessible for XPC by opening the DNA at the lesion to generate a helix-destabilizing substrate that is recognized by XPC [21, 22]. The recruitment of XPC is further dependent on direct protein–protein interactions with DDB2 [23, 24]. These findings suggest that DDB2 is needed to bring XPC in proximity of the lesion, but that the binding of XPC opposite of the DNA lesion requires the displacement of DDB2 to prevent steric clashes between the two damage-recognition proteins.
DNA damage detection in nucleosomes
The process of GG-NER has been fully reconstituted in vitro with recombinant purified components and is independent of DDB2 under these conditions [25, 26]. While reconstituted GG-NER operates well on naked DNA, genomic DNA is tightly wrapped around histones creating a barrier for DNA repair proteins to access DNA lesions buried in nucleosomal DNA [27, 28]. Earlier biochemical studies showed that chromatin remodelers can alleviate the chromatin barrier to repair proteins, thereby making lesions accessible to NER [29, 30]. Before any mechanisms of chromatin rearrangements were known, the repair of DNA lesions was envisioned to occur through an access-repair-restore model [31, 32]. It is now becoming clear that DDB2 has a key role in facilitating DNA lesion-recognition in a chromatin context [22, 33, 34]. DDB2 directly binds photolesions embedded in nucleosomal DNA (Fig. 1c) and mediates slide-assisted site exposure of buried lesions that face the nucleosome core [22, 35]. Additionally, as discussed extensively below, DDB2 plays a key role in regulating the recruitment and the activity of several chromatin remodelling and modifying enzymes to regulate downstream steps during GG-NER. These findings provide a mechanistic explanation for why DDB2 is essential for the repair of CPDs, while the repair of 6-4PPs is enhanced by, but not dependent on DDB2 [36].
The emerging picture is that the interplay between XPC and DDB2 is tightly regulated by post-translational modifications (PTMs) on the damage-recognition proteins themselves as well as on chromatin containing DNA lesions. The tight interplay between these DNA lesion-recognition proteins, their interconnection with PTMs such as ubiquitylation, SUMOylation, methylation, poly(ADP-ribos)ylation, acetylation, and the functional links with chromatin remodelling activities regulate not only the initial recognition of DNA lesions in chromatin, but also the downstream recruitment and necessary displacement of NER factors as repair progresses.
DNA damage-recognition proteins and their interconnection with ubiquitylation
The CRL4DDB2 ligase and histone H3 and H4 ubiquitylation in response to UV
The E3 ubiquitin ligase activity of the CRL4DDB2 complex has been linked to histone ubiquitylation during GG-NER. One study reported that the CRL4DDB2 complex mediates the UV-induced ubiquitylation of histone H3 and H4, resulting in a weakened interaction between histones and DNA thereby facilitating XPC recruitment (Fig. 2a, b) [37]. Although these findings suggest a link between H3 and H4 ubiquitylation and GG-NER, it will be important to identify the precise residues that are targeted for ubiquitylation and determine the mechanistic basis for XPC recruitment to these ubiquitylated histones.
The role of ubiquitylation and SUMOylation in GG-NER. a DDB2 is part of the CRL4DDB2 ubiquitin ligase complex together with DDB1, CUL4A and RBX1 that binds to photolesions. b The CRL4DDB2 ligase ubiquitylates H3 and H4 leading to chromatin decompaction through an unknown mechanism, which stimulates XPC recruitment. DDB2 also ubiquitylates XPC, which increases its affinity for DNA lesions. DDB2 becomes SUMOylated at K309, which stimulates XPC recruitment and promotes CPD repair. c DDB2 must dissociate to allow stable XPC binding to the DNA lesion. The displacement of DDB2 is stimulated by the recruitment of TFIIH by XPC. The ubiquitin-selective segregase VCP/p97 further stimulates the dissociation of DDB2 through extraction of ubiquitylated DDB2 from chromatin. XPC is SUMOylated at K81, K89 and K183, which was suggested to stimulate the handover between XPC and DDB2. d The SUMO-dependent E3 ubiquitin ligase RNF111 recognizes and ubiquitylates the SUMOylated form of XPC. e The ubiquitylated form of XPC may also be extracted from chromatin by VCP/p97 to enable efficient recruitment of the endonuclease XPG. This is important because XPG and XPC cannot coexist in the same NER complex
XPC and DDB2 ubiquitylation and SUMOylation facilitate DNA-lesions recognition
Regulating the interplay and handover between lesion-recognition proteins DDB2 and XPC is crucial to initiate GG-NER in chromatin. The catalytic activity of the CRL4DDB2 ubiquitin ligase complex has a key role during these early transactions. The CRL4DDB2 complex ubiquitylates XPC in response to UV irradiation (Fig. 2b). However, this does not result in its proteasomal degradation but rather stabilizes the association of the protein with DNA [23, 38]. CRL4DDB2 also auto-ubiquitylates DDB2 triggering its degradation [39, 40]. It is believed that the differential impact of ubiquitylation of the two damage sensors stimulates the handover from DDB2 to XPC, a process required for GG-NER progression. In addition, DDB2 becomes conjugated with SUMO-1 at lysine residue K309 in response to UV irradiation. This modification was shown to stimulate XPC recruitment and regulate efficient repair of CPDs [41].
DDB2 ubiquitylation regulates its chromatin extraction
The handover of DNA lesions from DDB2 to XPC is tightly regulated at multiple levels. First, the initial transient XPC-mediated recruitment of the TFIIH complex stimulates DDB2 dissociation thereby promoting the formation of a stable XPC-TFIIH complex [42] (Fig. 2c). Furthermore, the ubiquitin-selective segregase VCP/p97 is involved in extracting ubiquitylated DDB2 from damaged DNA to reduce its chromatin dwell time [43] (Fig. 2c). The inability to extract DDB2 from chromatin interferes with the stable binding of XPC and TFIIH to DNA lesions [42], suggesting that while the initial binding of DDB2 stimulates XPC recruitment, its prolonged binding actually inhibits subsequent GG-NER progression. The interaction between VCP/p97 and DDB2 is stimulated by the deacetylase SIRT6, suggesting that the UV-induced deacetylation of DDB2 promotes its ubiquitylation and subsequent extraction from chromatin [44]. Interestingly, the UV-induced SUMOylation of XPC at lysine residues K81, K89 and K183 was suggested to regulate the release of DDB2 in trans. More specifically, an XPC mutant that cannot be SUMOylated (3KR) shows a stronger UV-induced immobilization on chromatin and a more pronounced DNA repair defect, which was partially alleviated by the loss of DDB2 [45]. These findings suggest that XPC SUMOylation promotes efficient DDB2 dissociation and DNA damage handover to XPC. While this is an interesting possibility, an alternative—but not necessarily mutually exclusive—function for XPC SUMOylation is discussed in Section XPC extraction from chromatin requires SUMOylation .
XPC ubiquitylation may regulate its chromatin extraction
XPC is possibly also extracted from chromatin by VCP/p97, but conflicting data exists as siRNA-mediated depletion of VCP was found to increase XPC binding to local DNA damage in one study [43], while treatment of cells with VCP inhibitor was found to reduce XPC binding in another study [42]. Extraction of ubiquitylated XPC may facilitate the assembly of the NER pre-incision complex (Fig. 2d, e). In particular the XPG endonuclease and XPC cannot coexist in the same NER complex [46,47,48]. Importantly, ubiquitylated XPC needs to remain bound long enough to recruit the TFIIH complex, which may be regulated by two deubiquitylases, USP11 and USP7, that each interact with and deubiquitylate XPC to prevent its untimely extraction [49, 50].
XPC extraction from chromatin requires SUMOylation
Although DDB2 was reported to ubiquitylate XPC [23], another E3 ligase known as RNF111 (Arkadia) was also shown to act on XPC [47, 51]. RNF111 is a so-called SUMO-targeted ubiquitin E3 ligase (STUbL) that selectively ubiquitylates substrates that were previously conjugated with SUMO (Fig. 2d). Indeed, XPC is modified by SUMO-1 at lysine residues K81, K89, K183 and by SUMO-2 under unchallenged conditions [45, 47], although one study reported the UV-induced SUMOylation of XPC [52]. These studies showed that while SUMOylation of XPC did not affect its initial binding to lesions, it was required for the extraction of XPC from chromatin, in conjunction with ubiquitylation by RNF111 [43, 45, 47, 51]. Consistent with XPC and XPG being mutually exclusive in NER complexes, the RNF111-mediated ubiquitylation of XPC is required for efficient XPG recruitment [47] (Fig. 2e). As described above, an XPC-3KR SUMOylation-deficient mutant becomes strongly immobilized on chromatin after UV irradiation in a DDB2-dependent manner. This suggests that SUMOylated XPC may regulate the release of DDB2 in trans [45]. An alternative explanation, which is more in line with results from these other studies [47] is that the XPC-3KR mutant itself is not extracted from chromatin in a timely manner and blocks the NER reaction. It is possible that this effect is exacerbated by DDB2, which stimulates XPC recruitment to chromatin after UV irradiation [53], resulting in even higher levels of XPC on chromatin.
The CRL4DDB2 ligase and histone H2A ubiquitylation in response to UV
The ubiquitylation of histone H2A has also been linked to GG-NER [54,55,56,57], although general consensus about the underlying mechanism is lacking. One study observed a reduction of H2A ubiquitylation within the first 30 min after UV irradiation followed by a DDB2-mediated restoration of H2A mono-ubiquitylation at 2 h post UV to levels similar as before UV irradiation [56]. Whether this reflects the canonical H2A ubiquitylation at K119 [58] or perhaps another residue detected by the same antibody is currently unclear. Conceptually, it is not clear how reducing H2A ubiquitylation levels after UV and restoring these levels in a DDB2-dependent manner could facilitate GG-NER. Another study did not observe a decrease in H2A ubiquitylation levels, but did report increased levels in the first 30 min after UV in a manner dependent on DDB2 and the canonical H2A ligase RING1B [55].
To complicate matters further, not CRL4DDB2 but the E3 ubiquitin ligase RNF8 was shown to catalyze H2A ubiquitylation as a late DNA damage signalling event during GG-NER [57]. This is consistent with an earlier study showing that H2A ubiquitylation after UV is dependent on functional GG-NER and subsequent ATR activation [54, 59], which is required for H2AX phosphorylation and RNF8 recruitment [57]. These findings suggest a mechanism in which damage excision exposes single-stranded DNA that, probably following gap extension by exonuclease EXO1 [60], triggers ATR activation and subsequent DNA damage signalling that is similar to the DNA double-strand break (DSB) response. In the DSB response, RNF8 was shown to target histone H1 [61], while the subsequent recruitment of RNF168 targets histone H2A at K13/K15 [62]. Taken together, the available data suggests that H3/H4 ubiquitylation by CRL4DDB2 complex facilitates GG-NER [37] (see The CRL4DDB2 ligase and histone H3 and H4 ubiquitylation in response to UV), while a potential role of H2A ubiquitylation by CRL4DDB2 during early GG-NER remains more enigmatic.
An alternative E3 ubiquitin ligase complex containing DDB2
One study proposed that the ubiquitylation of H2A during early GG-NER is not carried out by the canonical CRL4DDB2 ubiquitin complex, but rather by an alternative E3 ubiquitin ligase complex consisting of DDB2-DDB1-CUL4B-RING1B (CUL4B/RING1BDDB2) [55]. RING1B is the catalytic subunit of the polycomb-repressive complex 1 involved in gene silencing during differentiation [63]. The initial recruitment of CUL4B/RING1BDDB2 to DNA lesions by DDB2 was suggested to deposit H2A ubiquitylation, which is recognized by the ubiquitin-binding domain of ZRF1. Upon recruitment to DNA lesions, ZRF1 was suggested to remodel the CUL4B/RING1BDDB2 complex and exchange CUL4B-RING1B with CUL4A-RBX1, to turn the CUL4B/RING1BDDB2 complex into the canonical CUL4A/RBX1DDB2 complex [55]. Instead of targeting histones, the CUL4A/RBX1DDB2 complex was found to ubiquitylate XPC [55], consistent with previous reports [23] (see XPC and DDB2 ubiquitylation and SUMOylation facilitate DNA-lesions recognition).
Although the involvement of ZRF1 and the potential remodelling of a DDB2 containing E3 ubiquitin ligase complex with two functional modules—CUL4B/RING1B and CUL4A/RBX1—in GG-NER is very intriguing, these findings have not been verified by other groups yet and also raise many conceptual questions. For instance, proteomics approaches have identified the presence of the CRL4ADDB2 complex containing RBX1 in unirradiated cells [20, 42, 56], which will be recruited to DNA lesions through DDB2. It is, therefore, unclear what the added advantage of localized remodelling of a CRL4DDB2 complex is. Also, how is the relative recruitment of the CUL4B/RING1BDDB2 and CUL4A/RBX1DDB2 complexes regulated? Answering these questions will provide a better understanding of the role of the E3 ubiquitin ligase complexes containing DDB2 during early GG-NER.
Chromatin remodelling during the DNA damage-recognition step in GG-NER
The binding of DDB2 triggers chromatin unfolding and opening in response to UV irradiation [33, 34], which is thought to facilitate XPC recruitment. Interestingly, while DDB2 recruitment occurs independently of ATP hydrolysis, the recruitment of XPC is inhibited when ATP is depleted [34], suggesting that chromatin accessibility is likely increased by the activity of ATP-dependent chromatin remodelers. In the following sections, we discuss the role of chromatin remodelers during the initiation of GG-NER.
The INO80 complex stimulates XPC recruitment
The INO80 remodeler consists of 10–15 polypeptides and exhibits ATP-dependent chromatin remodelling activity [64]. Besides its role in DSB repair [65] and possibly interstrand crosslink repair [66], the INO80 complex is also implicated in GG-NER [67]. Both the INO80 and the ARP5 subunits were shown to associate with and stimulate the removal of UV-induced DNA lesions. INO80 interacted with DDB1 and cells depleted of INO80 showed decreased XPC recruitment, suggesting that INO80 may be recruited by CRL4DDB2 upstream of XPC [67] (Fig. 3a). Because formal proof for this scenario is still lacking, it will be important to establish whether DDB2 is indeed required for INO80 recruitment. Interestingly, yeast INO80 interacts with Rad4—the yeast orthologue of XPC- and INO80-deficient yeast strains are sensitive to UV irradiation [68]. Nevertheless, in yeast INO80 was implicated in restoring chromatin after repair rather than facilitating lesion removal, making it currently unclear whether INO80 has an evolutionary conserved role or possibly multiple roles in GG-NER.
The role of chromatin remodelers and PARylation in GG-NER. a Lesion recognition by DDB2 may recruit the ATP-dependent chromatin remodeler INO80. While INO80 was shown to be recruited by DDB1, we speculate that this is also dependent on DDB2. b DDB2 interacts with PARP1 and stimulates its catalytic activity. Note that PARP1 binds to photolesions independently of DDB2 or XPC. PARP1 modifies itself and DDB2 with PAR chains. The PAR-binding chromatin remodeler ALC1 is recruited and stimulates GG-NER. PARP1 also interacts with XPC already in the absence of DNA damage and facilitates its recruitment to photolesions, particularly at low damage load. c XPC recruits the chromatin remodeler CHD1. d CHD1 facilitates the displacement of XPC to stimulate TFIIH recruitment
PARP1, DDB2 and XPC: A ménage à trois
Poly(ADP-ribose) polymerase 1 (PARP1) has been linked to the early stages of GG-NER through its interaction with both DDB2 [69, 70] and XPC [71] (Fig. 3b). PARP1 uses NAD + as a substrate to add poly-ADP-ribose (PAR) chains to target proteins. Such PAR chains can contain up to 200 ADP-ribose units [72] and form highly branched structures [73] thereby adding a strong negative charge to target proteins. Interestingly, PARP1 associates with UV-induced DNA lesions independently of XPC and DDB2 [71], suggesting that PARP1 may be a third independent sensor of photolesions [74, 75]. The interaction between PARP1 and DDB2 was suggested to stimulate the catalytic activity of PARP1 resulting in PARylation of DDB2, which increased its chromatin retention by inhibiting its ubiquitin-mediated proteasomal degradation [69]. By preventing untimely degradation of DDB2, the PARP1-dependent modification of DDB2 stimulates XPC recruitment to DNA lesions [69, 70]. This illustrates that the chromatin dwell time of DDB2 is tightly controlled to ensure that it is sufficiently long to stimulate XPC recruitment [69, 70], without inhibiting full XPC engagement and subsequent TFIIH recruitment [42, 43].
Independently of this PARP1-DDB2 mechanism, PARP1 also directly interacts with XPC in the nucleoplasm of unchallenged cells and stimulates its recruitment to DNA lesions. While the catalytic activity of PARP1 was not required to form the PARP1-XPC complex, it did stimulate the recruitment of XPC to DNA lesions in a DDB2-independent manner [71]. These findings reveal that PARP1 is tightly linked to early DNA damage recognition by both DDB2 and XPC (Fig. 3b). What the exact mechanism of PARP1 in damage recognition is, whether XPC is involved in stimulating the catalytic activity of PARP enzymes and whether other PARP enzymes, such as PARP2 and PARP3, are involved in GG-NER remain open questions for future research.
The poly-ADP-ribose-dependent chromatin remodeler ALC1 regulates GG-NER
The ATP-dependent chromatin remodeler ALC1, also called CHD1L, becomes activated upon binding PAR chains through its macrodomain [76], resulting in increased chromatin accessibility through nucleosome sliding [77]. ALC1 is recruited to UV-induced DNA lesions in a PARP1-dependent manner and stimulates CPD repair [69] (Fig. 3b). Given the intricate interplay between PARP1, DDB2 and XPC [34, 69,70,71], these DNA damage sensors are likely involved in regulating ALC1 recruitment or activation in response to UV irradiation (Fig. 3b). Depletion of DDB2 was indeed shown to affect the recruitment of ALC1 to sites of UV-induced DNA lesions in XPA-deficient cells [69]. It is important to note that the detection of the PAR response during GG-NER initiation in these studies often required the depletion of the PARG glycohydrolase, which catalyses removal of PAR chains, in GG-NER-deficient cells to boost PAR levels. Now that more sensitive tools have been developed in the last few years, such as recombinant antibody-like ADP-ribose binding proteins [78], it will be important to confirm these earlier findings and re-evaluate conclusions under more physiological settings.
CHD1 stimulates the XPC to TFIIH handover
CHD1 belongs to the CHD family of ATP-dependent chromatin remodelers and contains a central SNF2-like ATPase domain, a DNA-binding domain in its C-terminal and two tandem chromodomains in its N-terminus [79]. CHD1 was reported to be recruited to nucleosomes after UV irradiation in an XPC-dependent manner and to stimulate efficient XPC displacement and subsequent TFIIH recruitment [80] (Fig. 3d). Although clearly detectable, the impact on TFIIH recruitment was rather modest and resulted in delayed CPD repair kinetics in CHD1-depleted cells [80]. Although these findings suggest that CHD1 acts on XPC to promote its displacement or that subsequent TFIIH recruitment may require a different chromatin configuration, these ideas are difficult to reconcile with NER models in which XPC forms a stable DNA damage verification complex together with TFIIH [17, 42, 81]. Thus, the precise mechanism underlying CHD1 function in GG-NER and requirement of its ATP-dependent chromatin remodelling activity remain to be further verified and established. Also, whether other CHD family members, including CHD2, CHD3 and CHD4 which have been found to be important to DSB repair pathways in different chromatin environments [82,83,84,85,86,87,88], function in NER remains to be investigated.
The role of SWI/SNF remodelers in GG-NER: a confusing affair
The SWI/SNF chromatin remodelers incorporate either BRM or BRG1 as ATPase subunit to confer ATP-dependent chromatin remodelling activity [89]. The loss of either BRM or BRG1 results in a NER defect, highlighting an involvement in GG-NER [90,91,92,93]. One study reported a UV-induced interaction between BRG1 and DDB2 and suggested that BRG1 stimulates the recruitment of XPC to DNA lesions early during GG-NER [92]. Somewhat confusingly, BRG1 was found to accumulate at sites of UV-induced DNA lesions only at very late time-points after UV (8 h) when DDB2 and XPC are no longer bound to damage sites [92], arguing against direct recruitment of BRG1 by DDB2 to sites of DNA damage. Another study showed that BRG1 can interact with XPC in co-IP experiments and that BRG1 stimulates XPG recruitment without affecting XPC recruitment [93].
More recent work demonstrates that these remodelers likely affect GG-NER through an indirect mechanism [91]. The SWI/SNF ATPases BRM and BRG1 were found to promote the transcription of the GTF2H1 gene encoding the p62 core subunit of the TFIIH complex by binding to its promoter. Depletion of either BRM or BRG1 indeed downregulates p62 expression and therefore compromises TFIIH stability and the recruitment of GG-NER proteins that bind downstream of TFIIH, including XPG [91]. This is consistent with reduced XPA and XPG recruitment reported earlier [93, 94]. Importantly, the DNA damage sensitivity of BRM/BRG1-depleted cells correlates with p62 levels and re-expression of p62 restores their phenotype [91], revealing an indirect involvement of SWI/SNF chromatin remodelers rather than a direct role during DNA damage recognition in GG-NER. Loss of SWI/SNF subunits was also found to confer UV hypersensitivity in yeast and C. elegans, suggestive of functional evolutionary conservation [95, 96]. Although the reported interaction of two subunits with Rad4 in yeast may point to a more direct role in GG-NER in this species, mapping of genome-wide repair in yeast lacking SWI/SNF subunits shows that this complex is only required for GG-NER in a small subset of genes [97]. Instead, the related RSC ATP-dependent remodeling complex was found to promote GG-NER in both nucleosomal and non-nucleosomal DNA throughout the yeast genome.
DNA damage-recognition proteins and their interconnection with histone modifications
DDB2 triggers histone acetylation in response to UV irradiation
The acetylation of histones at various lysine residues is associated with increased chromatin accessibility [98] due to a weakened electrostatic interaction between DNA and histone tails [99, 100]. In response to UV irradiation there is a strong increase in global H3 and H4 acetylation, suggesting that this modification acts to stimulate DNA repair in chromatin [101, 102]. However, the precise roles of histone acetylation in response to UV irradiation are not yet fully understood. For instance, there is a strong increase in H3 and H4 acetylation immediately as well as several hours after UV [101, 103], while cycling cells also degrade acetylated histones independently of NER in response to replication stress [104].
Histone acetyltransferases (HATs) transfer an acetyl-group from acetyl-coenzyme A onto acceptor proteins such as histones. DDB2 interacts with a number of HATs and targets their histone acetyltransferase activity to chromatin containing DNA lesions. Earlier studies revealed that DDB2 interacts with the HATs p300 [105, 106] and the STAGA complex [107], containing the GCN5 catalytic subunit, which predominantly acetylates H3 [108] (Fig. 4). In addition, DDB1 was found to interact with a GCN5-containing complex that acetylates H3 [109]. Although GCN5 has been implicated in promoting NER via acetylation of H3K9 in both yeast and mammals [110,111,112], the exact roles of these HATs in GG-NER requires further investigation. Nonetheless, these findings clearly highlight the connection between DDB2 and histone acetyltransferase activities. In further support of such a connection, DDB2 itself was found to be acetylated [113] and deacetylated by SIRT6 in response to UV irradiation [44].
A model of the role of acetylation, deacetylation and methylation in GG-NER. a UV-induced lesions in chromatin are recognized by DDB2 resulting in the recruitment of three histone acetyltransferases (STAGA, p300 and phosphorylated HBO1). b These enzymes catalyse histone acetylation leading to increased chromatin accessibility. Phosphorylated HBO1 recruits the chromatin remodeler SNF2H. c Efficient recruitment of XPC also requires histone deacetylation, which is facilitated by DDB2 through the proteolytic degradation of p300 and HBO1, as well as through the recruitment of histone deacetylases (HDAC1-4). Finally, DDB2 also recruits the methyltransferase ASH1L and possibly DOT1L, which methylate H3K4 and H3K79, respectively. XPC preferentially associates with nucleosomes containing methylated histones
More recent findings suggest that DDB2 interacts with the histone acetyltransferase HBO1, also called KAT7, from the MYST family in a UV-dependent manner and facilitates its recruitment to CPDs [102]. Once recruited by DDB2, the HBO1 enzymatic activity stimulates acetylation of H4 and H3K14 and recruits the ATP-dependent chromatin remodeler ACF1-SNF2H through protein–protein interaction in response to UV irradiation, which facilitates XPC recruitment to photolesions [102] (Fig. 4). It should be mentioned, however, that an earlier study found that ACF1-SNF2H functions in TC-NER without an apparent role in regulating GG-NER efficiency [114]. DDB2 was suggested to specifically interact with and recruit phosphorylated HBO1 to sites of DNA damage, which is a substrate of the ATR protein kinase [115]. It is, however, unclear precisely how HBO1 can be phosphorylated by ATR. While ATR has been implicated during the damage recognition step [116, 117], multiple studies have shown that ATR activation is triggered later in NER in a manner that is dependent on dual incision [57, 59, 118,119,120]. One potential explanation could be that successful repair of 6-4PP triggers ATR activation, which would stimulate the HBO1-DDB2 interaction and facilitate CPD repair.
Histone deacetylation stimulates XPC recruitment
While DDB2 may stimulate UV-induced histone acetylation during early repair, DDB2 may also promote the reversal of this chromatin mark at later time-points by regulating the proteolytic degradation of HATs and the recruitment of histone deacetylases (HDACs). DDB2 is incorporated in the CRL4DDB2 E3 ubiquitin ligase complex [20] that ubiquitylates phosphorylated HBO1 leading to its proteasomal degradation after UV irradiation [115]. Similarly, p300 is also degraded by the proteasome in a UV-dependent manner [121], but to what extent this is regulated by DDB2 remains to be determined.
DDB2 was also reported to facilitate recruitment of histone deacetylases HDAC1 and HDAC2 to UV-induced DNA lesions resulting in deacetylation of H3K56 [122]. Indeed, acetylation levels of both H3K56 and H3K9 were reduced in response to UV irradiation [123]. At late time-points after UV H3K56 acetylation was increased, a step suggested to shut-down the UV-induced cell cycle checkpoint [124]. The precise function of H3K9 and H3K56 deacetylation during the early steps of GG-NER remains to be elucidated.
Besides HDAC1 and HDAC2 [122], additional histone deacetylation steps by HDAC3 and HDAC4 have been implicated in GG-NER. In fact, all four HDACs were found to stimulate recruitment of XPC to UV-induced DNA lesions [125,126,127], possibly by lowering the inhibitory impact of histone acetylation on XPC binding to nucleosomes [125] (Fig. 4). Although the precise recruitment mechanism of HDAC3 and HDAC4 and the potential involvement of DDB2 are currently unclear, HDAC3 was specifically linked to H3K14 deacetylation in response to UV irradiation, which was found to stimulate CPD repair in chromatin [126, 127]. How the HBO1-dependent H3K14 acetylation and the HDAC3-dependent H3K14 deacetylation are orchestrated and synergize to stimulate CPD repair remains to be elucidated. Although current literature suggests that DDB2 stimulates recruitment of HDAC1, HDAC2 and possibly other deacetylases resulting in local histone deacetylation (H3K9, H3K14, H3K56, H3K27) necessary for efficient XPC recruitment, further studies are needed to confirm these findings and provide a mechanistic basis for how histone deacetylation facilitates XPC binding. Importantly, it remains to be determined how a combinatorial chromatin code involving specific acetylated and deacetylated histone tails shapes the optimal chromatin landscape for GG-NER. Considering that UV irradiation also triggers replication stress that causes proteasomal degradation of acetylated histones [104] and that both DDB2 and XPC are rapidly recruited to DNA damage sites within seconds [40, 128], it will be important to determine the histone PTM code immediately after UV irradiation and independently of DNA replication.
Histone methylation stimulates the DDB2–XPC handover
Histone methylation is catalysed by histone methyltransferases that mono, di or tri-methylate histone tails [129]. DDB2 was found to interact with and recruit the ASH1L histone methyltransferase to UV-induced DNA lesions resulting in increased H3K4 tri-methylation levels in chromatin containing DNA lesions, which is required for the repair of CPDs [130]. H3K4 tri-methylation, in turn, stimulates the association of XPC with nucleosomes involving a short β-turn motif (XPC residues 741–757) located between the two well-characterized β-hairpin domains BHD2 and BHD3 involved in DNA binding [130] (see Fig. 1b). Conversely, DDB2 preferentially associates with unmethylated nucleosomes, suggesting that H3K4 tri-methylation may stimulate the DDB2–XPC handover at CPDs (Fig. 4). The tri-methylation of H3K4 is associated with active transcription and serves as a binding platform for chromatin remodelers [131]. Thus, UV-induced histone methylation could possibly trigger chromatin remodelling to facilitate GG-NER besides directly influencing XPC binding as well.
In addition to H3K4 tri-methylation, UV irradiation was also found to trigger increased H3K79 tri-methylation by methyltranferase DOT1L [132]. In contrast to K4 which is located in the H3 tail, the K79 residue is located in the H3 core. The action of DOT1L is thought to facilitate XPC recruitment in part through depositing H3K79 tri-methylation to trigger XPC binding and in part through a direct protein–protein interaction between XPC and DOT1L [132]. Similarly, yeast DOT1L was found to promote GG-NER via H3K79 tri-methylation [133, 134]. However, unlike in mammalian cells, which show increased H3K79 tri-methylation [132], UV irradiation does not appear to increase H3K79 tri-methylation in yeast [134, 135]. By contrast, another study in mouse embryonic fibroblasts challenged the view that DOT1L is important for GG-NER, suggesting it rather acts in transcription recovery after UV [136]. Interestingly, mice genetically deleted for DOT1L develop melanomas upon UV irradiation, consistent with the frequent deletion of DOT1L observed in human melanomas [132]. Thus, how the XPC-DOT1L interaction contributes to GG-NER and whether DNA damage detection by XPC is directly influenced by its interaction with histones needs confirmation and further investigation.
The spatial organization of GG-NER in distinct chromatin domains
The cell nucleus is a highly compartmentalized structure that contains distinct structural domains. Chromosomes consists of several dense chromatin domains of about 100–500 nm that each consist of several megabase pairs of DNA. An approximately 100-nm-wide shell at the surface of condensed chromatin domains—known as the perichromatin region—contains partly decondensed chromatin where GG-NER was shown to mainly take place [137, 138]. Electron microscopy experiments revealed that XPC is only moderately enriched in condensed chromatin domains, while both XPC and XPA became strongly enriched in the perichromatin region following UV irradiation. These findings suggest that DNA lesions are recognized in condensed chromatin domains and subsequently relocate to the perichromatin region to be repaired. Indeed, electron microscopy experiments show that UV-damaged chromatin domains undergo significant expansion, which might promote this translocation [138]. Similarly, DNA double-strand breaks in heterochromatin were also found to relocate to the periphery of condensed chromatin domains to be repaired [139, 140].
In line with these findings, the repair of CPDs in heterochromatin is slower than in euchromatin and strongly depends on DDB2 for efficient repair [141, 142]. Live-cell imaging revealed that DDB2 mediates extensive heterochromatin decompaction that is accompanied by linker histone displacement [143]. Interestingly, the UV-induced rapid heterochromatin decompaction occurred within 30 min, is fully compatible with the recruitment of GG-NER proteins within heterochromatin domains, and was followed by a much slower heterochromatin recompaction phase within 12 h [143].
While CPDs form in both eu- and heterochromatin, it appears that UV irradiation selectively triggers 6–4 PP formation in euchromatin [142], with a preference for internucleosomal regions over nucleosome core particles [144]. Interestingly, DDB2 preferentially associates with internucleosomal regions and directs XPC to these sites in an ubiquitin-dependent manner to suppress the association of XPC with nucleosome core particles [144]. According to this model, DDB2 prioritizes GG-NER in internucleosomal regions to ensure rapid repair of 6–4 PPs and CPDs in these genomic regions, while the repair of CPDs in nucleosome core particles is stimulated by protein–protein interactions between DDB2 and XPC in an ubiquitin-independent mechanism [144].
Concluding remarks
The last few years have witnessed the identification of many new links between chromatin modulators and GG-NER. This review focused on recent insights into the coordinated DDB2-dependent recruitment of histone acetyltransferases [102, 105] and histone methyltransferases [130] that together with the DDB2-associated E3 ubiquitin ligase [37, 55] extensively modify histone tails to create a local chromatin environment that facilitates early XPC recruitment. Identifying the specific histone tail residues that are modified during GG-NER and their interconnections will be important future goals, together with mechanistic studies to unravel how exactly histone PTMs influence the binding of XPC to DNA and its detection of DNA lesions. These events are aided by the association of a number of ATP-dependent chromatin remodelers that probably mediate further chromatin opening to facilitate not only early recognition of DNA lesions [67, 69, 102], but possibly also DNA damage handover to promote progression of the GG-NER reaction [80]. To better understand their precise involvement, it will be necessary to study histone and nucleosome occupancy and dynamics in response to UV-induced DNA lesions, which has, thus, far been difficult because NER lesions cannot be induced at a predefined location. Electron and fluorescence microscopic techniques have found clear evidence for chromatin expansion, histone eviction and chromatin restoration during and after completion of GG-NER [33, 34, 138, 145,146,147]. However, the specific ATP-dependent chromatin remodelers involve in mediating these GG-NER steps remain elusive. The development of techniques to map the nucleosomal landscape at single nucleotide resolution following UV irradiation in both yeast and mammalian cells [148, 149] will be a powerful new tool to better understand chromatin dynamics during GG-NER. A third seemingly independent DNA lesion-recognition protein—PARP1—also acts in GG-NER [34, 69,70,71], but its precise links with XPC and DDB2 need further exploration.
An emerging theme is that DNA lesion-recognition factors also need to dissociate in a timely fashion to prevent them from inhibiting subsequent repair steps. Timely removal from chromatin is tightly coordinated through ubiquitylation of both XPC and DDB2 and their subsequent ubiquitin-dependent extraction by the VCP segregase [42, 43]. These ubiquitylation events, in turn, are also subjected to tight regulation and require prior SUMOylation of XPC [47, 51] or can be prevented by competitive PARylation of DDB2 [69]. Powerful new methods including sensitive proteomic approaches [150] and genome-wide CRISPR screens [151] will not only identify the full repertoire of chromatin modulators of GG-NER, but will also facilitate subsequent structural studies of how GG-NER operates in nucleosomes by cryo-EM [22]. New developments now allow the study of GG-NER in intact organisms [152], providing insights into developmentally regulated chromatin modulators. A better understanding of how these post-translational modifications and remodelers progressively modify chromatin in a stepwise fashion during the different stages of repair will further reveal how GG-NER leaves it mark on chromatin.
Data availability
Not applicable.
References
Gates KS (2009) An overview of chemical processes that damage cellular DNA: spontaneous hydrolysis, alkylation, and reactions with radicals. Chem Res Toxicol 22:1747–1760
Hoeijmakers JH (2009) DNA damage, aging, and cancer. N Engl J Med 361:1475–1485
Ciccia A, Elledge SJ (2010) The DNA damage response: making it safe to play with knives. Mol Cell 40:179–204
Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH (2014) Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol 15:465–481
van den Heuvel D, van der Weegen Y, Boer DEC, Ogi T, Luijsterburg MS (2021) Transcription-coupled DNA Repair: from mechanism to human disorder. Trends Cell Biol. https://doi.org/10.1016/j.tcb.2021.02.007
Scharer OD (2013) Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol 5:a012609
Sugasawa K (2016) Molecular mechanisms of DNA damage recognition for mammalian nucleotide excision repair. DNA Repair (Amst) 44:110–117
Gregersen LH, Svejstrup JQ (2018) The cellular response to transcription-blocking DNA damage. Trends Biochem Sci 43:327–341
Lans H, Hoeijmakers JHJ, Vermeulen W, Marteijn JA (2019) The DNA damage response to transcription stress. Nat Rev Mol Cell Biol 20:766–784
Camenisch U, Trautlein D, Clement FC, Fei J, Leitenstorfer A, Ferrando-May E, Naegeli H (2009) Two-stage dynamic DNA quality check by xeroderma pigmentosum group C protein. EMBO J. https://doi.org/10.1038/emboj.2009.187
Maillard O, Solyom S, Naegeli H (2007) An aromatic sensor with aversion to damaged strands confers versatility to DNA repair. PLoS Biol 5:e79
Min JH, Pavletich NP (2007) Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature 449:570–575
Sugasawa K, Okamoto T, Shimizu Y, Masutani C, Iwai S, Hanaoka F (2001) A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev 15:507–521
Paul D, Mu H, Zhao H, Ouerfelli O, Jeffrey PD, Broyde S, Min JH (2019) Structure and mechanism of pyrimidine-pyrimidone (6–4) photoproduct recognition by the Rad4/XPC nucleotide excision repair complex. Nucleic Acids Res 47:6015–6028
Mu H, Zhang Y, Geacintov NE, Broyde S (2018) Lesion sensing during initial binding by yeast XPC/Rad4: toward predicting resistance to nucleotide excision repair. Chem Res Toxicol 31:1260–1268
Gunz D, Hess MT, Naegeli H (1996) Recognition of DNA adducts by human nucleotide excision repair. Evidence for a thermodynamic probing mechanism. J Biol Chem 271:25089–25098
Mu H, Geacintov NE, Broyde S, Yeo JE, Scharer OD (2018) Molecular basis for damage recognition and verification by XPC-RAD23B and TFIIH in nucleotide excision repair. DNA Repair (Amst) 71:33–42
Chen X, Velmurugu Y, Zheng G, Park B, Shim Y, Kim Y, Liu L, Van Houten B, He C, Ansari A, Min JH (2015) Kinetic gating mechanism of DNA damage recognition by Rad4/XPC. Nat Commun 6:5849
Groisman R, Kuraoka I, Chevallier O, Gaye N, Magnaldo T, Tanaka K, Kisselev AF, Harel-Bellan A, Nakatani Y (2006) CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes Dev 20:1429–1434
Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K, Nakatani Y (2003) The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113:357–367
Scrima A, Konickova R, Czyzewski BK, Kawasaki Y, Jeffrey PD, Groisman R, Nakatani Y, Iwai S, Pavletich NP, Thoma NH (2008) Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell 135:1213–1223
Matsumoto S, Cavadini S, Bunker RD, Grand RS, Potenza A, Rabl J, Yamamoto J, Schenk AD, Schubeler D, Iwai S, Sugasawa K, Kurumizaka H, Thoma NH (2019) DNA damage detection in nucleosomes involves DNA register shifting. Nature 571:79–84
Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, Mori T, Iwai S, Tanaka K, Hanaoka F (2005) UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 121:387–400
Yasuda G, Nishi R, Watanabe E, Mori T, Iwai S, Orioli D, Stefanini M, Hanaoka F, Sugasawa K (2007) In vivo destabilization and functional defects of the xeroderma pigmentosum C protein caused by a pathogenic missense mutation. Mol Cell Biol 27:6606–6614
Aboussekhra A, Biggerstaff M, Shivji MK, Vilpo JA, Moncollin V, Podust VN, Protic M, Hubscher U, Egly JM, Wood RD (1995) Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell 80:859–868
Mu D, Park CH, Matsunaga T, Hsu DS, Reardon JT, Sancar A (1995) Reconstitution of human DNA repair excision nuclease in a highly defined system. J Biol Chem 270:2415–2418
Gong F, Kwon Y, Smerdon MJ (2005) Nucleotide excision repair in chromatin and the right of entry. DNA Repair (Amst) 4:884–896
Hara R, Mo J, Sancar A (2000) DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Mol Cell Biol 20:9173–9181
Hara R, Sancar A (2002) The SWI/SNF chromatin-remodeling factor stimulates repair by human excision nuclease in the mononucleosome core particle. Mol Cell Biol 22:6779–6787
Ura K, Araki M, Saeki H, Masutani C, Ito T, Iwai S, Mizukoshi T, Kaneda Y, Hanaoka F (2001) ATP-dependent chromatin remodeling facilitates nucleotide excision repair of UV-induced DNA lesions in synthetic dinucleosomes. EMBO J 20:2004–2014
Polo SE, Almouzni G (2015) Chromatin dynamics after DNA damage: The legacy of the access-repair-restore model. DNA Repair (Amst) 36:114–121
Smerdon MJ, Lieberman MW (1978) Nucleosome rearrangement in human chromatin during UV-induced DNA-repair synthesis. Proc Natl Acad Sci USA 75:4238–4241
Adam S, Dabin J, Chevallier O, Leroy O, Baldeyron C, Corpet A, Lomonte P, Renaud O, Almouzni G, Polo SE (2016) Real-time tracking of parental histones reveals their contribution to chromatin integrity following DNA damage. Mol Cell 64:65–78
Luijsterburg MS, Lindh M, Acs K, Vrouwe MG, Pines A, van Attikum H, Mullenders LH, Dantuma NP (2012) DDB2 promotes chromatin decondensation at UV-induced DNA damage. J Cell Biol 197:267–281
Osakabe A, Tachiwana H, Kagawa W, Horikoshi N, Matsumoto S, Hasegawa M, Matsumoto N, Toga T, Yamamoto J, Hanaoka F, Thoma NH, Sugasawa K, Iwai S, Kurumizaka H (2015) Structural basis of pyrimidine-pyrimidone (6–4) photoproduct recognition by UV-DDB in the nucleosome. Sci Rep 5:16330
Moser J, Volker M, Kool H, Alekseev S, Vrieling H, Yasui A, van Zeeland AA, Mullenders LH (2005) The UV-damaged DNA binding protein mediates efficient targeting of the nucleotide excision repair complex to UV-induced photo lesions. DNA Repair (Amst) 4:571–582
Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument-Bromage H, Tempst P, Xiong Y, Zhang Y (2006) Histone H3 and H4 Ubiquitylation by the CUL4-DDB-ROC1 Ubiquitin Ligase Facilitates Cellular Response to DNA Damage. Mol Cell 22:383–394
Sugasawa K (2006) UV-induced ubiquitylation of XPC complex, the UV-DDB-ubiquitin ligase complex, and DNA repair. J Mol Histol. https://doi.org/10.1007/s10735-006-9044-7
El-Mahdy MA, Zhu Q, Wang QE, Wani G, Praetorius-Ibba M, Wani AA (2006) Cullin 4A-mediated proteolysis of DDB2 protein at DNA damage sites regulates in vivo lesion recognition by XPC. J Biol Chem. https://doi.org/10.1074/jbc.M511834200
Luijsterburg MS, Goedhart J, Moser J, Kool H, Geverts B, Houtsmuller AB, Mullenders LH, Vermeulen W, van Driel R (2007) Dynamic in vivo interaction of DDB2 E3 ubiquitin ligase with UV-damaged DNA is independent of damage-recognition protein XPC. J Cell Sci 120:2706–2716
Han C, Zhao R, Kroger J, He J, Wani G, Wang QE, Wani AA (2017) UV radiation-induced SUMOylation of DDB2 regulates nucleotide excision repair. Carcinogenesis 38:976–985
Ribeiro-Silva C, Sabatella M, Helfricht A, Marteijn JA, Theil AF, Vermeulen W, Lans H (2020) Ubiquitin and TFIIH-stimulated DDB2 dissociation drives DNA damage handover in nucleotide excision repair. Nat Commun 11:4868
Puumalainen MR, Lessel D, Ruthemann P, Kaczmarek N, Bachmann K, Ramadan K, Naegeli H (2014) Chromatin retention of DNA damage sensors DDB2 and XPC through loss of p97 segregase causes genotoxicity. Nat Commun 5:3695
Geng A, Tang H, Huang J, Qian Z, Qin N, Yao Y, Xu Z, Chen H, Lan L, Xie H, Zhang J, Jiang Y, Mao Z (2020) The deacetylase SIRT6 promotes the repair of UV-induced DNA damage by targeting DDB2. Nucleic Acids Res 48:9181–9194
Akita M, Tak YS, Shimura T, Matsumoto S, Okuda-Shimizu Y, Shimizu Y, Nishi R, Saitoh H, Iwai S, Mori T, Ikura T, Sakai W, Hanaoka F, Sugasawa K (2015) SUMOylation of xeroderma pigmentosum group C protein regulates DNA damage recognition during nucleotide excision repair. Sci Rep 5:10984
Riedl T, Hanaoka F, Egly JM (2003) The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J 22:5293–5303
van Cuijk L, van Belle GJ, Turkyilmaz Y, Poulsen SL, Janssens RC, Theil AF, Sabatella M, Lans H, Mailand N, Houtsmuller AB, Vermeulen W, Marteijn JA (2015) SUMO and ubiquitin-dependent XPC exchange drives nucleotide excision repair. Nat Commun 6:7499
Wakasugi M, Sancar A (1998) Assembly, subunit composition, and footprint of human DNA repair excision nuclease. Proc Natl Acad Sci USA 95:6669–6674
He J, Zhu Q, Wani G, Sharma N, Han C, Qian J, Pentz K, Wang QE, Wani AA (2014) Ubiquitin-specific protease 7 regulates nucleotide excision repair through deubiquitinating XPC protein and preventing XPC protein from undergoing ultraviolet light-induced and VCP/p97 protein-regulated proteolysis. J Biol Chem 289:27278–27289
Shah P, Qiang L, Yang S, Soltani K, He YY (2017) Regulation of XPC deubiquitination by USP11 in repair of UV-induced DNA damage. Oncotarget 8:96522–96535
Poulsen SL, Hansen RK, Wagner SA, van Cuijk L, van Belle GJ, Streicher W, Wikstrom M, Choudhary C, Houtsmuller AB, Marteijn JA, Bekker-Jensen S, Mailand N (2013) RNF111/Arkadia is a SUMO-targeted ubiquitin ligase that facilitates the DNA damage response. J Cell Biol 201:797–807
Wang QE, Zhu Q, Wani G, El-Mahdy MA, Li J, Wani AA (2005) DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation. Nucleic Acids Res 33:4023–4034
Nishi R, Alekseev S, Dinant C, Hoogstraten D, Houtsmuller AB, Hoeijmakers JH, Vermeulen W, Hanaoka F, Sugasawa K (2009) UV-DDB-dependent regulation of nucleotide excision repair kinetics in living cells. DNA Repair (Amst) 8:767–776
Bergink S, Salomons FA, Hoogstraten D, Groothuis TA, de Waard H, Wu J, Yuan L, Citterio E, Houtsmuller AB, Neefjes J, Hoeijmakers JH, Vermeulen W, Dantuma NP (2006) DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev 20:1343–1352
Gracheva E, Chitale S, Wilhelm T, Rapp A, Byrne J, Stadler J, Medina R, Cardoso MC, Richly H (2016) ZRF1 mediates remodeling of E3 ligases at DNA lesion sites during nucleotide excision repair. J Cell Biol 213:185–200
Kapetanaki MG, Guerrero-Santoro J, Bisi DC, Hsieh CL, Rapic-Otrin V, Levine AS (2006) The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.0511160103
Marteijn JA, Bekker-Jensen S, Mailand N, Lans H, Schwertman P, Gourdin AM, Dantuma NP, Lukas J, Vermeulen W (2009) Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J Cell Biol 186:835–847
Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y (2004) Role of histone H2A ubiquitination in Polycomb silencing. Nature 431:873–878
Hanasoge S, Ljungman M (2007) H2AX phosphorylation after UV irradiation is triggered by DNA repair intermediates and is mediated by the ATR kinase. Carcinogenesis 28:2298–2304
Sertic S, Pizzi S, Cloney R, Lehmann AR, Marini F, Plevani P, Muzi-Falconi M (2011) Human exonuclease 1 connects nucleotide excision repair (NER) processing with checkpoint activation in response to UV irradiation. Proc Natl Acad Sci USA 108:13647–13652
Thorslund T, Ripplinger A, Hoffmann S, Wild T, Uckelmann M, Villumsen B, Narita T, Sixma TK, Choudhary C, Bekker-Jensen S, Mailand N (2015) Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature 527:389–393
Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, Marteijn JA, Sixma TK (2012) RNF168 ubiquitinates K13–15 on H2A/H2AX to drive DNA damage signaling. Cell 150:1182–1195
Richly H, Rocha-Viegas L, Ribeiro JD, Demajo S, Gundem G, Lopez-Bigas N, Nakagawa T, Rospert S, Ito T, Di Croce L (2010) Transcriptional activation of polycomb-repressed genes by ZRF1. Nature 468:1124–1128
Jin J, Cai Y, Yao T, Gottschalk AJ, Florens L, Swanson SK, Gutierrez JL, Coleman MK, Workman JL, Mushegian A, Washburn MP, Conaway RC, Conaway JW (2005) A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J Biol Chem 280:41207–41212
van Attikum H, Fritsch O, Hohn B, Gasser SM (2004) Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119:777–788
Andreev V, Hristova R, Asparuhova M, Danovski G, Stoynov S, Gospodinov A (2019) Mammalian INO80 chromatin remodeler cooperates with FANCM to mediate DNA interstrand crosslink-induced checkpoint activation and repair. DNA Repair (Amst) 74:38–50
Jiang Y, Wang X, Bao S, Guo R, Johnson DG, Shen X, Li L (2010) INO80 chromatin remodeling complex promotes the removal of UV lesions by the nucleotide excision repair pathway. Proc Natl Acad Sci USA 107:17274–17279
Sarkar S, Kiely R, McHugh PJ (2010) The Ino80 chromatin-remodeling complex restores chromatin structure during UV DNA damage repair. J Cell Biol 191:1061–1068
Pines A, Vrouwe MG, Marteijn JA, Typas D, Luijsterburg MS, Cansoy M, Hensbergen P, Deelder A, de Groot A, Matsumoto S, Sugasawa K, Thoma N, Vermeulen W, Vrieling H, Mullenders L (2012) PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1. J Cell Biol 199:235–249
Robu M, Shah RG, Petitclerc N, Brind’amour J, Kandan-Kulangara F, Shah GM (2013) Role of poly(ADP-ribose) polymerase-1 in the removal of UV-induced DNA lesions by nucleotide excision repair. Proc Natl Acad Sci USA 110:1658–1663
Robu M, Shah RG, Purohit NK, Zhou P, Naegeli H, Shah GM (2017) Poly(ADP-ribose) polymerase 1 escorts XPC to UV-induced DNA lesions during nucleotide excision repair. Proc Natl Acad Sci USA 114:E6847–E6856
Gibson BA, Kraus WL (2012) New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol 13:411–424
Miwa M, Saikawa N, Yamaizumi Z, Nishimura S, Sugimura T (1979) Structure of poly(adenosine diphosphate ribose): identification of 2’-[1"-ribosyl-2"-(or 3"-)(1"’-ribosyl)]adenosine-5’,5",5"’-tris(phosphate) as a branch linkage. Proc Natl Acad Sci USA 76:595–599
Pines A, Mullenders LH, van Attikum H, Luijsterburg MS (2013) Touching base with PARPs: moonlighting in the repair of UV lesions and double-strand breaks. Trends Biochem Sci 38:321–330
Purohit NK, Robu M, Shah RG, Geacintov NE, Shah GM (2016) Characterization of the interactions of PARP-1 with UV-damaged DNA in vivo and in vitro. Sci Rep 6:19020
Singh HR, Nardozza AP, Moller IR, Knobloch G, Kistemaker HAV, Hassler M, Harrer N, Blessing C, Eustermann S, Kotthoff C, Huet S, Mueller-Planitz F, Filippov DV, Timinszky G, Rand KD, Ladurner AG (2017) A Poly-ADP-ribose trigger releases the auto-inhibition of a chromatin remodeling oncogene. Mol Cell 68:860-871 e867
Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, Owen-Hughes T, Boulton SJ (2009) Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 325:1240–1243
Gibson BA, Conrad LB, Huang D, Kraus WL (2017) Generation and characterization of recombinant antibody-like ADP-ribose binding proteins. Biochemistry 56:6305–6316
Marfella CG, Imbalzano AN (2007) The Chd family of chromatin remodelers. Mutat Res 618:30–40
Ruthemann P, Balbo Pogliano C, Codilupi T, Garajova Z, Naegeli H (2017) Chromatin remodeler CHD1 promotes XPC-to-TFIIH handover of nucleosomal UV lesions in nucleotide excision repair. EMBO J 36:3372–3386
Sugasawa K, Akagi J, Nishi R, Iwai S, Hanaoka F (2009) Two-step recognition of DNA damage for mammalian nucleotide excision repair: Directional binding of the XPC complex and DNA strand scanning. Mol Cell 36:642–653
Goodarzi AA, Kurka T, Jeggo PA (2011) KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat Struct Mol Biol 18:831–839
Klement K, Luijsterburg MS, Pinder JB, Cena CS, Del Nero V, Wintersinger CM, Dellaire G, van Attikum H, Goodarzi AA (2014) Opposing ISWI- and CHD-class chromatin remodeling activities orchestrate heterochromatic DNA repair. J Cell Biol 207:717–733
Larsen DH, Poinsignon C, Gudjonsson T, Dinant C, Payne MR, Hari FJ, Rendtlew Danielsen JM, Menard P, Sand JC, Stucki M, Lukas C, Bartek J, Andersen JS, Lukas J (2010) The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J Cell Biol 190:731–740
Luijsterburg MS, Acs K, Ackermann L, Wiegant WW, Bekker-Jensen S, Larsen DH, Khanna KK, van Attikum H, Mailand N, Dantuma NP (2012) A new non-catalytic role for ubiquitin ligase RNF8 in unfolding higher-order chromatin structure. EMBO J 31:2511–2527
Luijsterburg MS, de Krijger I, Wiegant WW, Shah RG, Smeenk G, de Groot AJ, Pines A, Vertegaal AC, Jacobs JJ, Shah GM, van Attikum H (2016) PARP1 Links CHD2-mediated chromatin expansion and H3.3 Deposition to DNA repair by non-homologous end-joining. Mol Cell 61:547–562
Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP (2010) Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J 29:3130–3139
Smeenk G, Wiegant WW, Vrolijk H, Solari AP, Pastink A, van Attikum H (2010) The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J Cell Biol 190:741–749
Pulice JL, Kadoch C (2016) Composition and Function of Mammalian SWI/SNF Chromatin Remodeling Complexes in Human Disease. Cold Spring Harb Symp Quant Biol 81:53–60
Gong F, Fahy D, Liu H, Wang W, Smerdon MJ (2008) Role of the mammalian SWI/SNF chromatin remodeling complex in the cellular response to UV damage. Cell Cycle 7:1067–1074
Ribeiro-Silva C, Aydin OZ, Mesquita-Ribeiro R, Slyskova J, Helfricht A, Marteijn JA, Hoeijmakers JHJ, Lans H, Vermeulen W (2018) DNA damage sensitivity of SWI/SNF-deficient cells depends on TFIIH subunit p62/GTF2H1. Nat Commun 9:4067
Zhang L, Zhang Q, Jones K, Patel M, Gong F (2009) The chromatin remodeling factor BRG1 stimulates nucleotide excision repair by facilitating recruitment of XPC to sites of DNA damage. Cell Cycle 8:3953–3959
Zhao Q, Wang QE, Ray A, Wani G, Han C, Milum K, Wani AA (2009) Modulation of nucleotide excision repair by mammalian SWI/SNF chromatin-remodeling complex. J Biol Chem 284:30424–30432
Watanabe R, Kanno SI, Mohammadi Roushandeh A, Ui A, Yasui A (2017) Nucleosome remodelling, DNA repair and transcriptional regulation build negative feedback loops in cancer and cellular ageing. Philos Trans R Soc Lond B Biol Sci 372:20160473
Gong F, Fahy D, Smerdon MJ (2006) Rad4-Rad23 interaction with SWI/SNF links ATP-dependent chromatin remodeling with nucleotide excision repair. Nat Struct Mol Biol 13:902–907
Lans H, Marteijn JA, Schumacher B, Hoeijmakers JH, Jansen G, Vermeulen W (2010) Involvement of global genome repair, transcription coupled repair, and chromatin remodeling in UV DNA damage response changes during development. PLoS Genet 6:e1000941
Bohm KA, Hodges AJ, Czaja W, Selvam K, Smerdon MJ, Mao P, Wyrick JJ (2021) Distinct roles for RSC and SWI/SNF chromatin remodelers in genomic excision repair. Genome Res 31:1047–1059
Grunstein M (1997) Histone acetylation in chromatin structure and transcription. Nature 389:349–352
Brower-Toland B, Wacker DA, Fulbright RM, Lis JT, Kraus WL, Wang MD (2005) Specific contributions of histone tails and their acetylation to the mechanical stability of nucleosomes. J Mol Biol 346:135–146
Kim J, Lee J, Lee TH (2015) Lysine acetylation facilitates spontaneous DNA dynamics in the nucleosome. J Phys Chem B 119:15001–15005
Kim MK, Shin JM, Eun HC, Chung JH (2009) The role of p300 histone acetyltransferase in UV-induced histone modifications and MMP-1 gene transcription. PLoS ONE 4:e4864
Niida H, Matsunuma R, Horiguchi R, Uchida C, Nakazawa Y, Motegi A, Nishimoto K, Sakai S, Ohhata T, Kitagawa K, Moriwaki S, Nishitani H, Ui A, Ogi T, Kitagawa M (2017) Phosphorylated HBO1 at UV irradiated sites is essential for nucleotide excision repair. Nat Commun 8:16102
Ramanathan B, Smerdon MJ (1986) Changes in nuclear protein acetylation in u.v.-damaged human cells. Carcinogenesis 7:1087–1094
Mandemaker IK, Geijer ME, Kik I, Bezstarosti K, Rijkers E, Raams A, Janssens RC, Lans H, Hoeijmakers JH, Demmers JA, Vermeulen W, Marteijn JA (2018) DNA damage-induced replication stress results in PA200-proteasome-mediated degradation of acetylated histones. EMBO Rep. https://doi.org/10.15252/embr.201745566
Datta A, Bagchi S, Nag A, Shiyanov P, Adami GR, Yoon T, Raychaudhuri P (2001) The p48 subunit of the damaged-DNA binding protein DDB associates with the CBP/p300 family of histone acetyltransferase. Mutat Res 486:89–97
Rapic-Otrin V, McLenigan MP, Bisi DC, Gonzalez M, Levine AS (2002) Sequential binding of UV DNA damage binding factor and degradation of the p48 subunit as early events after UV irradiation. Nucleic Acids Res 30:2588–2598
Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG (2001) Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol 21:6782–6795
Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, Allis CD (1996) Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383:269–272
Brand M, Moggs JG, Oulad-Abdelghani M, Lejeune F, Dilworth FJ, Stevenin J, Almouzni G, Tora L (2001) UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J 20:3187–3196
Guo R, Chen J, Mitchell DL, Johnson DG (2010) GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage. Nucleic Acids Res 39(4):1390–1397
Waters R, van Eijk P, Reed S (2015) Histone modification and chromatin remodeling during NER. DNA Repair (Amst) 36:105–113
Yu S, Evans K, van Eijk P, Bennett M, Webster RM, Leadbitter M, Teng Y, Waters R, Jackson SP, Reed SH (2016) Global genome nucleotide excision repair is organized into domains that promote efficient DNA repair in chromatin. Genome Res 26:1376–1387
Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325:834–840
Aydin OZ, Marteijn JA, Ribeiro-Silva C, Rodriguez Lopez A, Wijgers N, Smeenk G, van Attikum H, Poot RA, Vermeulen W, Lans H (2014) Human ISWI complexes are targeted by SMARCA5 ATPase and SLIDE domains to help resolve lesion-stalled transcription. Nucleic Acids Res 42:8473–8485
Matsunuma R, Niida H, Ohhata T, Kitagawa K, Sakai S, Uchida C, Shiotani B, Matsumoto M, Nakayama KI, Ogura H, Shiiya N, Kitagawa M (2016) UV damage-induced phosphorylation of HBO1 Triggers CRL4DDB2-mediated degradation to regulate cell proliferation. Mol Cell Biol 36:394–406
Ray A, Milum K, Battu A, Wani G, Wani AA (2013) NER initiation factors, DDB2 and XPC, regulate UV radiation response by recruiting ATR and ATM kinases to DNA damage sites. DNA Repair (Amst) 12:273–283
Ray A, Mir SN, Wani G, Zhao Q, Battu A, Zhu Q, Wang QE, Wani AA (2009) Human SNF5/INI1, a component of the human SWI/SNF chromatin remodeling complex, promotes nucleotide excision repair by influencing ATM recruitment and downstream H2AX phosphorylation. Mol Cell Biol 29:6206–6219
Marini F, Nardo T, Giannattasio M, Minuzzo M, Stefanini M, Plevani P, Muzi Falconi M (2006) DNA nucleotide excision repair-dependent signaling to checkpoint activation. Proc Natl Acad Sci USA 103:17325–17330
Matsumoto M, Yaginuma K, Igarashi A, Imura M, Hasegawa M, Iwabuchi K, Date T, Mori T, Ishizaki K, Yamashita K, Inobe M, Matsunaga T (2007) Perturbed gap-filling synthesis in nucleotide excision repair causes histone H2AX phosphorylation in human quiescent cells. J Cell Sci 120:1104–1112
Vrouwe MG, Pines A, Overmeer RM, Hanada K, Mullenders LH (2011) UV-induced photolesions elicit ATR-kinase-dependent signaling in non-cycling cells through nucleotide excision repair-dependent and -independent pathways. J Cell Sci 124:435–446
Wang QE, Han C, Zhao R, Wani G, Zhu Q, Gong L, Battu A, Racoma I, Sharma N, Wani AA (2013) p38 MAPK- and Akt-mediated p300 phosphorylation regulates its degradation to facilitate nucleotide excision repair. Nucleic Acids Res 41:1722–1733
Zhu Q, Battu A, Ray A, Wani G, Qian J, He J, Wang QE, Wani AA (2015) Damaged DNA-binding protein down-regulates epigenetic mark H3K56Ac through histone deacetylase 1 and 2. Mutat Res 776:16–23
Tjeertes JV, Miller KM, Jackson SP (2009) Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J 28:1878–1889
Battu A, Ray A, Wani AA (2011) ASF1A and ATM regulate H3K56-mediated cell-cycle checkpoint recovery in response to UV irradiation. Nucleic Acids Res 39:7931–7945
Kakumu E, Nakanishi S, Shiratori HM, Kato A, Kobayashi W, Machida S, Yasuda T, Adachi N, Saito N, Ikura T, Kurumizaka H, Kimura H, Yokoi M, Sakai W, Sugasawa K (2017) Xeroderma pigmentosum group C protein interacts with histones: regulation by acetylated states of histone H3. Genes Cells 22:310–327
Li S, Zhou M, Ze K, Sun X, Zhao C, Li Z, Lu H, Jiao Y, Wang T, Li S, Hua L, Cai H, Li X (2020) Protective role of histone deacetylase 4 from ultraviolet radiation-induced DNA lesions. Mol Carcinog 59:1292–1301
Nishimoto K, Niida H, Uchida C, Ohhata T, Kitagawa K, Motegi A, Suda T, Kitagawa M (2020) HDAC3 is required for XPC recruitment and nucleotide excision repair of DNA damage induced by UV irradiation. Mol Cancer Res 18:1367–1378
Hoogstraten D, Bergink S, Ng JM, Verbiest VH, Luijsterburg MS, Geverts B, Raams A, Dinant C, Hoeijmakers JH, Vermeulen W, Houtsmuller AB (2008) Versatile DNA damage detection by the global genome nucleotide excision repair protein XPC. J Cell Sci 121:2850–2859
Gong F, Miller KM (2019) Histone methylation and the DNA damage response. Mutat Res 780:37–47
Balbo Pogliano C, Gatti M, Ruthemann P, Garajova Z, Penengo L, Naegeli H (2017) ASH1L histone methyltransferase regulates the handoff between damage recognition factors in global-genome nucleotide excision repair. Nat Commun 8:1333
Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S (2005) Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 438:1181–1185
Zhu B, Chen S, Wang H, Yin C, Han C, Peng C, Liu Z, Wan L, Zhang X, Zhang J, Lian CG, Ma P, Xu ZX, Prince S, Wang T, Gao X, Shi Y, Liu D, Liu M, Wei W, Wei Z, Pan J, Wang Y, Xuan Z, Hess J, Hayward NK, Goding CR, Chen X, Zhou J, Cui R (2018) The protective role of DOT1L in UV-induced melanomagenesis. Nat Commun 9:259
Bostelman LJ, Keller AM, Albrecht AM, Arat A, Thompson JS (2007) Methylation of histone H3 lysine-79 by Dot1p plays multiple roles in the response to UV damage in Saccharomyces cerevisiae. DNA Repair (Amst) 6:383–395
Tatum D, Li S (2011) Evidence that the histone methyltransferase Dot1 mediates global genomic repair by methylating histone H3 on lysine 79. J Biol Chem 286:17530–17535
Rossodivita AA, Boudoures AL, Mecoli JP, Steenkiste EM, Karl AL, Vines EM, Cole AM, Ansbro MR, Thompson JS (2014) Histone H3 K79 methylation states play distinct roles in UV-induced sister chromatid exchange and cell cycle checkpoint arrest in Saccharomyces cerevisiae. Nucleic Acids Res 42:6286–6299
Oksenych V, Zhovmer A, Ziani S, Mari PO, Eberova J, Nardo T, Stefanini M, Giglia-Mari G, Egly JM, Coin F (2013) Histone methyltransferase DOT1L drives recovery of gene expression after a genotoxic attack. PLoS Genet 9:e1003611
Fakan S, van Driel R (2007) The perichromatin region: A functional compartment in the nucleus that determines large-scale chromatin folding. Semin Cell Dev Biol 18:676–681
Solimando L, Luijsterburg MS, Vecchio L, Vermeulen W, van Driel R, Fakan S (2009) Spatial organization of nucleotide excision repair proteins after UV-induced DNA damage in the human cell nucleus. J Cell Sci 122:83–91
Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH (2011) Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell 144:732–744
Tsouroula K, Furst A, Rogier M, Heyer V, Maglott-Roth A, Ferrand A, Reina-San-Martin B, Soutoglou E (2016) Temporal and Spatial Uncoupling of DNA Double Strand Break Repair Pathways within Mammalian Heterochromatin. Mol Cell 63:293–305
Adar S, Hu J, Lieb JD, Sancar A (2016) Genome-wide kinetics of DNA excision repair in relation to chromatin state and mutagenesis. Proc Natl Acad Sci USA 113:E2124-2133
Han C, Srivastava AK, Cui T, Wang QE, Wani AA (2016) Differential DNA lesion formation and repair in heterochromatin and euchromatin. Carcinogenesis 37:129–138
Fortuny A, Chansard A, Caron P, Chevallier O, Leroy O, Renaud O, Polo SE (2021) Imaging the response to DNA damage in heterochromatin domains reveals core principles of heterochromatin maintenance. Nat Commun 12:2428
Fei J, Kaczmarek N, Luch A, Glas A, Carell T, Naegeli H (2011) Regulation of nucleotide excision repair by UV-DDB: prioritization of damage recognition to internucleosomal DNA. PLoS Biol 9:e1001183
Adam S, Polo SE, Almouzni G (2013) Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell 155:94–106
Dinant C, Ampatziadis-Michailidis G, Lans H, Tresini M, Lagarou A, Grosbart M, Theil AF, van Cappellen WA, Kimura H, Bartek J, Fousteri M, Houtsmuller AB, Vermeulen W, Marteijn JA (2013) Enhanced chromatin dynamics by FACT promotes transcriptional restart after UV-induced DNA damage. Mol Cell 51:469–479
Duan MR, Smerdon MJ (2010) UV damage in DNA promotes nucleosome unwrapping. J Biol Chem 285:26295–26303
Nakata K, Iwata K, Kojima K, Kanai K (1989) Computed tomography of liver sarcoidosis. J Comput Assist Tomogr 13:707–708
van Eijk P, Nandi SP, Yu S, Bennett M, Leadbitter M, Teng Y, Reed SH (2019) Nucleosome remodeling at origins of global genome-nucleotide excision repair occurs at the boundaries of higher-order chromatin structure. Genome Res 29:74–84
Branon TC, Bosch JA, Sanchez AD, Udeshi ND, Svinkina T, Carr SA, Feldman JL, Perrimon N, Ting AY (2018) Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol 36:880–887
Olivieri M, Cho T, Alvarez-Quilon A, Li K, Schellenberg MJ, Zimmermann M, Hustedt N, Rossi SE, Adam S, Melo H, Heijink AM, Sastre-Moreno G, Moatti N, Szilard RK, McEwan A, Ling AK, Serrano-Benitez A, Ubhi T, Feng S, Pawling J, Delgado-Sainz I, Ferguson MW, Dennis JW, Brown GW, Cortes-Ledesma F, Williams RS, Martin A, Xu D, Durocher D (2020) A genetic map of the response to DNA damage in human cells. Cell 182:481-496 e421
Sabatella M, Thijssen KL, Davo-Martinez C, Vermeulen W, Lans H (2021) Tissue-specific DNA repair activity of ERCC-1/XPF-1. Cell Rep 34:108608
Funding
MSL was supported by an ENW-M (OCENW.KLEIN.090) and ALW-VIDI grants (ALW.016.161.320) from the Dutch Research Council (NWO). HL is funded by CancerGenomiCs.nl and the Oncode Institute, which is partly financed by the Dutch Cancer Society. ODS was supported by the Korean Institute of Basic Science (IBS-R022-A1) and the US National Cancer Institute (P01-CA092584, R01CA218315).
Author information
Authors and Affiliations
Contributions
KA, HL, ODS and MSL conceived and discussed the outline of the review. KA wrote the first draft of the manuscript. MSL edited and finalized the manuscript. HL and ODS edited the manuscript. KA and MSL generated all figures and tables. ODS generated all structures.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Apelt, K., Lans, H., Schärer, O.D. et al. Nucleotide excision repair leaves a mark on chromatin: DNA damage detection in nucleosomes. Cell. Mol. Life Sci. 78, 7925–7942 (2021). https://doi.org/10.1007/s00018-021-03984-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-021-03984-7