Abstract
The walls surrounding the cells of all land-based plants provide mechanical support essential for growth and development as well as protection from adverse environmental conditions like biotic and abiotic stress. Composition and structure of plant cell walls can differ markedly between cell types, developmental stages and species. This implies that wall composition and structure are actively modified during biological processes and in response to specific functional requirements. Despite extensive research in the area, our understanding of the regulatory processes controlling active and adaptive modifications of cell wall composition and structure is still limited. One of these regulatory processes is the cell wall integrity maintenance mechanism, which monitors and maintains the functional integrity of the plant cell wall during development and interaction with environment. It is an important element in plant pathogen interaction and cell wall plasticity, which seems at least partially responsible for the limited success that targeted manipulation of cell wall metabolism has achieved so far. Here, we provide an overview of the cell wall polysaccharides forming the bulk of plant cell walls in both monocotyledonous and dicotyledonous plants and the effects their impairment can have. We summarize our current knowledge regarding the cell wall integrity maintenance mechanism and discuss that it could be responsible for several of the mutant phenotypes observed.
Similar content being viewed by others
Introduction
Plants represent essential sources of food and produce feedstocks for biofuel and fine chemical production [1]. To improve food crop performance and facilitate bioenergy production, significant efforts have been made to modify plant cell wall composition. However, these efforts have yielded mixed results [2, 3]. The reason seems to be that plant cell walls are extremely plastic and effects of the genetically induced changes are limited by the modification of other cell wall components. This seems to be comparable to processes taking place during development and interaction with the environment, where perception of physical and chemical stimuli leads to highly adaptive and tightly controlled changes in cell wall composition and structure. The tight control is essential because turgor pressure levels in plant cells are often equivalent to pressure levels in car tires, meaning that uncontrolled/accidental weakening of cell walls leads to cell bursting and cell death. Currently it seems that the plant cell wall integrity (CWI) maintenance mechanism forms an essential component of cell wall plasticity by being responsible for perceiving stimuli indicating CWI impairment and initiating adaptive responses in cellular and cell wall metabolism leading to changes in wall composition and structure.
Different polysaccharides and proteins form the primary and secondary cell walls surrounding land plant cells. Primary cell walls of Arabidopsis thaliana (Arabidopsis, serving here as representative example for dicotyledonous plants) contain mostly pectins [rhamnogalacturonan-I (RG-I 11%), rhamnogalacturonan-II (RG-II 8%), and homogalacturonan (HG 23%)] followed by 24% hemicellulose and 14% cellulose [4]. Primary cell walls of grasses, here representing monocotyledonous plant species, contain mostly hemicellulose (20–45%; including different types of xylans), cellulose (20–30%), pectins (5–10%) and in Poaceae and related families, mixed-linkage glucans (MLGs), which are absent in dicots [5, 6]. Secondary cell walls are deposited once cells have terminally differentiated and consist in Arabidopsis mainly of cellulose (40–80%), lignin (5–25%) and hemicellulose (10–40%) [7]. In addition, cell wall-specific proteins like extensins, expansins, hydroxyproline- and glycine-rich proteins and dynamically formed polysaccharides like callose can be also found in cell walls and will not be covered here in detail since they have already been recently reviewed [7, 8]. This simplified global overview summarizes the main components, which form the bulk of the primary and secondary cell walls in plants and could therefore be relevant in the context of CWI maintenance.
Here, we will initially review processes giving rise to the main cell wall components and assess the consequences their impairment has on plant growth, development and stress responses. Since knowledge about primary cell walls is most extensive in Arabidopsis, we will use them as baseline for comparison purposes with knowledge on cell walls in other plant species (both mono- and dicots). This will also enable us to provide perspective about components possibly involved in or affected by CWI maintenance. We will finish by summarizing the current knowledge about the CWI maintenance mechanism and discuss concepts for its mode of action.
Cellulose biosynthesis in dicots
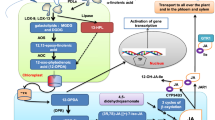
The primary cell wall in Arabidopsis consists of a framework of cellulose microfibrils cross-linked by xyloglucans and embedded in a matrix of acid-rich pectic polysaccharides [8, 9]. Primary cell walls are produced right after cell division and during cell elongation, highlighting the involvement of the walls in cell morphogenesis and the need for extensibility. Cellulose is the most abundant water-insoluble polymer found in nature. This linear polymer consists of β (1 → 4) linked d-glucose units and is synthesized by the plasma membrane-localized cellulose synthase complexes (CSCs). CSCs are transmembrane structures consisting of several CELLULOSE SYNTHASE A (CESA) proteins organized in a rosette shape associated with a large number of other proteins [10,11,12] (Fig. 1; Table 1). Ten CESA proteins have been identified in Arabidopsis. AtCESA1, AtCESA3 and AtCESA6 are involved in cellulose synthesis during primary cell wall, while AtCESA4, AtCESA7 and AtCESA8 are active during secondary cell wall establishment [13,14,15]. AtCESA6 can be replaced to some extent by AtCESA2, AtCESA5 and AtCESA9, suggesting partially redundant roles in primary cell wall CSCs [16, 17], while the biological role of AtCESA10 remains unclear [18]. Detailed structural analyses showed that all AtCESAs have eight transmembrane domains (TMDs), with two being located near the N-terminal region and six near the C-terminus [19]. Between TMD2 and TMD3 resides a large, highly conserved cytosolic region, responsible for uridine diphosphate (UDP) glucose binding and catalysis. By now a large number of mutations in AtCESA genes have been isolated, providing insights into the importance of the different domains within the CESA proteins through the mutant phenotypes caused (ranging from very mild to radial cell swelling and stunted growth) [20]. While knockout (KO) alleles for AtCESA1 and AtCESA3 lead to lethality, plants with AtCESA6 KO alleles are viable and exhibit only limited cell elongation defects (i.e., AtCESA6prc1−1 to AtCESA6prc1−12) [17, 20]. More recently, cellulose biosynthesis inhibitors (CBIs) have been combined with genetic approaches to analyze CESA activity as well as the responses to cell wall damage (CWD) induced by cellulose biosynthesis inhibition [21]. This is exemplified by isoxaben (ISX, a CBI), which was used to isolate AtCESA3ixr1−1, characterized by an amino acid substitution at Glycine-998 located at the start of TMD8, causing resistance to ISX. The mutation leads also to reduced levels of relative cellulose crystallinity but does not cause major growth defects [22, 23]. Impairing cellulose biosynthesis induces a variety of responses in plants. This is illustrated by the AtCESA1any1, AtCESA6eli1 and AtCESA6cev1 mutations [24,25,26]. AtCESA1any1 was originally implicated in microtubule organization in epidermal cells. AtCESA6eli1 was implicated in cell morphogenesis because of ectopic lignin deposition, whereas AtCESA6cev1 seemed required for pathogen response since it causes constitutive expression of VEGETATIVE STORAGE PROTEIN1 (VSP1, implicated in pathogen defense) and production of jasmonic acid (JA) [24, 26]. ISX, which inhibits cellulose biosynthesis in primary cell walls, triggers responses similar to the ones induced by the mutations described. Responses include induction of defense gene expression, phytohormone accumulation such as salicylic acid (SA)/JA (as observed in AtCESA6cev1) and lignin accumulation (as in AtCESA6eli1) [24, 26,27,28,29]. Observing these phenotypes led to the suggestion that a CWI maintenance mechanism, similar to the one described in yeast, may exist in plants [30]. This mechanism involves constant monitoring of the functional integrity of the cell wall and triggering of apparently compensatory responses to maintain integrity during development and interaction with the environment [28].
Global overview of proteins mediating primary (PCW) and secondary cell wall (SCW) formation in dicots (Arabidopsis) and monocots (Oryza). Demonstrated or putative (indicated by ?) protein localizations are displayed in panel a based on data mostly derived from Arabidopsis. Enzymes such as PMEIs, PAEs and PMEs, which have been found in the apoplast, are listed outside the cells in panels a and b. Ellipses represent Golgi bodies/stacks with enzymes located to highlight their positions either inside the Golgi or associated with Golgi membranes. Circles containing enzymes like GALS1/2/3 or CTL1 represent Golgi-derived vesicles. Grey, dashed circles near the nucleus (black ellipse) represent the endoplasmic reticulum. b Summarizes putative localizations of proteins in Oryza. Abbreviations are explained in main text. Protein localizations are based on the following references: [16, 34, 45, 47, 68, 74, 104, 125, 138, 155, 156, 160, 161, 163, 164, 167, 168, 171, 180, 255,256,257,258,259,260,261,262,263,264,265,266,267,268,269,270]
Loss of cellulose integrity affects also microtubule dynamics [31]. While it is not clear how ISX or genetic inhibition of cellulose synthesis affects microtubule orientation, the importance of the cytoskeleton in delivering CSCs to/recycling them at the PM has been reported [32,33,34] (Fig. 1). Golgi-derived vesicles containing CSCs interact with cortical microtubules through adaptors/docking sites, involving several proteins (CELLULOSE SYNTHASE INTERACTIVE 1 (CSI1/POM2), COMPANION OF CELLULOSE SYNTHASE (CC), PATROL1, STELLO1/2 and the EXOCYST COMPLEX) [32, 34,35,36]. These components control CSC delivery rate and allow CSC-containing vesicles to fuse with the plasma membrane. Mechanical distortion of cell walls and/or loss of CWI (i.e., osmotic stress or CBI treatment) trigger CESA re-localization to intracellular compartments [37, 38]. These have been termed small CESA containing compartments (SmaCCs) and when associated with cortical microtubules are referred to as microtubule-associated cellulose synthase compartments (MASCs) [35]. CSI1 is apparently the molecular linker between CSCs and cortical microtubules, since loss of CSI1 induces disassociation of CSCs from microtubules and results in cellulose deficiency [39]. CSI1 apparently connects CSCs to microtubules through the CC1/2 N-terminal domain, which also influences microtubule stability under salt stress [36, 40]. Tagged CSI1 protein labels both SmaCCs and MASCs, suggesting that it might be required for both CSC delivery and recycling [33]. While CBIs like ISX or thaxtomin A induce CSC internalization, other CBIs like dichlobenil inhibit CESA motility and induce localized accumulation at the plasma membrane [41]. The differential effects of CBIs on CESA dynamics are useful tools to dissect cellular processes affecting CSC activity. Even though certain ISX-induced responses (gene expression changes, JA/SA and lignin accumulation) can be prevented by osmoticum co-treatments (sorbitol or mannitol application), CESA internalization cannot be prevented by these treatments [42]. This suggests that CSC displacement itself is not the stimulus responsible for osmo-sensitive responses activated by the CWI maintenance mechanism.
The actin cytoskeleton has also been implicated in cellulose biosynthesis, since actin manipulation leads to cellulose deficiency and defective CESA trafficking [43]. This led to the hypothesis that the actin cytoskeleton mediates delivery of CESAs to the PM, where CSC adaptors interact with the CSCs and cortical microtubules to ensure correct CSC positioning in the PM. Actin cytoskeleton dynamics seem to respond differentially to different types of stress, since plasmolysis induces thicker actin bundles while CBI application leads to actin filament disruption [44]. In addition to actin and microtubules both motor proteins and cytoskeleton-associated proteins have been implicated in cellulose biosynthesis and response to stress. Loss of the Arabidopsis kinesin FRAGILE FIBER1 (FRA1) alters cellulose microfibril organization, implicating kinesins in CESA trafficking and/or positioning [45, 46]. Similarly, other CSC/cytoskeleton interacting proteins like the endo (1 → 4) β-d-glucanase KORRIGAN1 (KOR1), CHITINASE-LIKE 1 (CTL1/POM1) and CTL2 have been implicated in cellulose synthesis [47,48,49] (Fig. 1a). KOR1 was found to be associated with primary CESAs and seems required for both glucan chain synthesis and regulation of intracellular CSC dynamics [50, 51], while absence of CTL1/POM1 and CTL2 impairs motility of CSCs. Similar to cellulose mutants ctl1 seedlings exhibit both root cell swelling and ectopic lignin deposition [49, 52, 53]. It has been reported that the expression of CTL1 is controlled by GENERAL CONTROL NON‐REPRESSED PROTEIN 5 (GCN5) which encodes a histone acetyltransferase [54]. Plants lacking GCN5 exhibit reduced cellulose levels, salt and ISX-hypersensitivity, providing further evidence that GCN5 is involved in regulating cell wall metabolism. These recent observations are exciting since epigenetic control has not been associated before with the regulation of CWI and salt stress responses.
CSCs are also required for cellulose biosynthesis during secondary cell wall formation, where they consist of AtCESA 4, 7 and 8 [55] (Fig. 1a). As in the case of primary cell wall CESAs, secondary cell wall CESAs were identified through genetic screens that identified irregular xylem (irx) mutants (AtCESA8irx1, AtCESA7irx3 and AtCESA4irx5), illustrating the importance of cellulose in stem development and stability [13, 15]. By creating catalytically inactive isoforms of the three CESAs, their individual activities during cellulose biosynthesis in developing secondary cell walls were determined [56]. Intriguingly, KO alleles for AtCESA4, 7 and 8 do not cause lethality, thus allowing to study the effects of reduced cellulose production on pathogen responses. Interestingly, AtCESA8irx1, AtCESA7irx3, and AtCESA4irx5 confer resistance to Ralstonia solanacearum and Plectosphaerella cucumerina [57]. The resistance seems to be mediated by abscisic acid (ABA) signaling being constitutively activated by the defects in secondary cell wall formation [57]. This is in contrast to the production of SA and JA in response to primary cellulose inhibition, suggesting that differences in the responses to cellulose impairment exist between primary and secondary cell walls [24, 29].
Cellulose biosynthesis in monocots
While similarities exist regarding the molecular components involved in cellulose biosynthesis in mono and dicots, there are also differences. All dicots and most monocots have type I cell walls while species belonging to the Poaceae family have type II cell walls [58]. Many cultivated crops such as maize, rice (Oryza sativa L.) and wheat belong to the Poaceae family. Both type I and type II cell walls contain cellulose produced by cellulose synthases of the CESA gene family [59]. However, in contrast to type I walls, pectins contribute only to a small extent to the matrix polymers in type II walls. Moreover, in type II cell walls, cellulose microfibrils are crosslinked by glucuronoarabinoxylans instead of hemicellulose-pectin as in type I cell walls [58].
In monocot species, like barley (Hordeum vulgare, HvCESA), rice (Oryza sativa, OsCESA) and mais (Zea mays, ZmCESA), CESAs have been identified primarily based on homologies to Arabidopsis CESA genes [60]. In parallel brittle culm (bc) mutants were isolated, which exhibit reduced amounts of cellulose and decreased breaking strength [61, 62]. The rice brittle culm mutant S1-24 consists of a point mutation in OsCESA7, which causes reduced cellulose production in sclerenchyma cells thus leading to lower mechanical strength in the sclerenchyma cell walls [63]. Another bc mutant, bc5, leads to cell wall formation defects in sclerenchyma nodes, likely due to altered expression of OsCESA4, 7 and 9 as well as reductions in cellulose and hemicellulose production [64]. In rice and Sorghum bicolor, bc1 mutations have been recently shown to reside in genes encoding homologues of Arabidopsis COBRA-like proteins [65, 66] (Fig. 1a, b). COBRA-like proteins are GPI-anchored proteins, which are required for cellulose biosynthesis both during primary and secondary cell wall formation [66, 67]. Just like in Atcobl4 secondary cell walls, cellulose content is altered in both Sbbc1 and Osbc1, causing increased stem brittleness [65, 66]. Similar to Arabidopsis FRA1, rice FRA1 homologue (OsBC12) has been associated with orientation of cellulose microfibrils [68]. The existing evidence indicates pronounced similarities regarding the mode of action of cellulose biosynthesis in monocots and dicots, but our knowledge regarding the presence of a CWI maintenance mechanism in monocots is very limited. For example, treatment of Poa annua with the CBI Indaziflam inhibits growth, triggers root cell wall swelling and ectopic lignification, responses previously observed upon ISX treatment in dicots [69]. More targeted research activities will be required to assess similarities and differences of CBI responses in dicots and monocots.
Cellulose synthase-like enzymes in monocots and dicots
Cellulose synthase-like enzymes (CSLs) are divided into 9 subfamilies (CSLA–H and CSLJ), all belonging to the large glycosyltransferase 2 (GT2) superfamily [70, 71]. Comparative genome studies suggest that several of these families are restricted to dicots, like CSLG, while others, like CSLF and CSLH seem restricted to grass species [71]. Members of the CSLA, CSLC, and CSLD families have been identified in all land plants where genome sequence information is available, including the moss Physcomitrella patens and the lycophyte Selaginella moellendorffii [72, 73]. Based on genome sequence analysis, a super family of CSL proteins, grouped in 6 families named from CSLA–G (no F), has been identified in Arabidopsis [71]. Several mutants have been isolated in Arabidopsis, but only a small number of them exhibit pronounced phenotypes, which are not associated with cellulose biosynthesis. CSLD mutants such as csld2 and csld3 (the latter known as kojak) display defective root hair development in primary roots and csld2/csld3/csld5 plants exhibit reduced growth, suggesting the three proteins might be part of a multimeric complex [74,75,76]. Members of the CSLA and CSLD families have been implicated in the synthesis of mannans and glucomannans, while members of the CSLC family are required for synthesis of xyloglucans and other polysaccharides related to hemicellulose synthesis [75,76,77]. Intriguingly, the csld3 phenotype is rescued by expressing a modified version of CSLD3 protein containing the CESA6 catalytic domain, which suggests functional redundancy between CSL proteins and primary CESAs [78]. Mutants have been isolated for several members of the CSLA family, providing insights into their respective biological and biochemical functions. In Arabidopsis, mutations in CSLA9 (encoding a glucomannan synthase) cause reduced lateral root formation while mutations in CSLA7 cause embryo lethality [79, 80]. Notably, the csla7 phenotype can be rescued by overexpressing CSLA9, suggesting that CSLA7 and CSLA9 have similar biochemical functions and differences in activity might be caused by differential gene expression [80]. In Arabidopsis, mannans are present at low levels in primary cell walls and in greater abundance in secondary cell walls [81,82,83]. Arabidopsis csla2/csla3/csla9 triple mutants, lacking detectable glucomannans in stems, do not show alterations in stem development and structure [80]. However, if glucomannan content is enhanced (by over expressing CSLAs) plant development is affected [80]. This suggests that glucomannans are not growth limiting cell wall components and their absence can be compensated. However, increasing their levels has significant consequences, with the reasons to be determined.
A major difference between Arabidopsis and grass cell wall metabolism can be found in the functions of the CSLs, likely reflecting their role in the synthesis of monocot specific cell wall saccharides [84]. In fact, several CSLs are involved in the synthesis of mixed-linkage glucans (MLGs) and act as (1 → 3)/(1 → 4)-β-glucan synthases. The difference is further illustrated by the fact that in rice the CESA/CSLs superfamily has 45 members [85]. MLGs have been detected in large amounts in endosperms and therefore a storage function was proposed for them [86]. In maize and barley coleoptiles they are present in elongating tissues, while in several other grass species (including rice, Miscanthus and B. distachyon) they have been also observed in dry stems [87]. These observations suggest that MLGs are incorporated both into primary and secondary cell walls. Members of the CSLF family in rice as well as of the CSLH and J families in barley are involved in the biosynthesis of MLGs [71, 88]. Loss of function alleles of rice CSLF6 (cslf6-1 and cslf6-2) exhibit a 99% reduction of MLGs in developing leaves, while mature stems have weaker and more brittle cell walls, but mutant plants were able to grow [89] (Fig. 1b). This is surprising since these MLGs were supposed to have a function in type II cell walls similar to pectins in type I cell walls, i.e., forming a gel-like matrix in which cellulose microfibrils are embedded [90,91,92]. However, detailed analysis of cell wall composition found that cslf6-1 and cslf6-2 mutants exhibit moderate reduction in cellulose content compared to wildtype controls and suggested that MLGs may influence the spatial distribution of cellulose microfibrils [89, 93]. These observations also imply that manipulation of MLG production affect cell wall organisation, with the resulting phenotypes not easily explained by functional redundancies, supporting the existence of a CWI maintenance mechanism in these plant species.
Hemicellulose metabolism and effects of its impairment
Historically, non-cellulosic and non-pectic polysaccharides have been collectively named hemicelluloses. The classic features of hemicelluloses are β (1 → 4) glycosidic bonds, which form the dominant backbone linkage in an equatorial configuration [5]. Following this classification only a limited number of polymers (xyloglucan, xylans, mannans, glucomannans, and MLGs that are restricted to different grass families) will be discussed here with mannan and MLG biosynthesis being excluded since these topics have been covered in the previous section.
Xyloglucan in primary cell walls of dicots
Xyloglucan (XyG) consists of β (1 → 4)-linked glucans sometimes partially substituted with xylosyl substituents at the O-6 position or with species-specific side chain decorations. In graminaceous monocots, xylans and MLGs form major components of both primary and secondary cell walls [94]. While xyloglucan (XyG) comprises only 2–5% of the primary cell wall in monocots, it is the most abundant hemicellulose of primary cell walls in dicots (10–20%) and a minor component of secondary cell walls [90, 94,95,96]. Interestingly, in dicots XyG establishes interactions between cellulose microfibrils and pectins [97]. XyG turnover seems to influence cell elongation and cell wall remodeling [98]. In Arabidopsis, these events are mediated by XyG endotransglucosylases/hydrolases, (belonging to CAZy family GH16) [99, 100]. Similar to pectin and cellulose fragments, XyG breakdown products play a role in signaling in different plant species [101,102,103]. The complexity of XyG biosynthesis and sidechains means that many different enzymes are required for the synthesis of glucan chains and xylosyl, galactosyl or fucosyl substitutions. Members of the CSLC family (like CSLC4, CSLC5, and CSLC6) act as β (1 → 4)glucan synthases (using UDP-glucose as substrate) contributing to the production of the XyG backbone [104] (Fig. 1a). Xylosyl substitution is carried out by XyG xylosyltransferases (XXTs), which in Arabidopsis belong to glycosyltransferase (GT) family 34 and are exemplified by XXT1 and XXT2 [96]. xxt1xxt2 seedlings display aberrant and swollen root hairs and shorter hypocotyls. Adult xxt1xxt2 plants lack detectable XyG and display mild growth phenotypes characterized by reduced rosette size and shorter petioles with enhanced mechanical extensibility compared to wildtype controls, indicating a role for XyG in modulating cell wall strength [105]. These rather mild mutant phenotypes raise questions regarding the importance of XyG for plant growth under standard growth conditions [96]. XXT activity seems highly conserved, since expression of OsXXT1 in Arabidopsis rescues the root hair defects of Arabidopsis xxt1xxt2 plants [106]. In Arabidopsis, gene expression analysis showed that XXT1 and CSLC4 are expressed in the same tissues and developmental stages, suggesting that XXT1 and CSLC4 might be both active in glucan synthesis. Moreover, XXT1, XXT2, XXT5 and CSLC4 seem to be localized in the Golgi, where they add xylosyl groups to the glucan backbone [104] (Fig. 1a). XYLOGLUCAN L-SIDECHAIN GALACTOSYLTRANSFERASE 2 (XLT2) and MURUS3 (MUR3) were both shown to act as galactosyltransferases in Arabidopsis [107, 108]. They are required for adding d‐galactopyranosyl (β-d-Galp) to an internal d‐xylopyranosyl (α-d-Xylp) or an α-d-Xylp adjacent to an unbranched β-d-Glc. Interestingly, MUR3 KO plants (mur3-3) exhibit dramatically reduced growth, display cabbage-like leaves and are constitutively resistant to Hyaloperonospora parasitica infections [107, 109]. XyG in mur3 is characterized by reduced levels of galactosyl substitutions and almost no fucosyl residues. XXTs and MUR3 might have tissue-specific functions, because mur3 does not exhibit root hair phenotypes as observed in xxt1 xxt2 [108, 110]. Interestingly, MUR3 mutant phenotypes are rescued by knocking out xylosyltransferases (such as XXT1, 2 and 5) [107]. Double and triple mutant plants (xxt1/xxt2/mur3-3, xxt2/mur3-3 and xxt5/mur3-3) resemble wildtype plants both with respect to growth phenotypes and on the transcriptomic level, even though XyG deficiency is not reversed in the double or triple mutant backgrounds [111]. As in the case of glucomannans it seems that plants can adapt to the absence of XyG, while dysfunctional XyG structures have deleterious effects [107]. Intriguingly, plants lacking XLT2 are phenotypically normal, lacking the cabbage-like phenotype of mur3 plants [107]. This implies that the severe phenotypes are caused by specific changes in galactosylation of XyG mediated by MUR3 and that these have a biological function and/or their presence is detected by the plant causing responses [107]. Since MUR3, like XXT1, localizes to the Golgi cis and medial cisternae (Fig. 1a), the existence of a dedicated XyG synthesis complex has been proposed [112]. The biological role of XyG fucosylation remains to be fully determined since l-fucosylated oligosaccharins inhibit pea epicotyls elongation and fucosylated XyG bind cellulose in vitro even though cellulose–xyloglucan binding does not require XyG fucosylation [113, 114]. These observations indicate that plants can recognize specific changes in XyG structures, compensate for deviations from the normal state and that the effects are specific as illustrated by differences between XLT2 and MUR3. However, we do not know the stimuli indicating deviations, the detection mechanism in place or the mode of action of the downstream processes bringing about changes in other cell wall polysaccharides.
Xylan formation and modification in monocots and dicots
Xylans include all the hemicellulose polymers containing (l → 4)-linked β-d-xylopyranosyl units as backbone, like glucuronoxylan, arabinoxylan, glucuronoarabinoxylan, and l-arabino-(4-O-methyl-d-glucurono)-xylan. Xylans are important components of cell walls and produced by a group of enzymes [115]. Roughly 10–20% of the xylose sugars in the xylan backbone are substituted. Xylan side chains frequently contain arabinose, which either forms a single substituent or connects other sugars like xylose, galactose and 4-O-methyl-d-glucuronic acid to the xylan backbone [116]. In contrast, methylated glucuronic acid (GlcA) can directly bind xylose, without the need for an arabinose linker [5]. In Arabidopsis the degree of polymerization (DP) of xylan in secondary cell walls can reach up to 150 xylose units while in grasses (like wheat) the DP can reach up to 4000 xylose repetitions [117]. In grasses, xylans frequently exhibit arabinose side chains and are extensively decorated with α-(1 → 2)- and α-(1 → 3)-linked arabinofuranose molecules. Moreover, in these plants xylans are crosslinked via ferulic acid residues to lignin [5]. Even though xylans are the second most abundant biopolymers after cellulose, xylan biosynthesis is still poorly understood at the biochemical level. In Arabidopsis, a family of GT43 glycosyltransferases (IRX9/IRX9-L, IRX14/IRX14-L) has been implicated in the synthesis of xylan backbones during secondary cell wall formation [118, 119] (Fig. 1a). In monocots (specifically Poales), glucuronoarabinoxylan is the main hemicellulose in primary cell walls while xylan side chains interact with lignin in secondary cell walls to enhance cell wall strength [120, 121]. In the rice genome, ten GT43 encoding genes were identified but the involvement in xylan biosynthesis has not been confirmed for all of them yet [116]. Similarly to XyG, xylan biosynthesis seems to also occur in the Golgi based on localization data for OsGT43 proteins [116]. In Arabidopsis, both IRREGULAR XYLEM9 and 14 (IRX9, IRX14) have xylosyltransferase activity and are required to extend xylan backbones [122]. IRX9 and IRX14 mutant plants are slightly smaller than wildtype controls, exhibit thinner xylem vessel cell walls and have reduced levels of xylose [123]. IRX9-LIKE (IRX9-L) and IRX14-LIKE (IRX14-L), respective homologues of IRX9 and IRX14, are also members of the GT43 family and loss of function alleles result in no obvious mutant phenotypes [124]. However, irx9-2/irx9-L and irx14/irx14-L plants exhibit stunted growth with small leaves and reduced xylan levels [124]. Heterologous expression of the rice OsGT43A and OsGT43E in Atirx9 or OsGT43J in Atirx14 rescues the mutant phenotypes, indicating that OsGT43A and OsGT43E represent rice homologs of the Arabidopsis genes and that their enzymatic activities are conserved [116]. Glucuronoxylan is made of a backbone of β (l → 4)-linked xylose residues decorated with α (1 → 2)-linked GlcA or 4-O-methyl-GlcA (MeGlcA) [125]. IRX10 and IRX10-L, belonging to the Arabidopsis GT47 family, are required for its synthesis [126, 127] (Fig. 1a). However, while irx10 and irx10-l single mutants are phenotypically normal, suggesting functional redundancy between the GT47 family members, irx10/irx10-l double mutants are severely dwarfed, exhibit thinner cell walls in xylem vessels and produce infertile flowers [126]. The results suggest that for certain types of cell wall modifications either no functional redundancy exists or no CWI monitoring mechanism is in place to initiate compensatory responses.
Bioinformatics-based analyses of grass genomes suggest that GT47 and GT61 glycosyltransferase families may encode xylan arabinosyltransferases (XATs) [128]. Heterologous expression of either wheat or rice GT61s in Arabidopsis leads to mono-arabinosylation of xylan chains, supporting a role for GT61s as α-(1 → 3)-arabinosyltransferases [128]. Phenotypes observed in wheat (Triticum aestivum) where TaXAT1 expression was suppressed by RNAi suggest that TaXAT1 mediates α-(1 → 3)-arabinose mono substitution of xylose backbones [129]. Other members of the GT61 family (like rice OsXAT2, OsXAT3 and wheat TaXAT2) function also as arabinosyltransferases, since they complement mutant phenotypes of homologous Arabidopsis genes [128]. In addition, it was shown that the OsXYXT1 function as xylosyltransferase [130]. Intriguingly, a forward genetic screen for mutations affecting saccharification performance in Brachypodium distachyon identified a mutation, which seems to reside in a GT61 family member and improves saccharification performance of the plant material while not having obvious impacts on plant growth and interaction with the environment [131]. Members of the GT8 family function as glucuronyltransferases. For α (1 → 2)-linked GlcA SUBSTITUTION OF XYLAN 1, 2 and 4 (GUX1, 2, 4), xylan glucuronosyltransferase activities have been demonstrated [132]. However, even if gux1 gux2 plants show reduced glucuronoxylan levels, they do not exhibit growth defects. At the same time, gux1/gux2/gux3 triple mutant plants are smaller than controls and exhibit reduced resistance to breaking force in stems [133]. In the triple mutant plants, interaction between xylan and cellulose seems impaired, leading to reduction in secondary cell wall thickness and mechanical strength [125].
Glucuronoarabinoxylans have been detected in rice and other monocots in large amounts in both primary and secondary cell walls and are modified in different ways [134]. Examples for such modifications include acetylation and methylation. In non-woody plants 30% of GlcA is not methylated while in woody plants nearly all of the GlcA is [135]. The degree of glucuronoarabinoxylan methylation together with the amount of arabinose substitutions differs between monocot species and can also vary between tissues from the same species [136]. An important structural component of glucuronoarabinoxylans in grasses is the presence of ferulic and coumaric acid, which is thought to provide additional cell wall strength [137]. In dicots, glucuronoxylan methylation is carried out by DUF579 domain-containing proteins, like the GLUCURONOXYLAN METHYLTRANSFERASE1 (GXMT1) [138]. gxmt1-1 mutants show reduced glucuronoxylan methylation coupled with enhanced lignin methylation [138]. Interestingly, no mechanical defects are observed in their stems suggesting that either changes in methylation do not affect the mechanical cell wall properties or the effects of their modification are neutralized. Acetylation is performed by AXY9 (ALTERED XYLOGLUCAN9), REDUCED WALL ACETYLATION (RWA) and DUF231 domain-containing proteins [139,140,141,142]. Mutations in the four RWAs reduce stem resistance to mechanical force, suggesting that xylan acetylation modulates mechanical stability. Our current knowledge regarding glucuronoxylan formation and modification is still rather limited as evidenced by the overview provided here. However, combining bioinformatics studies with heterologous expressions in Arabidopsis will help to fill in the knowledge gaps. In several cases, exemplified by IRX9/IRX9-L, IRX10/IRX10-L IRX14/IRX14-L and GUX1, 2, 3, the results obtained suggest that functional redundancy between enzymes with similar activities is responsible for the mutant phenotypes observed. In other cases like the methylation of glucuronoxylan, the corresponding changes observed in lignin methylation suggest adaptations in cell wall metabolism take place, but the mode of action of the mechanism responsible remains to be determined. To summarize, with respect to CWI maintenance and hemicelluloses, there is currently no clear evidence that hemicellulose integrity is monitored in a similar manner as described for cellulose. This might indicate either the absence of dedicated monitoring mechanisms or simply a lack of knowledge due to limited research activity in the area.
Pectin metabolism and effects of its impairment
While cellulose and hemicellulose are major components of load-bearing polysaccharide networks in both primary and secondary cell walls, pectins are mostly found in primary walls and play a role in the context of development, growth and adaptation to changing environments. Biochemically, pectins are usually defined as galacturonic acid (GalA)-containing polysaccharides [143]. Homogalacturonan (HG) is a polysaccharide consisting of α-(l → 4)-linked GalA residues and is covalently bound to RG-I and RG-II [143]. RG-II consists of a GalA backbone decorated with complex, yet conserved, side chains that contribute to cell adhesion and rigidity by complexing borate as dimer [143]. The RG-I backbone consists of alternating GalA and rhamnose residues, decorated with arabinan, galactan and arabinogalactan side chains that differ depending on species and developmental stage [143].
Pectins are major components of the early cell plate during cytokinesis and primary cell walls, where they contribute to the regulation of cell-to-cell adhesion and cell elongation [144, 145]. Seeds of flowering plants produce pectin-rich mucilage, which is important for seed hydration, germination and adherence [146]. Regulation of these different processes is attributed to pectin methylesterases (PMEs), which control the degree of methylesterification (DM) in HG and influence HG cross-linking through Ca2+-bridges [147]. The controlled modification and degradation of pectin is crucial for cell separation processes, such as leaf abscission and fruit ripening [144, 148]. While pectins have previously been regarded as a gel, in which a hemicellulose-tethered cellulose scaffold is embedded, more recent research indicated that direct pectin-cellulose interactions exist [149]. It is currently not clear how this interaction works and which pectin polymers are involved, questions that have important implications for potential load-bearing functions of pectin. Compared to other cell wall components, pectin seems to be more readily degraded upon environmental stress. Examples are pathogen-induced breakdown of pectin enhancing infection success [150] or mobilization of carbohydrates from pectin during starvation periods [151]. Interestingly both intact pectin and pectin fragments have been shown to bind to plasma membrane-localized receptor kinases, suggesting that pectin integrity is closely monitored by plant cells [152, 153].
Pectin in primary cell walls of dicots
Pectin polymers are formed in the Golgi from nucleotide sugars. While the chemical complexity of the different pectin polymers implicates a large number of transferases in pectin formation, only few transferase proteins have been characterized in detail. HG is synthesized by GALACTURONOSYLTRANSFERASE1 (GAUT1), which is anchored to the Golgi by GAUT7 [154, 155] (Fig. 1a). The GAUT1:GAUT7 complex seems to include, or associate with, several additional cell wall biosynthesis proteins, such as the putative HG- methyltransferase QUASIMODO3 (QUA3) and KOR1 [155, 156]. GAUT8/QUA1 is required for HG biosynthesis and cell adhesion [157, 158] but also for normal xylan biosynthesis [159]. Interestingly, loss of putative HG-methyltransferases like QUA2 and GAUT8/QUA1 causes similar mutant phenotypes including reduced HG content, impaired cell adhesion and stunted growth [160]. Similarly, the QUA2-unrelated putative methyltransferases COTTON GOLGI-RELATED2 (CGR2) and CGR3 are required for normal pectin composition and growth, further highlighting the importance of pectin modification for plant development [161]. A recent proteomic analysis, characterizing auxin-dependent alterations in root tissue, identified GAUT10 as positive regulator of the root apical meristem length and cell number in a process that seems to be sucrose-sensitive [162]. CWD on the other hand negatively affects root apical meristem length and cell number, emphasizing the importance of CWI maintenance for root development [42]. Xylosylation of HG is mediated by XYLOGALACTURONAN DEFICIENT1 (XGD1), but the biological function of xylogalacturonan remains to be determined [163] (Fig. 1a). While the transferases involved in RG-I backbone syntheses are unknown, the synthesis of RG-I side chains is understood much better. β-(l → 4)-galactan is synthesized by GALACTAN SYNTHASE1 (GALS1) and putatively its two homologs, GALS2 and GALS3, which exhibit weaker overall expression and distinctly specific expression patterns [164]. ARABINAN DEFICIENT1 (ARAD1) and ARAD2 have been identified as non-redundant putative α-(l → 5)-arabinosyltransferases, which seem to form a complex at least during transient overexpression [165, 166]. Surprisingly, despite their impact on RG-I side chain abundance, no growth phenotypes have been reported for gals1 and arad1/arad2 mutant plants [164, 166]. Recently, GAUT11 and MUCILAGE-RELATED70 (MUCI70) were shown to be essential for RG-I production in seed mucilage [143, 167]. While the biochemical activity of MUCI70 is unknown, GAUT11 has been shown to have HG:GalA transferase activity, suggesting that HG and RG-I elongation in mucilage might require mutual enzyme activities [167]. The only transferase activity so far reported for RG-II synthesis is mediated by the RHAMNOGALACTURONAN XYLOSYLTRANSFERASEs (RGXTs), RGXT1, RGXT2, RGXT3 and RGXT4 [168,169,170]. While mutants for RGXT1 and RGXT2 show no obvious phenotypes and rgxt3 mutants have not been studied yet, loss of RGXT4 leads to defective pollen tube and root growth [168,169,170]. The decoration of RG-II with galactose can be reduced by RNA interference-induced suppression of GDP-GALACTOSE TRANSPORTER1 (GGLT1) expression, leading to reduced RG-II dimerization and pronounced growth defects [171] (Fig. 1a). Impaired RG-II dimerization and growth have been previously described in mur1, where fucosylation of several polysaccharides is affected [172, 173]. Congruently, both mur1 and gglt1 mutants are impaired in growth recovery after salt stress [153, 171]. General growth defects in mur1 and gglt1 and root growth defects in rgxt4 mutants can be rescued by borate supplementation, illustrating the importance of RG-II dimerization for cell wall function [171, 172]. Moreover, pectin-crosslinking by combined borate- and Ca2+-treatment can rescue impaired CWI during growth recovery after salt stress [153]. Similar to hemicellulose, HG and RG-I can be acetylated by members of the RWA and TRICHOME BIREFRINGENCE-LIKE (TBL) protein families [174]. While RWA2 is required for acetylation of both xyloglucan and pectin, TBL10 is specifically required for RG-I acetylation [142, 175]. Mutants for TBL10 do not display a visible growth phenotype but exhibited increased drought stress tolerance [175]. Loss of TBL10 does not affect pathogen resistance, which is surprising, since overexpression of Aspergillus nidulans RG-I acetylesterase or loss of RWA2 lead to increased resistance to the necrotrophic fungal pathogen Botrytis cinerea [142, 176]. TBL44/POWDERY MILDEW RESISTANT5 (PMR5) seems to acetylate HG and mutants are affected in cell wall acetylation, pectin and cellulose content [177, 178]. pmr5 mutants exhibit increased resistance to biotrophic powdery mildew fungi and the hemibiotrophic fungus Colletotrichum higginsianum [151, 177]. Powdery mildew resistance of pmr5 is partially suppressed by a mutation in RWA2, whereas rwa2 plants are hypersusceptible to powdery mildew infection [178]. Hyphae from powdery mildew and C. higginsianum appeared deformed or shrunken on infected pmr5 tissues, indicating that pectin modification is critical for host–pathogen compatibility and hyphal expansion. This defect in fungal growth is aggravated on pmr5/pmr6 double mutant plants, which show increased pectin content (PMR6 encodes a putative pectate lyase) and penetration resistance to C. higginsianum [151, 177]. In contrast, mur8 mutants, exhibiting reduced RG-I content in leaves, display strongly reduced penetration resistance to this fungus [151, 179].
The above-mentioned mutant phenotypes illustrate the impact of intact pectin biosynthesis and modification on CWI during growth, development and (a)biotic stress. Modification of the degree of acetylation and methylesterification is achieved by the activities of pectin acetyl esterases (PAEs) and PMEs, respectively. PAEs and PMEs form large gene families and their functions have been reviewed in detail elsewhere [180]. Here, we will only present selected examples to highlight the importance of pectin acetylation and methylesterification for functional integrity of plant cell walls. Heterologous expression of a Vigna radiata PAE in potato tubers caused a strong decrease in pectin acetylation and increased stiffness of cell walls [181]. Overexpression of Populus trichocarpa PECTIN ACETYLESTERASE1 (PtPAE1) in tobacco caused defects in cell elongation of floral filaments and pollen tube growth, suggesting that regulation of pectin acetylation might be critical for plant reproduction [182]. PAE activity is dependent on the DM of HG and has direct implications for the formation of Ca2+ “egg box” structures, indicating synergism between PAEs and PMEs [180]. Similar to the pathogen response phenotypes reported for rwa and tbl mutants, several studies demonstrated pronounced effects of loss or overexpression of PME or PME inhibitor (PMEI) genes on resistance to bacterial, fungal and viral pathogens [183,184,185,186,187,188]. The DM is critical for regulation of cell wall elasticity and this is mechanistically achieved by the activities of PMEs and their endogenous inhibitors [189]. Research into the cell wall mechanics of Arabidopsis plants overexpressing PME5 or PMEI3 indicated that asymmetric de-methylesterification underlies growth symmetry breaking and anisotropic growth [145]. Apparently, loss or inhibition of PME activity can have a direct and severe impact on the mechanical strength of tissue, as reported for pme35 mutant and PMEI5 over-expressing plants [190, 191]. Growth defects are probably in large part caused by brassinosteroid (BR) signalling, which is activated by impaired pectin structure [191]. Activation of BR signalling is mediated by RECEPTOR-LIKE PROTEIN44 (RLP44), which directly interacts with the BR receptor BRASSINOSTEROID INSENSITIVE1 (BRI1) and its co-receptor BRI1-ASSOCIATED KINASE1 (BAK1) [192, 193]. RLP44 also promotes interaction of BAK1 with PHYTOSULFOKINE-RECEPTOR1 (PSKR1) to control xylem cell fate and switching between BR and PSK pathways seems to be regulated through RLP44 phosphorylation [193, 194]. It would be interesting to understand how biochemical (pectin methylesterification status) or functional (status of cell wall integrity) information is perceived by RLP44.
Sensing of alterations in pectin polymerisation and structure is better understood for a family of RECEPTOR-LIKE KINASEs (RLKs), the WALL-ASSOCIATED KINASEs (WAKs). WAK1 and WAK2 bind to pectin oligomers in vitro, with a preference for de-esterified and short oligogalacturonides (OGs) [152]. The available literature on WAKs was reviewed recently and a model suggested, in which WAK-mediated signalling is modulated by the degree of HG polymerisation (intact pectin vs. OGs), thus representing a monitoring mechanism capable of detecting the extent of pectin degradation [152].
Using an elegant bioengineering approach, Benedetti et al. demonstrated that polygalacturonase-dependent in planta production of OGs induces defence responses and provides increased resistance to bacterial and fungal pathogens [195]. Since growth and development are negatively regulated by OG production, authors from the same lab investigated a regulatory mechanism inactivating OG-dependent signalling. The discovery of oxidized OGs by mass spectrometry lead them to the identification of OG OXIDASE1 (OGOX1), a berberine bridge enzyme-like protein, possibly required for regulation of growth-defence trade-off [196]. Recently, the characterization of in vivo produced OGs during Botrytis cinerea infection was accomplished [197]. Unexpectedly, OGs were mostly generated by the activity of fungal pectin lyases, yielding acetyl- and methylesterified OGs, instead of un-methylesterified OGs derived from polygalacturonase activity [197]. Polygalacturonase-dependent OGs did not occur in a degree of polymerization previously found to be elicitor-active, but only in shorter fragments that were oxidized late in the infection process [197]. Responses induced by elicitor-active un-methylesterified OGs seem to be distinct from responses induced through the osmo-sensitive CWI maintenance mechanism [198]. The latter causes JA accumulation in response to enzymatic degradation of Arabidopsis cell walls by polygalacturonase, a response absent in OG-treated Arabidopsis seedlings. Intriguingly, CWI impairment in Arabidopsis seedlings caused by CBI is correlated with increased cell wall pectin content, suggesting that pectin might accumulate to compensate for loss of cellulose in primary cell walls [199, 200].
Pectin in monocots
Pectins are generally less abundant in monocot species compared to dicots and particularly lowly abundant in grasses [120]. Nonetheless, interesting phenotypes have been reported for mutants affected in pectin biosynthesis or modification. Downregulation of the HG:GalA transferase GAUT4 in switchgrass leads to reduced HG and RG-II content, increased growth and cell wall extractability, thus improving properties for biofuel production from lignocellulosic biomass [201]. Further research indicated that reduced recalcitrance to cell wall extraction also stems from alterations in hemicellulose and lignin structure. In particular, reduced lignin-carbohydrate complex cross-linkages seem to facilitate lignin degradation during pre-treatment of lignocellulosic biomass [202].
PME activity has been demonstrated to be of crucial importance in grasses during development and for pathogen resistance. For example, recent research showed that pollen- and pistil-expressed PMEs control cross-incompatibility in maize [203, 204]. Similar to findings in dicots, modulation of PME activity by host plants or fungal pathogens contributes to the outcome of plant-pathogen interaction in wheat, demonstrating the importance of HG DM despite the low overall abundance of pectin in cell walls of grasses [205, 206]. Pectin degradation induced by overexpression of the polygalacturonase subunit OsBURP16 in rice-impaired cell adhesion and increased sensitivity to cold, salinity and drought stress, indicating a critical role of intact pectin structure for abiotic stress tolerance [207]. Aluminum toxicity represents another economically important cause of abiotic stress that is strongly influenced by pectin abundance and modification. The DM of pectin determines the binding capacity of root cell walls for aluminum in maize, rice and wheat [208,209,210]. In wheat, exposure to aluminum induces nitric oxide accumulation, which in turn leads to increased PME activity and a reduced DM of pectin [210]. While no direct role for nitric oxide in CWI signaling has been shown to date, recent evidence in Arabidopsis supported the idea that nitrate reductase-dependent nitric oxide formation might be required for the initiation of cell wall damage responses [42]. These representative examples illustrate that even if a cell wall compound like pectin in monocot cell walls is only lowly abundant it still can have profound impact on cell wall composition, structure and performance during growth, development, interaction with the environment and economically relevant feedstock traits. It also reinforces the question how changes in cell wall structure and composition are detected and the resulting changes brought about.
The plant cell wall integrity maintenance mechanism in dicots
The overview provided above illustrates comprehensively the plasticity of plant cell walls and how their composition and structure change in highly dynamic ways in different plant species. The changes, induced by developmental cues, interactions with the environment or genetic manipulation, ensure that cell walls can perform their biological functions in different situations. In addition to these functions, CWI has to be maintained constantly as well since otherwise the high turgor pressure levels, prevalent in plant cells, will lead to cell bursting and death. The available evidence, deriving often from research on primary cell walls in dicots, supports the existence of a dedicated plant CWI maintenance mechanism [191, 211, 212]. This mechanism is monitoring the functional integrity of cell walls and initiating adaptive changes in cell wall and cellular metabolism to maintain integrity. Impairment of CWI can occur during growth and development, interaction between plants and their environment (including exposure to biotic and abiotic stress) and also through artificial measures like inhibitor treatments (CBIs) or genetic manipulation of cell wall metabolism. The extent of impairment probably varies, ranging from increased strain during etiolation in elongating hypocotyls to physical CWD and cell wall rupture during exposure to abiotic or biotic stress like drought, high salinity or pathogen infection [150, 213, 214]. Here, we will focus on the knowledge available regarding CWI maintenance in primary cell walls, discuss how CWI impairment may be perceived in plant cells and review how CWI signalling is integrated with other processes to regulate downstream responses exemplified by integration of CWI signalling with signals deriving from pattern triggered immunity (PTI). We will not incorporate the available knowledge regarding CWI maintenance in pollen tube growth since this has been recently reviewed comprehensively [215].
The existence of a CWI maintenance mechanism in plants was postulated after observing that reduction in cellulose production in the root elongation zone of Arabidopsis seedlings either by genetic means or CBI treatment leads to ectopic deposition of lignin (as a putatively compensatory measure) and activation of defence responses (including production of JA and induction of defence genes like VSP1, PDF1.2) [24, 26,27,28]. Follow-up studies showed that CBI-induced changes in cell wall composition also include enhanced callose production and modifications of neutral cell wall sugars [199, 200]. When cell wall-degrading enzymes (like pectinase and cellulase) are used to induce CWD, lignin and callose deposition are also detectable in cotyledons, suggesting that the induction of the responses is not limited to elongating cells in seedling roots [198]. Intriguingly, several of the responses (including gene expression, JA/SA production, lignin/callose deposition, necrosis and metabolic changes) are sensitive to osmotic manipulation, i.e., co-treatments with osmotica reduce the extent of the responses profoundly [29, 198, 199, 216]. Simultaneously, treatments with supernatants from wildtype seedlings exposed to CWD-inducing agents or with known elicitors such as OGs or flg22 do not induce phytohormone production or defence gene expression in the same manner as CWD-induced CWI impairment does [198]. These results suggest that cell wall-derived fragments are probably not primarily responsible for the effects observed after CBI treatments. Intriguingly, in Saccharomyces cerevisiae most, if not all, effects induced by CWI impairment are also suppressed by osmotic co-treatments, i.e., mild hyper-osmotic stress [30]. These observations suggest that turgor-sensitive processes are relevant for detection of CWI impairment. Based on the available data, different stimuli are conceivable as indicators of CWI impairment and consequently different sensors could be involved. Recently, a biophysical model has been developed, which provides a conceptual framework for investigating the responsible mechanisms [217]. One possibility is a change in the surface tension of cell walls, leading to conformational changes in CWI sensors and activation of signalling cascades. Similar mechanisms have been described in yeast and animal cells involving mechanosensitive proteins [218, 219]. Other possibilities are that CWI impairment affects organisation of the cytoskeleton, allows displacement of plasma membrane versus cell wall or causes distortion of the plasma membrane itself, events that may serve as indirect signals. These possibilities could be plant specific or similar to mechanisms contributing to detection of CWI impairment in yeast [30, 220]. The first one could involve plasma membrane localized proteins connecting the cytoskeleton with the cell wall, the second consists of the regular turgor-monitoring processes while the third one involves a plasma membrane-localized, stretch-activated calcium channel complex like MID1 CCH1 in yeast. Interestingly, the Arabidopsis-derived plasma membrane-localised calcium channels MID1-COMPLEMENTING ACTIVITY1 (MCA1) and MCA2 are capable of partially complementing MID1 CCH1 deficient yeast strains (Table 2) [221, 222]. MCA1 is required for the response to CWI impairment in Arabidopsis while both MCA1 and 2 have been implicated in mechano- and hypo-osmotic stress-perception [198, 222]. MECHANOSENSITIVE CHANNEL OF SMALL CONDUCTANCE-LIKE 2 (MSL2) and MSL3, chloroplast-localized ion channels also required for hypo-osmotic stress perception have also been implicated in CWI maintenance as well while ARABIDOPSIS HISTIDINE KINASES 1–4, required for perception of hyper-osmotic stress, seem not necessary for detection of CWI impairment [198, 223, 224]. This observation reinforces the notion that shrinking of the plasma membrane activates signalling processes distinctly different from those activated by plasma membrane expansion. Importantly, sudden plasma membrane expansion does not occur in plant cells under non-stress conditions due to the rigid cell walls.
Results from genetic analyses suggest that MCA1, the RLKs FEI2 and MALE DISCOVERER1-INTERACTING RLK 2 (MIK2) act downstream of THESEUS1 (THE1) in initiating CWD responses [198, 225]. THE1 belongs to the CrRLK1L (Catharanthus roseus RLK1-like) family of receptor kinases with 18 members, which are characterized by a conserved structure consisting of extracellular malectin-like domains, a transmembrane region and a cytoplasmic kinase domain [226, 227]. Homologs of THE1 have been found throughout the plant kingdom including “ancient” plants like Marchantia polymorpha indicating that THE1-mediated processes are widespread [226, 228]. Increasing or decreasing THE1 activity using loss- or gain-of-function alleles results in corresponding changes in CWD responses induced by CBI treatment or cell wall-degrading enzymes, implicating THE1 in CWI maintenance [198, 229]. In animals, malectin domains bind to components of the extracellular matrix [226]. For another CrRLK1L family member, FERONIA (FER), the available data suggest that FER can bind pectic polysaccharides through its malectin-like domain in vitro [153, 230]. However, the situation seems to differ in vivo since there FER binding to the cell wall apparently occurs via LEUCINE-RICH REPEAT EXTENSIN (LRX) proteins [231]. THE1, FER and several LRX proteins bind different RAPID ALKALIZATION FACTORs (RALFs) in a pH dependent manner [232,233,234,235] RALFs are small peptides, which induce changes in apoplastic pH and might modulate binding preferences of CrRLK1L proteins [233]. Recently emerged evidence suggests that the pH in the apoplast is changing dynamically in response to pathogen infection and could contribute to initiating stress-specific defence responses [236]. In contrast to THE1, loss of FER activity has profound effects on a large number of processes including biotic/abiotic stress responses, gametophytic development, mechano-perception, Ca2+-signalling and carbohydrate metabolism [153, 226, 237]. Based on these observations and results from biochemical studies it was proposed that FER might act as scaffold, which regulates interactions/integrates different CWI-related signals to control and coordinate downstream responses [232]. While the exact mode of action of FER remains to be determined, under standardized conditions FER and THE1 have opposite effects on CWD-induced responses like JA production and lignin deposition [198]. This implies that FER and THE1 have different functions during CWI maintenance, which is further supported by results from the functional characterization of FER indicating that FER is required during recovery from CWI impairment [153]. The authors have proposed that FER is required for confirmation that CWI has been re-established and growth can commence again. Intriguingly FER has recently been shown to regulate organization of the vacuole, reinforcing the notion that changes in the state of the cell wall might be coordinated with changes in turgor levels via FER and vacuole organization [231].
While FER and THE1 are currently the CrRLK1Ls with the strongest connections to CWI maintenance, it remains to be determined if other CWI sensors exist. The involvement of several other CrRLK1L family members in CWI maintenance has been investigated under the same conditions as FER and THE1 [198]. HERCULES1 and HERCULES2 loss of function alleles have similar but less pronounced effects than a FER knock down allele, while loss of ERULUS activity results in no significant differences to wildtype controls [198]. These results suggest that different family members may have evolved specialized functions. However, none of the CrRLK1Ls examined (including THE1 loss of function seedlings) exhibited complete loss of phytohormone production and lignin deposition in response to CBI-induced CWD [198]. This suggests that either redundancy occurs within the family or that other CWI sensors exist. A functional analysis of genes identified through a combination of transcriptomics data and FTIR-based cell wall analysis has implicated several new molecular components in the CWI maintenance [238]. Loss of the WALL STRESS RESPONSE 4 (WSR4) RLK affected cellulose levels and ISX-induced JA and lignin accumulation, indicating a role of WSR4 in CWI maintenance. In parallel, loss of the dirigent-like protein WSR1 caused mild alterations in cell wall structure, impaired ISX-induced SA accumulation and increased susceptibility to the necrotrophic fungus Plectosphaerella cucumerina [238].
CWI during plant development
Contributions of CWI to plant development has been reviewed competently recently. Therefore, we will discuss here only a particular aspect [211, 226]. While ploidy levels affect cell wall formation, knowledge regarding the influence of the cell wall on plant cell cycle activity is very limited [42, 239]. Recently the impact of CBI treatments on cell cycle progression was assessed in Arabidopsis to determine if the state of the cell wall is coordinated with cell cycle activity [42]. Activity was inhibited within hours after start of treatment, while osmotic co-treatments reduced the inhibitor effects on expression of the cytokinin-regulated cell cycle regulator CYCD3;1 and CBI-induced changes in cytokinin metabolism. Interestingly, none of the genes previously implicated in the CBI-induced responses (including THE1) are required for the changes observed in cell cycle gene expression. Instead NIA1 NIA2-dependent processes seem to be essentially required for the effects on the cell cycle. NIA1 NIA2 encode nitrate reductases, which are required for nitrogen metabolism and their mutant alleles are frequently used as genetic tools to reduce NO production [240]. Interestingly, CBI-induced phytohormone production and lignin deposition, classic hallmark responses of CWI impairment, are completely absent in nia1 nia2 seedlings. Results from a genetic analysis place NIA1 NIA2 downstream from THE1 [42]. The observations also imply that additional, unknown CWI sensors exist, which detect CWI impairment arising during growth, development and interaction with the environment. Calcium-based signal transduction seems to be involved in CWI signalling, and the signals generated seem to feed into NIA1 NIA2-mediated processes, which lead to activation of downstream responses including lignin deposition and phytohormone production through additional to be determined molecular components (Fig. 2).
Overview of CWI signaling components, their interactions and coordination between CWI and PTI signaling. Mechanical distortion of the CW induces CWI responses through THE1 and leads to production of elicitor peptides PEP1 and PEP3, which can suppress CWI responses and growth via PEPR1/2 (represented here by PEPR1) [198]. In parallel to mechanical distortion, the action of THE1 can be regulated by RALF34, which binds to THE1 at high apoplastic pH, leading to further alkalinisation of the apoplast [233]. THE1 activates CWI responses via MCA1 and FEI2 and NIA1 NIA2-dependent processes, which repress growth actively [198]. RALF1 and RALF23 induce alkalinization of the apoplast in a FER-dependent manner [247, 250]. Both of them also affect hormone signaling pathways through FER: RALF23 by inhibiting the de-stabilizing effect of FER on the transcription factor MYC2, a master regulator of JA signaling; RALF1 by activating ABI2, a repressor of ABA signaling [247,248,249]. In addition, RALF23 has been shown to inhibit FER’s scaffold activity for pattern-recognition receptors (PRR) and their co-receptor BAK1, thus reducing sensitivity of the plant to respective PAMPs [232]. In addition to being involved in chemical signaling through PAMPs, RALFs and phytohormones, FER is capable of sensing physical signals from the CW, possibly through LRX proteins linking FER to CW [234]. Brown bars represents the cell wall (CW), grey bars represent the plasma membrane (PM). The white space in between the CW and PM represents the CW–PM interphase, where solutes can diffuse freely and changes in the mechanical forces are being sensed. Abbreviations are explained in main text
A model for the plant CWI maintenance mechanism
Signals generated by the CWI maintenance mechanism have to be coordinated with signals deriving from other processes to ensure that plants respond to changes in their environment in an integrated and adaptive manner. This can also involve integration of signals generated in response to chemical and physical stimuli. While our knowledge on this topic is very limited, we will use here the process responsible for integrating signals derived from CWI maintenance and PTI as example since it provides food for thought how signal integration may work (Fig. 2). PTI is responsible for activation and modulation of defence responses in case a plant gets infected by a pathogen [241, 242]. This process involves chemical signals in the form of pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs) derived from the plant itself [150]. Examples for DAMPs are cell wall fragments like xyloglucan or cellobiose, extracellular ATP or small peptides like PEP1, which can all induce defence responses in unharmed cells [103, 243, 244]. PEP1 acts in PTI as enhancer of immune responses and is activated by the calcium-regulated METACASPASE4 in response to cell integrity impairment [245, 246]. Intriguingly CWI signalling is inhibited by PEP1 and impaired PTI signalling leads to enhanced CWI signalling [198]. The relevance of CWI signalling in pathogen response is indicated by loss of THE1 or FER affecting resistance to Botrytis cinerea and Fusarium oxysporum [225, 232]. These results suggest that CWI and PTI signalling are coordinated during pathogen infection and CWI monitoring could function as backup system to induce stress responses in case PTI is impaired. In fact, it has been proposed that FER might function as scaffold protein at least in two different complexes active during PTI. This creates the possibility that FER forms the key element integrating CWI and PTI signalling [232]. In one complex, the interaction between FER and BAK1, a key regulator of PTI, modulates activation of PTI controlled responses. FER seems to be released from this complex through interaction with RALF23. This interaction controls simultaneously FER-dependent regulation of MYC2, which is responsible for control of JA and SA production [247]. In parallel, FER interaction with RALF1 regulates ABA INSENSITIVE2 (ABI2) activity, a key regulator of ABA-based signaling processes [248, 249]. Last but not least RALF1-mediated, FER-dependent inhibition of the PM-localized H+-ATPase PLASMA MEMBRANE PROTON ATPASE 2 (AHA2), could lead to changes in apoplastic pH [250]. Changes in apoplastic pH in turn can affect other signaling processes exemplified by the pH-dependent binding of RALF34 to THE1, possibly activating CWI signaling [233]. In the second complex FER seems to interact with LRX proteins and RALF22/23 to regulate responses to salt stress and reorganization of the vacuole [231, 234]. These results suggest that FER has a key function in integrating inputs from multiple cell wall-plasma membrane associated signalling processes and regulating downstream responses represented here by different phytohormones. More importantly this possibility would explain why FER in Arabidopsis and FER homologs in other species are involved in such a large number of biological processes [251,252,253,254].
Concluding remarks
To summarize it has become obvious that plant cell walls in different plant species are extremely plastic. They modify their composition and structure in a highly adaptive manner and often the phenotypes (or the lack thereof) observed cannot be explained by simple functional redundancy between family members. These observations have led to proposals that in plants, like in other organisms such as yeast, a CWI maintenance mechanism exists. Recent years have seen a dramatic increase in our knowledge regarding the mode of action of the plant CWI maintenance mechanism in Arabidopsis, including the identification of several key molecular components involved. In parallel it has become obvious that homologs of these components exist in other plant species, implying that the mechanism exists there as well. This is further supported by mutant phenotypes observed, which cannot be easily explained using arguments such as genetic redundancy. While we still struggle to understand the molecular mechanisms underlying CWI maintenance, the speed of discoveries on the topic in recent years strongly suggests we head towards a thorough understanding of its mode of action in model organisms like Arabidopsis. This will enable us to apply the knowledge generated to improve the performance of food and bioenergy crops in the future in a knowledge-based manner to facilitate adaptation to a rapidly changing environment.
References
Ragauskas AJ, Williams CK, Davison BH, Britovsek G, Cairney J, Eckert CA, Frederick WJ, Hallett JP, Leak DJ, Liotta CL, Mielenz JR, Murphy R, Templer R, Tschaplinski T (2006) The path forward for biofuels and biomaterials. Science 311:484–489
Doblin MS, Johnson KL, Humphries J, Newbigin EJ, Bacic A (Tony) (2013) Are designer plant cell walls a realistic aspiration or will the plasticity of the plant’s metabolism win out? Curr Opin Biotechnol 26:108–114
Mahon EL, Mansfield SD (2019) Tailor-made trees: engineering lignin for ease of processing and tomorrow’s bioeconomy. Curr Opin Biotechnol 56:147–155
Zablackis E, Huang J, Müller B, Darvill AG, Albersheim P (1995) Characterization of the cell-wall polysaccharides of Arabidopsis thaliana leaves. Plant Physiol 107:1129–1138
Scheller HV, Ulvskov P (2010) Hemicelluloses. Annu Rev Plant Biol 61:263–289
Cosgrove DJ (1997) Assembly and enlargement of the primary cell wall in plants. Annu Rev Cell Dev Biol 13:171–201
Kumar M, Campbell L, Turner S (2016) Secondary cell walls: biosynthesis and manipulation. J Exp Bot 67:515–531
Höfte H, Voxeur A (2017) Plant cell walls. Curr Biol 27:R865–R870
Penning BW, Hunter CT, Tayengwa R, Eveland AL, Dugard CK, Olek AT, Vermerris W, Koch KE, McCarty DR, Davis MF, Thomas SR, Mccann MC, Carpita NC (2009) Genetic resources for maize cell wall biology. Plant Physiol 151:1703–1728
Kumar M, Turner S (2015) Plant cellulose synthesis: CESA proteins crossing kingdoms. Phytochemistry 112:91–99
McFarlane HE, Döring A, Persson S (2014) The cell biology of cellulose synthesis. Annu Rev Plant Biol 65:69–94
Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM, Poindexter P, Khitrov N, Auer M, Somerville CR (1996) Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci 93:12637–12642
Taylor NG, Scheible WR, Cutler S, Somerville CR, Turner SR, Brown R, Baulcombe D, Fincher GB, Vernhettes S, Höfte H (1999) The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11:769–780
Taylor NG, Laurie S, Turner SR (2000) Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12:2529–2539
Turner SR, Somerville CR (1997) Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9:689–701
Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci U S A 104:15572–15577
Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, Khitrov N, Auer M, Somerville CR (2007) Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci U S A 104:15566–15571
Beeckman T, Przemeck GKH, Stamatiou G, Lau R, Terryn N, De Rycke R, Inzé D, Berleth T (2002) Genetic complexity of cellulose synthase A gene function in Arabidopsis embryogenesis. Plant Physiol 130:1883–1893
Li S, Bashline L, Lei L, Gu Y (2014) Cellulose synthesis and its regulation. Arab B 12:e0169
Hu H, Zhang R, Tao Z, Li X, Li Y, Huang J, Li X, Han X, Feng S, Zhang G, Peng L (2018) Cellulose synthase mutants distinctively affect cell growth and cell wall integrity for plant biomass production in Arabidopsis. Plant Cell Physiol 59:1144–1157
Tateno M, Brabham C, Debolt S (2016) Cellulose biosynthesis inhibitors—a multifunctional toolbox. J Exp Bot 67:533–542
Shim I, Law R, Kileeg Z, Stronghill P, Northey JGB, Strap JL, Bonetta DT (2018) Alleles causing resistance to isoxaben and flupoxam highlight the significance of transmembrane domains for CESA protein function. Front Plant Sci 9:1152
Scheible WR, Eshed R, Richmond T, Delmer D, Somerville C (2001) Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proc Natl Acad Sci U S A 98:10079–10084
Ellis C, Karafyllidis I, Wasternack C, Turner JG (2002) The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14:1557–1566
Woodley M, Mulvihill A, Fujita M, Wasteneys GO (2018) Exploring microtubule-dependent cellulose-synthase-complex movement with high precision particle tracking. Plants 7:53
Cano-Delgado AI, Metzlaff K, Bevan MW (2000) The eli1 mutation reveals a link between cell expansion and secondary cell wall formation in Arabidopsis thaliana. Development 127:3395–3405
Ellis C, Turner JG (2001) The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13:1025–1033
Caño-Delgado A, Penfield S, Smith C, Catley M, Bevan M (2003) Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J 34:351–362
Denness L, McKenna JF, Segonzac C, Wormit A, Madhou P, Bennett M, Mansfield J, Zipfel C, Hamann T (2011) Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species- and jasmonic acid-dependent process in Arabidopsis. Plant Physiol 156:1364–1374
Levin DE (2011) Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189:1145–1175
Panteris E, Adamakis IDS, Daras G, Hatzopoulos P, Rigas S (2013) Differential responsiveness of cortical microtubule orientation to suppression of cell expansion among the developmental zones of Arabidopsis thaliana root apex. PLoS One 8:e82442
Zhu X, Li S, Pan S, Xin X, Gu Y (2018) CSI1, PATROL1, and exocyst complex cooperate in delivery of cellulose synthase complexes to the plasma membrane. Proc Natl Acad Sci U S A 115:E3578–E3587
Lei L, Li S, Gu Y (2012) Cellulose synthase interactive protein 1 (CSI1) mediates the intimate relationship between cellulose microfibrils and cortical microtubules. Plant Signal Behav 7:714–718
Zhang Y, Nikolovski N, Sorieul M, Vellosillo T, McFarlane HE, Dupree R, Kesten C, Schneider R, Driemeier C, Lathe R, Lampugnani E, Yu X, Ivakov A, Doblin MS, Mortimer JC, Brown SP, Persson S, Dupree P (2016) Golgi-localized STELLO proteins regulate the assembly and trafficking of cellulose synthase complexes in Arabidopsis. Nat Commun 7:11656
Lei L, Li S, Gu Y (2012) Cellulose synthase complexes: composition and regulation. Front Plant Sci. 3:75
Endler A, Kesten C, Schneider R, Zhang Y, Ivakov A, Froehlich A, Funke N, Persson S (2015) A mechanism for sustained cellulose synthesis during salt stress. Cell 162:1353–1364
Gutierrez R, Lindeboom JJ, Paredez AR, Emons AMC, Ehrhardt DW (2009) Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol 11:797–806
Crowell EF, Bischoff V, Desprez T, Rolland A, Stierhof YD, Schumacher K, Gonneau M, Höfte H, Vernhettes S (2009) Pausing of golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21:1141–1154
Li S, Lei L, Yingling YG, Gu Y (2015) Microtubules and cellulose biosynthesis: the emergence of new players. Curr Opin Plant Biol 28:76–82
Speicher TL, Li PZ, Wallace IS (2018) Phosphoregulation of the plant cellulose synthase complex and cellulose synthase-like proteins. Plants 7:52
DeBolt S, Gutierrez R, Ehrhardt DW, Somerville C (2007) Nonmotile cellulose synthase subunits repeatedly accumulate within localized regions at the plasma membrane in Arabidopsis hypocotyl cells following 2,6-dichlorobenzonitrile treatment. Plant Physiol 145:334–338
Gigli-Bisceglia N, Engelsdorf T, Strnad M, Vaahtera L, Khan GA, Yamoune A, Alipanah L, Novák O, Persson S, Hejatko J, Hamann T (2018) Cell wall integrity modulates Arabidopsis thaliana cell cycle gene expression in a cytokinin- and nitrate reductase-dependent manner. Dev 145:dev166678
Sampathkumar A, Gutierrez R, McFarlane HE, Bringmann M, Lindeboom J, Emons AM, Samuels L, Ketelaar T, Ehrhardt DW, Persson S (2013) Patterning and lifetime of plasma membrane-localized cellulose synthase is dependent on actin organization in Arabidopsis interphase cells. Plant Physiol 162:675–688
Tolmie F, Poulet A, McKenna J, Sassmann S, Graumann K, Deeks M, Runions J (2017) The cell wall of Arabidopsis thaliana influences actin network dynamics. J Exp Bot 68:4517–4527
Zhong R, Burk DH, Morrison WH, Ye Z-H (2002) A kinesin-like protein is essential for oriented deposition of cellulose microfibrils and cell wall strength. Plant Cell Online 14:3101–3117
Ganguly A, DeMott L, Dixit R (2017) The Arabidopsis kinesin-4, FRA1, requires a high level of processive motility to function correctly. J Cell Sci 130:1232–1238
Zuo J, Niu QW, Nishizawa N, Wu Y, Kost B, Chua NH (2000) KORRIGAN, an Arabidopsis endo-1,4-β-glucanase, localizes to the cell plate by polarized targeting and is essential for cytokinesis. Plant Cell 12:1137–1152
Pagant S, Bichet A, Sugimoto K, Lerouxel O, Desprez T, McCann M, Lerouge P, Vernhettes S, Höfte H (2002) KOBITO1 encodes a novel plasma membrane protein necessary for normal synthesis of cellulose during cell expansion in Arabidopsis. Plant Cell 14:2001–2013
Sanchez-Rodriguez C, Bauer S, Hematy K, Saxe F, Ibanez AB, Vodermaier V, Konlechner C, Sampathkumar A, Ruggeberg M, Aichinger E, Neumetzler L, Burgert I, Somerville C, Hauser M-T, Persson S (2012) CHITINASE-LIKE1/POM-POM1 and its homolog CTL2 are glucan-interacting proteins important for cellulose biosynthesis in Arabidopsis. Plant Cell 24:589–607
Vain T, Crowell EF, Timpano H, Biot E, Desprez T, Mansoori N, Trindade LM, Pagant S, Robert S, Höfte H, Gonneau M, Vernhettes S (2014) The cellulase KORRIGAN is part of the cellulose synthase complex. Plant Physiol 165:1521–1532
Mansoori N, Timmers J, Desprez T, Kamei CLA, Dees DCT, Vincken J-P, Visser RGF, Höfte H, Vernhettes S, Trindade LM, Trindade LM (2014) KORRIGAN1 interacts specifically with integral components of the cellulose synthase machinery. PLoS ONE 9:e112387
Hauser MT, Morikami A, Benfey PN (1995) Conditional root expansion mutants of Arabidopsis. Dev 121:1237–1252
Zhong R, Kays SJ, Schroeder BP, Ye ZH (2002) Mutation of a chitinase-like gene causes ectopic deposition of lignin, aberrant cell shapes, and overproduction of ethylene. Plant Cell 14:165–179
Zheng M, Liu X, Lin J, Liu X, Wang Z, Xin M, Yao Y, Peng H, Zhou DX, Ni Z, Sun Q, Hu Z (2019) Histone acetyltransferase GCN5 contributes to cell wall integrity and salt stress tolerance by altering the expression of cellulose synthesis genes. Plant J 97:587–602
Zhong R, Ye ZH (2015) Secondary cell walls: biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol 56:195–214
Kumar M, Mishra L, Carr P, Pilling M, Gardner P, Mansfield SD, Turner S (2018) Exploiting CELLULOSE SYNTHASE (CESA) class specificity to probe cellulose microfibril biosynthesis. Plant Physiol 177:151–167
Hernández-Blanco C, Feng DX, Hu J, Sánchez-Vallet A, Deslandes L, Llorente F, Berrocal-Lobo M, Keller H, Barlet X, Sánchez-Rodríguez C, Anderson LK, Somerville S, Marco Y, Molinaa A (2007) Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 19:890–903
Yokoyama R, Nishitani K (2004) Genomic basis for cell-wall diversity in plants. A comparative approach to gene families in rice and Arabidopsis. Plant Cell Physiol 45:1111–1121
Li L, Hey S, Liu S, Liu Q, McNinch C, Hu HC, Wen TJ, Marcon C, Paschold A, Bruce W, Schnable PS, Hochholdinger F (2016) Characterization of maize roothairless6 which encodes a D-type cellulose synthase and controls the switch from bulge formation to tip growth. Sci Rep 6:34395
Kaur S, Dhugga KS, Gill K, Singh J (2016) Novel structural and functional motifs in cellulose synthase (CesA) genes of bread wheat (Triticum aestivum, L.). PLoS One 11:e0147046
Kokubo A, Sakurai N, Kuraishi S, Takeda K (1991) Culm brittleness of barley (Hordeum vulgare L.) mutants is caused by smaller number of cellulose molecules in cell wall. Plant Physiol 97:509–514
Kokubo A, Kuraishi S, Sakurai N (1989) Culm strength of Barley. Plant Physiol 91:876–882
Wang D, Qin Y, Fang J, Yuan S, Peng L, Zhao J, Li X (2016) A missense mutation in the zinc finger domain of OsCESA7 deleteriously affects cellulose biosynthesis and plant growth in rice. PLoS One 11:e0153993
Aohara T, Kotake T, Kaneko Y, Takatsuji H, Tsumuraya Y, Kawasaki S (2009) Rice BRITTLE CULM 5 (BRITTLE NODE) is involved in secondary cell wall formation in the sclerenchyma tissue of nodes. Plant Cell Physiol 50:1886–1897
Li P, Liu Y, Tan W, Chen J, Zhu M, Lv Y, Liu Y, Yu S, Zhang W, Cai H (2019) Brittle Culm 1 Encodes a COBRA-like protein involved in secondary cell wall cellulose biosynthesis in Sorghum. Plant Cell Physiol 60:788–801
Li Y, Qian Q, Zhou Y, Yan M, Sun L, Zhang M, Fu Z, Wang Y, Han B, Pang X, Chen M, Li J (2003) BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell 15:2020–2031
Roudier F, Schindelman G, DeSalle R, Benfey PN (2002) The COBRA family of putative GPI-anchored proteins in Arabidopsis. A new fellowship in expansion. Plant Physiol 130:538–548
Zhang M, Zhang B, Qian Q, Yu Y, Li R, Zhang J, Liu X, Zeng D, Li J, Zhou Y (2010) Brittle Culm 12, a dual-targeting kinesin-4 protein, controls cell-cycle progression and wall properties in rice. Plant J 63:312–328
Brabham C, Lei L, Gu Y, Stork J, Barrett M, DeBolt S (2014) Indaziflam herbicidal action: a potent cellulose biosynthesis inhibitor. Plant Physiol 166:1177–1185
Richmond TA, Somerville CR (2000) The cellulose synthase superfamily. Plant Physiol 124:495–498
Kaur S, Dhugga KS, Beech R, Singh J (2017) Genome-wide analysis of the Cellulose synthase-like (Csl) gene family in bread wheat (Triticum aestivum L.). BMC Plant Biol 17:193
Yin Y, Huang J, Xu Y (2009) The cellulose synthase superfamily in fully sequenced plants and algae. BMC Plant Biol 9:99
Roberts AW, Bushoven JT (2007) The cellulose synthase (CESA) gene superfamily of the moss Physcomitrella patens. Plant Mol Biol 63:207–219
Favery B, Ryan E, Foreman J, Linstead P, Boudonck K, Steer M, Shaw P, Dolan L (2001) KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev 15:79–89
Yin L, Verhertbruggen Y, Oikawa A, Manisseri C, Knierim B, Prak L, Jensen JK, Knox JP, Auer M, Willats WGT, Scheller HV (2011) The cooperative activities of CSLD2, CSLD3, and CSLD5 are required for normal Arabidopsis development. Mol Plant 4:1024–1037
Verhertbruggen Y, Yin L, Oikawa A, Scheller HV (2011) Mannan synthase activity in the CSLD family. Plant Signal Behav 6:1620–1623
Zabotina OA (2012) Xyloglucan and its biosynthesis. Front Plant Sci 3:134
Park S, Szumlanski AL, Gu F, Guo F, Nielsen E (2011) A role for CSLD3 during cell-wall synthesis in apical plasma membranes of tip-growing root-hair cells. Nat Cell Biol 13:973–980
Zhu Y, Nam J, Carpita NC, Matthysse AG, Gelvin SB (2003) Agrobacterium-mediated root transformation is inhibited by mutation of an Arabidopsis Cellulose Synthase-Like gene. Plant Physiol 133:1000–1010
Goubet F, Barton CJ, Mortimer JC, Yu X, Zhang Z, Miles GP, Richens J, Liepman AH, Seffen K, Dupree P (2009) Cell wall glucomannan in Arabidopsis is synthesised by CSLA glycosyltransferases, and influences the progression of embryogenesis. Plant J 60:527–538
Schröder R, Atkinson RG, Redgwell RJ (2009) Re-interpreting the role of endo-β-mannanases as mannan endotransglycosylase/hydrolases in the plant cell wall. Ann Bot 104:197–204
Handford MG, Baldwin TC, Goubet F, Prime TA, Miles J, Yu X, Dupree P (2003) Localisation and characterisation of cell wall mannan polysaccharides in Arabidopsis thaliana. Planta 218:27–36
Marcus SE, Blake AW, Benians TAS, Lee KJD, Poyser C, Donaldson L, Leroux O, Rogowski A, Petersen HL, Boraston A, Gilbert HJ, Willats WGT, Paul Knox J (2010) Restricted access of proteins to mannan polysaccharides in intact plant cell walls. Plant J 64:191–203
Burton RA, Fincher GB (2009) (1,3;1,4)-β-d-glucans in cell walls of the Poaceae, lower plants, and fungi: a tale of two linkages. Mol Plant 2:873–882
Wang L, Guo K, Li Y, Tu Y, Hu H, Wang B, Cui X, Peng L (2010) Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol 10:282
Vega-Sánchez ME, Verhertbruggen Y, Scheller HV, Ronald PC (2013) Abundance of mixed linkage glucan in mature tissues and secondary cell walls of grasses. Plant Signal Behav 8:e23143
Kulkarni AR, Pattathil S, Hahn MG, York WS, O’Neill MA (2012) Comparison of arabinoxylan structure in bioenergy and model grasses. Ind Biotechnol 8:222–229
Doblin MS, Pettolino FA, Wilson SM, Campbell R, Burton RA, Fincher GB, Newbigin E, Bacic A (2009) A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-β-d-glucan synthesis in transgenic Arabidopsis. Proc Natl Acad Sci U S A 106:5996–6001
Vega-Sánchez ME, Verhertbruggen Y, Christensen U, Chen X, Sharma V, Varanasi P, Jobling SA, Talbot M, White RG, Joo M, Singh S, Auer M, Scheller HV, Ronald PC (2012) Loss of Cellulose synthase-like F6 function affects mixed-linkage glucan deposition, cell wall mechanical properties, and defense responses in vegetative tissues of rice. Plant Physiol 159:56–69
Carpita NC, Gibeaut DM (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3:1–30
Fincher GB (2009) Revolutionary times in our understanding of cell wall biosynthesis and remodeling in the grasses. Plant Physiol 149:27–37
Kiemle SN, Zhang X, Esker AR, Toriz G, Gatenholm P, Cosgrove DJ (2014) Role of (1,3)(1,4)-β-glucan in cell walls: interaction with cellulose. Biomacromolecules 15:1727–1736
Smith-Moritz AM, Hao Z, Fernández-Niño SG, Fangel JU, Verhertbruggen Y, Holman H-YY, Willats WGT, Ronald PC, Scheller HV, Heazlewood JL, Vega-Sánchez ME (2015) Structural characterization of a mixed-linkage glucan deficient mutant reveals alteration in cellulose microfibril orientation in rice coleoptile mesophyll cell walls. Front Plant Sci 6:1–13
O’Neill MA, York WS (2018) The composition and structure of plant primary cell walls. Annual plant reviews. Wiley, Chichester, pp 1–54
Fry SC (1989) The structure and functions of xyloglucan. J Exp Bot 40:1–11
Cavalier DM, Lerouxel O, Neumetzler L, Yamauchi K, Reinecke A, Freshour G, Zabotina OA, Hahn MG, Burgert I, Pauly M, Raikhel NV, Keegstra K (2008) Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20:1519–1537
Hayashi T, Marsden MPF, Delmer DP (1987) Pea xyloglucan and cellulose. Plant Physiol 83:384–389
Cosgrove DJ (2016) Catalysts of plant cell wall loosening. F1000Research 5 eCollection 2016
Nishitani K, Tominaga R (1992) Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J Biol Chem 267:21058–21064
Strohmeier M, Hrmova M, Fischer M, Harvey AJ, Fincher GB, Pleiss J (2009) Molecular modeling of family GH16 glycoside hydrolases: potential roles for xyloglucan transglucosylases/hydrolases in cell wall modification in the Poaceae. Protein Sci 13:3200–3213
Ferrari S, Savatin DV, Sicilia F, Gramegna G, Cervone F, De Lorenzo G (2013) Oligogalacturonides: plant damage-associated molecular patterns and regulators of growth and development. Front. Plant Sci. 4:49
Locci F, Benedetti M, Pontiggia D, Citterico M, Caprari C, Mattei B, Cervone F, De Lorenzo G (2019) An Arabidopsis berberine bridge enzyme-like protein specifically oxidizes cellulose oligomers and plays a role in immunity. Plant J 98:540–554
Claverie J, Balacey S, Lemaître-Guillier C, Brulé D, Chiltz A, Granet L, Noirot E, Daire X, Darblade B, Héloir MC, Poinssot B (2018) The cell wall-derived xyloglucan is a new DAMP triggering plant immunity in Vitis vinifera and Arabidopsis thaliana. Front Plant Sci 871:1725
Chou YH, Pogorelko G, Zabotina OA (2012) Xyloglucan xylosyltransferases XXT1, XXT2, and XXT5 and the glucan synthase CSLC4 form Golgi-localized multiprotein complexes. Plant Physiol 159:1355–1366
Park YB, Cosgrove DJ (2012) Changes in cell wall biomechanical properties in the xyloglucan-deficient xxt1/xxt2 mutant of Arabidopsis. Plant Physiol 158:465–475
Wang C, Li S, Ng S, Zhang B, Zhou Y, Whelan J, Wu P, Shou H (2014) Mutation in xyloglucan 6-xylosytransferase results in abnormal root hair development in Oryza sativa. J Exp Bot 65:4149–4157
Kong Y, Peña MJ, Renna L, Avci U, Pattathil S, Tuomivaara ST, Li X, Reiter WD, Brandizzi F, Hahn MG, Darvill AG, York WS, O’Neill MA (2015) Galactose-depleted xyloglucan is dysfunctional and leads to dwarfism in Arabidopsis. Plant Physiol 167:1296–1306
Madson M, Dunand C, Li X, Verma R, Vanzin GF, Caplan J, Shoue DA, Carpita NC, Reiter WD (2003) The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15:1662–1670
Tedman-Jones JD, Lei R, Jay F, Fabro G, Li X, Reiter WD, Brearley C, Jones JDG (2008) Characterization of Arabidopsis mur3 mutations that result in constitutive activation of defence in petioles, but not leaves. Plant J 56:691–703
Peña MJ, Ryden P, Madson M, Smith AC, Carpita NC (2004) The galactose residues of xyloglucan are essential to maintain mechanical strength of the primary cell walls in Arabidopsis during growth. Plant Physiol 134:443–451
Xu Z, Wang M, Shi D, Zhou G, Niu T, Hahn MG, O’Neill MA, Kong Y (2017) DGE-seq analysis of MUR3-related Arabidopsis mutants provides insight into how dysfunctional xyloglucan affects cell elongation. Plant Sci 258:156–169
Nikolovski N, Rubtsov D, Segura MP, Miles GP, Stevens TJ, Dunkley TPJ, Munro S, Lilley KS, Dupree P (2012) Putative glycosyltransferases and other plant Golgi apparatus proteins are revealed by LOPIT proteomics. Plant Physiol 160:1037–1051
Perrin RM, Jia Z, Wagner TA, O’Neill MA, Sarria R, York WS, Raikhel NV, Keegstra K (2003) Analysis of xyloglucan fucosylation in Arabidopsis. Plant Physiol 132:768–778
Côté F, Hahn MG (1994) Oligosaccharins: structures and signal transduction. Plant Mol Biol 26:1379–1411
Mortimer JC, Faria-Blanc N, Yu X, Tryfona T, Sorieul M, Ng YZ, Zhang Z, Stott K, Anders N, Dupree P (2015) An unusual xylan in Arabidopsis primary cell walls is synthesised by GUX3, IRX9L, IRX10L and IRX14. Plant J 83:413–426
Saulnier L, Vigouroux J, Thibault JF (1995) Isolation and partial characterization of feruloylated oligosaccharides from maize bran. Carbohydr Res 272:241–253
Freeman J, Lovegrove A, Wilkinson MD, Saulnier L, Shewry PR, Mitchell RAC (2016) Effect of suppression of arabinoxylan synthetic genes in wheat endosperm on chain length of arabinoxylan and extract viscosity. Plant Biotechnol J 14:109–116
Faik A (2010) Xylan biosynthesis: news from the grass. Plant Physiol 153:396–402
Persson S, Caffall KH, Freshour G, Hilley MT, Bauer S, Poindexter P, Hahn MG, Mohnen D, Somerville C (2007) The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. Plant Cell 19:237–255
Carpita NC (1996) Structure and biogenesis of the cell walls of grasses. Annu Rev Plant Physiol Plant Mol Biol 47:445–476
Grabber JH (2005) How do lignin composition, structure, and cross-linking affect degradability? A review of cell wall model studies. Crop Sci 45(3):820–831
Lee C, Zhong R, Ye ZH (2012) Arabidopsis family GT43 members are xylan xylosyltransferases required for the elongation of the xylan backbone. Plant Cell Physiol 53:135–143
Keppler BD, Showalter AM (2010) IRX14 and IRX14-LIKE, two glycosyl transferases involved in glucuronoxylan biosynthesis and drought tolerance in Arabidopsis. Mol Plant 3:834–841
Wu AM, Hörnblad E, Voxeur A, Gerber L, Rihouey C, Lerouge P, Marchant A (2010) Analysis of the Arabidopsis IRX9/IRX9-L and IRX14/IRX14-L pairs of glycosyltransferase genes reveals critical contributions to biosynthesis of the hemicellulose glucuronoxylan. Plant Physiol 153:542–554
Mortimer JC, Miles GP, Brown DM, Zhang Z, Segura MP, Weimar T, Yu X, Seffen KA, Stephens E, Turner SR, Dupree P (2010) Absence of branches from xylan in Arabidopsis gux mutants reveals potential for simplification of lignocellulosic biomass. Proc Natl Acad Sci U S A 107:17409–17414
Wu AM, Rihouey C, Seveno M, Hörnblad E, Singh SK, Matsunaga T, Ishii T, Lerouge P, Marchant A (2009) The Arabidopsis IRX10 and IRX10-LIKE glycosyltransferases are critical for glucuronoxylan biosynthesis during secondary cell wall formation. Plant J 57:718–731
Brown DM, Zhang Z, Stephens E, Dupree P, Turner SR (2009) Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis. Plant J 57:732–746
Anders N, Wilkinson MD, Lovegrove A, Freeman J, Tryfona T, Pellny TK, Weimar T, Mortimer JC, Stott K, Baker JM, Defoin-Platel M, Shewry PR, Dupree P, Mitchell RAC (2012) Glycosyl transferases in family 61 mediate arabinofuranosyl transfer onto xylan in grasses. Proc Natl Acad Sci U S A 109:989–993
Freeman J, Ward JL, Kosik O, Lovegrove A, Wilkinson MD, Shewry PR, Mitchell RAC (2017) Feruloylation and structure of arabinoxylan in wheat endosperm cell walls from RNAi lines with suppression of genes responsible for backbone synthesis and decoration. Plant Biotechnol J 15:1429–1438
Zhong R, Cui D, Phillips DR, Ye ZH (2018) A novel rice xylosyltransferase catalyzes the addition of 2-O-xylosyl side chains onto the xylan backbone. Plant Cell Physiol 59:554–565
Marriott PE, Sibout R, Lapierre C, Fangel JU, Willats WGT, Hofte H, Gómez LD, McQueen-Mason SJ (2014) Range of cell-wall alterations enhance saccharification in Brachypodium distachyon mutants. Proc Natl Acad Sci U S A 111:14601–14606
Rennie EA, Hansen SF, Baidoo EEK, Hadi MZ, Keasling JD, Scheller HV (2012) Three members of the Arabidopsis glycosyltransferase family 8 are xylan glucu-ronosyltransferases. Plant Physiol 159:1408–1417
Lee C, Teng Q, Zhong R, Ye ZH (2012) Arabidopsis GUX proteins are glucuronyltransferases responsible for the addition of glucuronic acid side chains onto xylan. Plant Cell Physiol 53:1204–1216
Vogel J (2008) Unique aspects of the grass cell wall. Curr Opin Plant Biol 11:301–307
Ebringerová A (2006) Structural diversity and application potential of hemicelluloses. Macromolecular symposia. Wiley, New York, pp 1–12
Oh CS, Kim H, Lee C (2013) Rice cell wall polysaccharides: structure and biosynthesis. J. Plant Biol. 56:274–282
Marcia MM (2009) Feruloylation in grasses: current and future perspectives. Mol Plant 2:861–872
Urbanowicz BR, Peña MJ, Ratnaparkhe S, Avci U, Backe J, Steet HF, Foston M, Li H, O’Neill MA, Ragauskas AJ, Darvill AG, Wyman C, Gilbert HJ, York WS (2012) 4-O-methylation of glucuronic acid in Arabidopsis glucuronoxylan is catalyzed by a domain of unknown function family 579 protein. Proc Natl Acad Sci U S A 109:14253–14258
Yuan Y, Teng Q, Zhong R, Haghighat M, Richardson EA, Ye ZH (2016) Mutations of Arabidopsis TBL32 and TBL33 affect xylan acetylation and secondary wall deposition. PLoS One 11:e0146460
Gille S, de Souza A, Xiong G, Benz M, Cheng K, Schultink A, Reca IB, Pauly M (2011) O-acetylation of Arabidopsis hemicellulose xyloglucan requires AXY4 or AXY4L, proteins with a TBL an DUF231 domain. Plant Cell 23:4041–4053
Schultink A, Naylor D, Dama M, Pauly M (2015) The role of the plant-specific altered xyloglucan9 protein in Arabidopsis cell wall polysaccharide o-acetylation. Plant Physiol 167:1271–1283
Manabe Y, Nafisi M, Verhertbruggen Y, Orfila C, Gille S, Rautengarten C, Cherk C, Marcus SE, Somerville S, Pauly M, Knox JP, Sakuragi Y, Scheller HV (2011) Loss-of-function mutation of REDUCED WALL ACETYLATION2 in Arabidopsis leads to reduced cell wall acetylation and increased resistance to Botrytis cinerea. Plant Physiol 155:1068–1078
Caffall KH, Mohnen D (2009) The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr Res 344:1879–1900
Daher FB, Braybrook SA (2015) How to let go: pectin and plant cell adhesion. Front. Plant Sci. 6:523
Peaucelle A, Wightman R, Höfte H (2015) The control of growth symmetry breaking in the Arabidopsis hypocotyl. Curr Biol 25:1746–1752
Western TL (2012) The sticky tale of seed coat mucilages: production, genetics, and role in seed germination and dispersal. Seed Sci. Res. 22:1–25
Levesque-Tremblay G, Pelloux J, Braybrook SA, Müller K (2015) Tuning of pectin methylesterification: consequences for cell wall biomechanics and development. Planta 242:791–811
Wang D, Yeats TH, Uluisik S, Rose JKC, Seymour GB (2018) Fruit softening: revisiting the role of pectin. Trends Plant Sci 23:302–310
Wang T, Park YB, Cosgrove DJ, Hong M (2015) Cellulose-pectin spatial contacts are Inherent to never-dried Arabidopsis primary cell walls: evidence from solid-state nuclear magnetic resonance. Plant Physiol 168:871–884
Bacete L, Mélida H, Miedes E, Molina A (2018) Plant cell wall-mediated immunity: cell wall changes trigger disease resistance responses. Plant J 93:614–636
Engelsdorf T, Will C, Hofmann J, Schmitt C, Merritt BB, Rieger L, Frenger MS, Marschall A, Franke RB, Pattathil S, Voll LM (2017) Cell wall composition and penetration resistance against the fungal pathogen Colletotrichum higginsianum are affected by impaired starch turnover in Arabidopsis mutants. J Exp Bot 68:701–713
Kohorn BD, Kobayashi M, Johansen S, Riese J, Huang LF, Koch K, Fu S, Dotson A, Byers N (2006) An Arabidopsis cell wall-associated kinase required for invertase activity and cell growth. Plant J 46:307–316
Feng W, Kita D, Peaucelle A, Cartwright HN, Doan V, Duan Q, Liu MC, Maman J, Steinhorst L, Schmitz-Thom I, Yvon R, Kudla J, Wu HM, Cheung AY, Dinneny JR (2018) The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr Biol 28:666–675.e5
Sterling JD, Atmodjo MA, Inwood SE, Kolli VSK, Quigley HF, Hahn MG, Mohnen D (2006) Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc Natl Acad Sci U S A 103:5236–5241
Atmodjo MA, Sakuragi Y, Zhu X, Burrell AJ, Mohanty SS, Atwood JA, Orlando R, Scheller HV, Mohnen D (2011) Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:galacturonosyltransferase complex. Proc Natl Acad Sci U S A 108:20225–20230
Miao Y, Li HY, Shen J, Wang J, Jiang L (2011) QUASIMODO 3 (QUA3) is a putative homogalacturonan methyltransferase regulating cell wall biosynthesis in Arabidopsis suspension-cultured cells. J Exp Bot 62:5063–5078
Bouton S, Leboeuf E, Mouille G, Leydecker M-T, Talbotec J, Granier F, Lahaye M, Höfte H, Truong H-N (2002) QUASIMODO1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis and cell adhesion in Arabidopsis. Plant Cell 14:2577–2590
Leboeuf E, Guillon F, Thoiron S, Lahaye M (2005) Biochemical and immunohistochemical analysis of pectic polysaccharides in the cell walls of Arabidopsis mutant QUASIMODO 1 suspension-cultured cells: implications for cell adhesion. J Exp Bot 56:3171–3182
Orfila C, Sørensen SO, Harholt J, Geshi N, Crombie H, Truong HN, Reid JSG, Knox JP, Scheller HV (2005) QUASIMODO1 is expressed in vascular tissue of Arabidopsis thaliana inflorescence stems, and affects homogalacturonan and xylan biosynthesis. Planta 222:613–622
Mouille G, Ralet MC, Cavelier C, Eland C, Effroy D, Hématy K, McCartney L, Truong HN, Gaudon V, Thibault JF, Marchant A, Höfte H (2007) Homogalacturonan synthesis in Arabidopsis thaliana requires a Golgi-localized protein with a putative methyltransferase domain. Plant J 50:605–614
Kim SJ, Held MA, Zemelis S, Wilkerson C, Brandizzi F (2015) CGR2 and CGR3 have critical overlapping roles in pectin methylesterification and plant growth in Arabidopsis thaliana. Plant J 82:208–220
Pu Y, Walley JW, Shen Z, Lang MG, Briggs SP, Estelle M, Kelley DR (2019) Quantitative early auxin root proteomics identifies GAUT10, a galacturonosyltransferase, as a novel regulator of root meristem maintenance. Mol Cell Proteom 18:1157–1170
Jensen JK, Sørensen SO, Harholt J, Geshi N, Sakuragi Y, Møller I, Zandleven J, Bernal AJ, Jensen NB, Sørensen C, Pauly M, Beldman G, Willats WGT, Scheller HV (2008) Identification of a xylogalacturonan xylosyltransferase involved in pectin biosynthesis in Arabidopsis. Plant Cell 20:1289–1302
Madrid Liwanag AJ, Ebert B, Verhertbruggen Y, Rennie EA, Rautengarten C, Oikawa A, Andersen MCF, Clausen MH, Scheller HV (2013) Pectin biosynthesis: GALS1 in Arabidopsis thaliana Is a β-1,4-Galactan β-1,4-Galactosyltransferase. Plant Cell 24:5024–5036
Harholt J, Jensen JK, Sørensen SO, Orfila C, Pauly M, Scheller HV (2006) ARABINAN DEFICIENT 1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis. Plant Physiol 140:49–58
Harholt J, Jensen JK, Verhertbruggen Y, Søgaard C, Bernard S, Nafisi M, Poulsen CP, Geshi N, Sakuragi Y, Driouich A, Knox JP, Scheller HV (2012) ARAD proteins associated with pectic Arabinan biosynthesis form complexes when transiently overexpressed in planta. Planta 236:115–128
Voiniciuc C, Engle KA, Günl M, Dieluweit S, Schmidt MHW, Yang JY, Moremen KW, Mohnen D, Usadel B (2018) Identification of key enzymes for pectin synthesis in seed mucilage. Plant Physiol 178:1045–1064
Egelund J, Damager I, Faber K, Olsen CE, Ulvskov P, Petersen BL (2008) Functional characterisation of a putative rhamnogalacturonan II specific xylosyltransferase. FEBS Lett 582:3217–3222
Egelund J, Petersen BL, Motawia MS, Damager I, Faik A, Olsen CE, Ishii T, Clausen H, Ulvskov P, Geshi N (2006) Arabidopsis thaliana RGXT1 and RGXT2 encode golgi-localized (1,3)-α-d-xylosyltransferases involved in the synthesis of pectic rhamnogalacturonan-II. Plant Cell 18:2593–2607
Liu XL, Liu L, Niu QK, Xia C, Yang KZ, Li R, Chen LQ, Zhang XQ, Zhou Y, Ye D (2011) MALE GAMETOPHYTE DEFECTIVE 4 encodes a rhamnogalacturonan II xylosyltransferase and is important for growth of pollen tubes and roots in Arabidopsis. Plant J 65:647–660
Sechet J, Htwe S, Urbanowicz B, Agyeman A, Feng W, Ishikawa T, Colomes M, Kumar KS, Kawai-Yamada M, Dinneny JR, O’Neill MA, Mortimer JC (2018) Suppression of Arabidopsis GGLT1 affects growth by reducing the l-galactose content and borate cross-linking of rhamnogalacturonan-II. Plant J 96:1036–1050
O’Neill MA, Eberhard S, Albersheim P, Darvill AG (2001) Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science 294:846–849
Reiter WD, Chapple CCS, Somerville CR (1993) Altered growth and cell walls in a fucose-deficient mutant of Arabidopsis. Science 261:1032–1035
Gille S, Pauly M (2012) O-acetylation of plant cell wall polysaccharides. Front. Plant Sci. 3:12
Stranne M, Ren Y, Fimognari L, Birdseye D, Yan J, Bardor M, Mollet JC, Komatsu T, Kikuchi J, Scheller HV, Sakuragi Y (2018) TBL10 is required for O-acetylation of pectic rhamnogalacturonan-I in Arabidopsis thaliana. Plant J 96:772–785
Pogorelko G, Lionetti V, Fursova O, Sundaram RM, Qi M, Whitham SA, Bogdanove AJ, Bellincampi D, Zabotina OA (2013) Arabidopsis and Brachypodium distachyon transgenic plants expressing Aspergillus nidulans acetylesterases have decreased degree of polysaccharide acetylation and increased resistance to pathogens. Plant Physiol 162:9–23
Vogel JP, Raab TK, Somerville CR, Somerville SC (2004) Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J 40:968–978
Chiniquy D, Underwood W, Corwin J, Ryan A, Szemenyei H, Lim CC, Stonebloom SH, Birdseye DS, Vogel J, Kliebenstein D, Scheller HV, Somerville S (2019) PMR5, an acetylation protein at the intersection of pectin biosynthesis and defense against fungal pathogens. Plant J tpj.14497
Mertz RA, Olek AT, Carpita NC (2012) Alterations in cell-wall glycosyl linkage structure of Arabidopsis murus mutants. Carbohydr Polym 89(2):331–339
Sénéchal F, Wattier C, Rustérucci C, Pelloux J (2014) Homogalacturonan-modifying enzymes: structure, expression, and roles in plants. J Exp Bot 65:5125–5160
Orfila C, Degan FD, Jørgensen B, Scheller HV, Ray PM, Ulvskov P (2012) Expression of mung bean pectin acetyl esterase in potato tubers: effect on acetylation of cell wall polymers and tuber mechanical properties. Planta 236:185–196
Gou JY, Miller LM, Hou G, Yu XH, Chen XY, Liu CJ (2012) Acetylesterase-mediated deacetylation of pectin impairs cell elongation, pollen germination, and plant reproduction. Plant Cell 24:50–65
Lionetti V, Raiola A, Camardella L, Giovane A, Obel N, Pauly M, Favaron F, Cervone F, Bellincampi D (2007) Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol 143:1871–1880
Lionetti V, Raiola A, Cervone F, Bellincampi D (2014) Transgenic expression of pectin methylesterase inhibitors limits tobamovirus spread in tobacco and Arabidopsis. Mol Plant Pathol 15:265–274
Lionetti V, Fabri E, De Caroli M, Hansen AR, Willats WGT, Piro G, Bellincampi D (2017) Three pectin methylesterase inhibitors protect cell wall integrity for Arabidopsis immunity to Botrytis. Plant Physiol 173:1844–1863
Bethke G, Grundman RE, Sreekanta S, Truman W, Katagiri F, Glazebrook J (2014) Arabidopsis PECTIN METHYLESTERASEs contribute to immunity against Pseudomonas syringae. Plant Physiol 164:1093–1107
Liu N, Sun Y, Pei Y, Zhang X, Wang P, Li X, Li F, Hou Y (2018) A pectin methylesterase inhibitor enhances resistance to Verticillium wilt. Plant Physiol 176:2202–2220
Raiola A, Lionetti V, Elmaghraby I, Immerzeel P, Mellerowicz EJ, Salvi G, Cervone F, Bellincampi D (2011) Pectin methylesterase is induced in Arabidopsis upon infection and is necessary for a successful colonization by necrotrophic pathogens. Mol Plant-Microbe Interact 24:432–440
Peaucelle A, Braybrook SA, Le Guillou L, Bron E, Kuhlemeier C, Höfte H (2011) Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr Biol 21:1720–1726
Hongo S, Sato K, Yokoyama R, Nishitani K (2012) Demethylesterification of the primary wall by PECTIN METHYLESTERASE35 provides mechanical support to the Arabidopsis stem. Plant Cell 24:2624–2634
Wolf S, Mravec J, Greiner S, Mouille G, Höfte H (2012) Plant cell wall homeostasis is mediated by brassinosteroid feedback signaling. Curr Biol 22:1732–1737
Wolf S, Van Der Does D, Ladwig F, Sticht C, Kolbeck A, Schürholz AK, Augustin S, Keinath N, Rausch T, Greiner S, Schumacher K, Harter K, Zipfel C, Höfte H (2014) A Receptor-Like Protein mediates the response to pectin modification by activating brassinosteroid signaling. Proc Natl Acad Sci U S A 111:15261–15266
Holzwart E, Huerta AI, Glöckner N, Garnelo Gómez B, Wanke F, Augustin S, Askani JC, Schürholz AK, Harter K, Wolf S (2018) BRI1 controls vascular cell fate in the Arabidopsis root through RLP44 and phytosulfokine signaling. Proc Natl Acad Sci U S A 115:11838–11843
Gomez BG, Lozano-Duran R, Wolf S (2019) Phosphorylation-dependent routing of RLP44 towards brassinosteroid or phytosulfokine signalling. bioRxiv 527754
Benedetti M, Pontiggia D, Raggi S, Cheng Z, Scaloni F, Ferrari S, Ausubel FM, Cervone F, De Lorenzo G (2015) Plant immunity triggered by engineered in vivo release of oligogalacturonides, damage-associated molecular patterns. Proc Natl Acad Sci U S A 112(17):5533–5538
Benedetti M, Verrascina I, Pontiggia D, Locci F, Mattei B, De Lorenzo G, Cervone F (2018) Four Arabidopsis berberine bridge enzyme-like proteins are specific oxidases that inactivate the elicitor-active oligogalacturonides. Plant J 94:260–273
Voxeur A, Habrylo O, Guénin S, Miart F, Soulié M-C, Rihouey C, Pau-Roblot C, Domon J-M, Gutierrez L, Pelloux J, Mouille G, Fagard M, Höfte H, Vernhettes S (2019) Oligogalacturonide production upon Arabidopsis thaliana–Botrytis cinerea interaction. Proc Natl Acad Sci U S A 116:201900317
Engelsdorf T, Gigli-Bisceglia N, Veerabagu M, McKenna JF, Vaahtera L, Augstein F, Van der Does D, Zipfel C, Hamann T (2018) The plant cell wall integrity maintenance and immune signaling systems cooperate to control stress responses in Arabidopsis thaliana. Sci Signal 11:eaao3070
Hamann T, Bennett M, Mansfield J, Somerville C (2009) Identification of cell-wall stress as a hexose-dependent and osmosensitive regulator of plant responses. Plant J 57:1015–1026
Manfield IW, Orfila C, McCartney L, Harholt J, Bernal AJ, Scheller HV, Gilmartin PM, Mikkelsen JD, Knox JP, Willats WGT (2004) Novel cell wall architecture of isoxaben-habituated Arabidopsis suspension-cultured cells: global transcript profiling and cellular analysis. Plant J 40:260–275
Biswal AK, Atmodjo MA, Li M, Baxter HL, Yoo CG, Pu Y, Lee YC, Mazarei M, Black IM, Zhang JY, Ramanna H, Bray AL, King ZR, Lafayette PR, Pattathil S, Donohoe BS, Mohanty SS, Ryno D, Yee K et al (2018) Sugar release and growth of biofuel crops are improved by downregulation of pectin biosynthesis. Nat Biotechnol 36:249–257
Li M, Yoo CG, Pu Y, Biswal AK, Tolbert AK, Mohnen D, Ragauskas AJ (2019) Downregulation of pectin biosynthesis gene GAUT4 leads to reduced ferulate and lignin-carbohydrate cross-linking in switchgrass. Commun Biol 2:22
Zhang Z, Zhang B, Chen Z, Zhang D, Zhang H, Wang H, Zhang Y, Cai D, Liu J, Xiao S, Huo Y, Liu J, Zhang L, Wang M, Liu X, Xue Y, Zhao L, Zhou Y, Chen H (2018) A PECTIN METHYLESTERASE gene at the maize Ga1 locus confers male function in unilateral cross-incompatibility. Nat Commun 9:3678
Lu Y, Hokin SA, Kermicle JL, Hartwig T, Evans MMS (2019) A pistil-expressed pectin methylesterase confers cross-incompatibility between strains of Zea mays. Nat Commun 10:2304
Volpi C, Janni M, Lionetti V, Bellincampi D, Favaron F, D’Ovidio R (2011) The ectopic expression of a pectin methyl esterase inhibitor increases pectin methyl esterification and limits fungal diseases in wheat. Mol Plant-Microbe Interact 24:1012–1019
Sella L, Castiglioni C, Paccanaro MC, Janni M, Schäfer W, D’Ovidio R, Favaron F (2016) Involvement of fungal pectin methylesterase activity in the interaction between Fusarium graminearum and wheat. Mol Plant-Microbe Interact 29:258–267
Liu H, Ma Y, Chen N, Guo S, Liu H, Guo X, Chong K, Xu Y (2014) Overexpression of stress-inducible OsBURP16, the β subunit of polygalacturonase 1, decreases pectin content and cell adhesion and increases abiotic stress sensitivity in rice. Plant Cell Environ 37:1144–1158
Eticha D, Stass A, Horst WJ (2005) Cell-wall pectin and its degree of methylation in the maize root-apex: significance for genotypic differences in aluminium resistance. Plant Cell Environ 28:1410–1420
Yang XY, Zeng ZH, Yan JY, Fan W, Bian HW, Zhu MY, Yang JL, Zheng SJ (2012) Association of specific pectin methylesterases with Al-induced root elongation inhibition in rice. Physiol Plant 148:502–511
Sun C, Lu L, Yu Y, Liu L, Hu Y, Ye Y, Jin C, Lin X (2016) Decreasing methylation of pectin caused by nitric oxide leads to higher aluminium binding in cell walls and greater aluminium sensitivity of wheat roots. J Exp Bot 67:979–989
Voxeur A, Höfte H (2016) Cell wall integrity signaling in plants: “To grow or not to grow that’s the question”. Glycobiology 26:950–960
Herger A, Dünser K, Kleine-Vehn J, Ringli C (2019) Leucine-Rich repeat extensin proteins and their role in cell wall sensing. Curr Biol 29:R851–R858
Bou Daher F, Chen Y, Bozorg B, Clough J, Jönsson H, Braybrook SA (2018) Anisotropic growth is achieved through the additive mechanical effect of material anisotropy and elastic asymmetry. Elife 7:e38161
Novaković L, Guo T, Bacic A, Sampathkumar A, Johnson KL (2018) Hitting the wall—sensing and signaling pathways involved in plant cell wall remodeling in response to abiotic stress. Plants 7:89
Ge Z, Cheung AY, Qu LJ (2019) Pollen tube integrity regulation in flowering plants: insights from molecular assemblies on the pollen tube surface. New Phytol 222:687–693
Wormit A, Butt SM, Chairam I, McKenna JF, Nunes-Nesi A, Kjaer L, O’Donnelly K, Fernie AR, Woscholski R, Barter LMC, Hamann T (2012) Osmosensitive changes of carbohydrate metabolism in response to cellulose biosynthesis inhibition. Plant Physiol 159:105–117
Fruleux A, Verger S, Boudaoud A (2019) Feeling stressed or strained? A biophysical model for cell wall mechanosensing in plants. Front Plant Sci 10:757
Zhao Q, Zhou H, Li X, Xiao B (2019) The mechanosensitive Piezo1 channel: a three-bladed propeller-like structure and a lever-like mechanogating mechanism. FEBS J 286:2461–2470
Dupres V, Alsteens D, Wilk S, Hansen B, Heinisch JJ, Dufrêne YF (2009) The yeast Wsc1 cell surface sensor behaves like a nanospring in vivo. Nat Chem Biol 5:857–862
Hamant O, Inoue D, Bouchez D, Dumais J, Mjolsness E (2019) Are microtubules tension sensors? Nat Commun 10:2360
Nakagawa Y, Katagiri T, Shinozaki K, Qi Z, Tatsumi H, Furuichi T, Kishigami A, Sokabe M, Kojima I, Sato S, Kato T, Tabata S, Iida K, Terashima A, Nakano M, Ikeda M, Yamanaka T, Iida H (2007) Arabidopsis plasma membrane protein crucial for Ca2+ influx and touch sensing in roots. Proc Natl Acad Sci U S A 104:3639–3644
Yamanaka T, Nakagawa Y, Mori K, Nakano M, Imamura T, Kataoka H, Terashima A, Iida K, Kojima I, Katagiri T, Shinozaki K, Iida H (2010) MCA1 and MCA2 that mediate Ca2+ uptake have distinct and overlapping roles in Arabidopsis. Plant Physiol 152:1284–1296
Veley KM, Marshburn S, Clure CE, Haswell ES (2012) Mechanosensitive channels protect plastids from hypoosmotic stress during normal plant growth. Curr Biol 22:408–413
Haswell ES, Peyronnet R, Barbier-Brygoo H, Meyerowitz EM, Frachisse JM (2008) Two MscS homologs provide mechanosensitive channel activities in the Arabidopsis root. Curr Biol 18:730–734
Van der Does D, Boutrot F, Engelsdorf T, Rhodes J, McKenna JF, Vernhettes S, Koevoets I, Tintor N, Veerabagu M, Miedes E, Segonzac C, Roux M, Breda AS, Hardtke CS, Molina A, Rep M, Testerink C, Mouille G, Höfte H et al (2017) The Arabidopsis leucine-rich repeat receptor kinase MIK2/LRR-KISS connects cell wall integrity sensing, root growth and response to abiotic and biotic stresses. PLoS Genet 13:e1006832
Franck CM, Westermann J, Boisson-Dernier A (2018) Plant malectin-like receptor kinases: from cell wall integrity to immunity and beyond. Annu Rev Plant Biol 69:301–328
Hematy K, Hofte H (2008) Novel receptor kinases involved in growth regulation. Curr Opin Plant Biol 11:321–328
Honkanen S, Jones VAS, Morieri G, Champion C, Hetherington AJ, Kelly S, Proust H, Saint-Marcoux D, Prescott H, Dolan L (2016) The mechanism forming the cell surface of tip-growing rooting cells is conserved among land plants. Curr Biol 26:3238–3244
Merz D, Richter J, Gonneau M, Sanchez-Rodriguez C, Eder T, Sormani R, Martin M, Hématy K, Höfte H, Hauser MT (2017) T-DNA alleles of the receptor kinase THESEUS1 with opposing effects on cell wall integrity signaling. J Exp Bot 68:4583–4593
Lin W, Tang W, Anderson C, Yang Z (2018) FERONIA’s sensing of cell wall pectin activates ROP GTPase signaling in Arabidopsis. bioRxiv 269647
Dünser K, Gupta S, Herger A, Feraru MI, Ringli C, Kleine-Vehn J (2019) Extracellular matrix sensing by FERONIA and Leucine-rich repeat extensins controls vacuolar expansion during cellular elongation in Arabidopsis thaliana. EMBO J 38(7):e100353
Stegmann M, Monaghan J, Smakowska-Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, Zipfel C (2017) The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355:287–289
Gonneau M, Desprez T, Martin M, Doblas VG, Bacete L, Miart F, Sormani R, Hématy K, Renou J, Landrein B, Murphy E, Van De Cotte B, Vernhettes S, De Smet I, Höfte H (2018) Receptor kinase THESEUS1 is a rapid alkalinization factor 34 receptor in Arabidopsis. Curr Biol 28:2452–2458.e4
Zhao C, Zayed O, Yu Z, Jiang W, Zhu P, Hsu CC, Zhang L, Andy Tao W, Lozano-Durán R, Zhu JK (2018) Leucine-rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis. Proc Natl Acad Sci U S A 115:13123–13128
Mecchia MA, Santos-Fernandez G, Duss NN, Somoza SC, Boisson-Dernier A, Gagliardini V, Martínez-Bernardini A, Fabrice TN, Ringli C, Muschietti JP, Grossniklaus U (2017) RALF4/19 peptides interact with LRX proteins to control pollen tube growth in Arabidopsis. Science 358:1600–1603
Kesten C, Gámez-Arjona FM, Scholl S, Menna A, Dora S, Huerta AI, Huang H-Y, Tintor N, Kinoshita T, Rep M, Krebs M, Schumacher K, Sánchez-Rodríguez C (2019) Pathogen-induced pH changes regulate the growth-defense balance of plants. bioRxiv 1–49
Shih HW, Miller ND, Dai C, Spalding EP, Monshausen GB (2014) The receptor-like kinase FERONIA is required for mechanical signal transduction in Arabidopsis seedlings. Curr Biol 24(16):1887–1892
Engelsdorf T, Kjaer L, Gigli-Bisceglia N, Vaahtera L, Bauer S, Miedes E, Wormit A, James L, Chairam I, Molina A, Hamann T (2019) Functional characterization of genes mediating cell wall metabolism and responses to plant cell wall integrity impairment. BMC Plant Biol 19:320
Bhosale R, Maere S, De Veylder L (2019) Endoreplication as a potential driver of cell wall modifications. Curr Opin Plant Biol 51:58–65
Chamizo-Ampudia A, Sanz-Luque E, Llamas A, Galvan A, Fernandez E (2017) Nitrate reductase regulates plant nitric oxide homeostasis. Trends Plant Sci 22:163–174
Boutrot F, Zipfel C (2017) Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu Rev Phytopathol 55:257–286
Hou S, Liu Z, Shen H, Wu D (2019) Damage-associated molecular pattern-triggered immunity in plants. Front. Plant Sci. 10:646
de Azevedo Souza C, Li S, Lin AZ, Boutrot F, Grossmann G, Zipfel C, Somerville SC (2017) Cellulose-derived oligomers act as damage-associated molecular patterns and trigger defense-like responses. Plant Physiol 173:2383–2398
Huffaker A, Pearce G, Ryan CA (2006) An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc Natl Acad Sci U S A 103:10098–10103
Bartels S, Boller T (2015) Quo vadis, Pep? Plant elicitor peptides at the crossroads of immunity, stress, and development. J Exp Bot 66:5183–5193
Hander T, Fernández-Fernández ÁD, Kumpf RP, Willems P, Schatowitz H, Rombaut D, Staes A, Nolf J, Pottie R, Yao P, Gonçalves A, Pavie B, Boller T, Gevaert K, Van Breusegem F, Bartels S, Stael S (2019) Damage on plants activates Ca2+ -dependent metacaspases for release of immunomodulatory peptides. Science 363:7486
Guo H, Nolan TM, Song G, Liu S, Xie Z, Chen J, Schnable PS, Walley JW, Yin Y (2018) FERONIA receptor kinase contributes to plant immunity by suppressing jasmonic acid signaling in Arabidopsis thaliana. Curr Biol 28:3316–3324.e6
Chen J, Yu F, Liu Y, Du C, Li X, Zhu S, Wang X, Lan W, Rodriguez PL, Liu X, Li D, Chen L, Luan S (2016) FERONIA interacts with ABI2-type phosphatases to facilitate signaling cross-talk between abscisic acid and RALF peptide in Arabidopsis. Proc Natl Acad Sci 113:E5519–E5527
Yu F, Qian L, Nibau C, Duan Q, Kita D, Levasseur K, Li X, Lu C, Li H, Hou C, Li L, Buchanan BB, Chen L, Cheung AY, Li D, Luan S (2012) FERONIA receptor kinase pathway suppresses abscisic acid signaling in Arabidopsis by activating ABI2 phosphatase. Proc Natl Acad Sci 109:14693–14698
Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR (2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343:408–411
Richter J, Ploderer M, Mongelard G, Gutierrez L, Hauser MT (2017) Role of CrRLK1L cell wall sensors HERCULES1 and 2, THESEUS1, and FERONIA in growth adaptation triggered by heavy metals and trace elements. Front Plant Sci 8:1554
Podgórska A, Burian M, Gieczewska K, Ostaszewska-Bugajska M, Zebrowski J, Solecka D, Szal B (2017) Altered cell wall plasticity can restrict plant growth under ammonium nutrition. Front Plant Sci 8:1344
Jia M, Ding N, Zhang Q, Xing S, Wei L, Zhao Y, Du P, Mao W, Li J, Li B, Jia W (2017) A FERONIA-like receptor kinase regulates strawberry (Fragaria × ananassa) fruit ripening and quality formation. Front Plant Sci 8:1099
Jia M, Du P, Ding N, Zhang Q, Xing S, Wei L, Zhao Y, Mao W, Li J, Li B, Jia W (2017) Two FERONIA-like receptor kinases regulate apple fruit ripening by modulating ethylene production. Front Plant Sci 8:1406
von Schaewen A, Rips S, Jeong IS, Koiwa H (2015) Arabidopsis thaliana KORRIGAN1 protein: N-glycan modification, localization, and function in cellulose biosynthesis and osmotic stress responses. Plant Signal Behav 10:1–4
Rips S, Bentley N, Jeong IS, Welch JL, von Schaewen A, Koiwa H (2014) Multiple n-glycans cooperate in the subcellular targeting and functioning of Arabidopsis KORRIGAN1. Plant Cell 26:3792–3808
Hermans C, Porco S, Vandenbussche F, Gille S, de Pessemier J, van der Straeten D, Verbruggen N, Bush DR (2011) Dissecting the role of CHITINASE-LIKE1 in nitrate-dependent changes in root architecture. Plant Physiol 157:1313–1326
Bernal AJ, Yoo CM, Mutwil M, Jensen JK, Hou G, Blaukopf C, Sørensen I, Blancaflor EB, Scheller HV, Willats WGT (2008) Functional analysis of the cellulose synthase-like genes CSLD1, CSLD2, and CSLD4 in tip-growing Arabidopsis cells. Plant Physiol 148:1238–1253
Liepman AH, Cavalier DM (2012) The CELLULOSE SYNTHASE-LIKE A and CELLULOSE SYNTHASE-LIKE C families: recent advances and future perspectives. Front. Plant Sci. 3:109
Davis J, Brandizzi F, Liepman AH, Keegstra K (2010) Arabidopsis mannan synthase CSLA9 and glucan synthase CSLC4 have opposite orientations in the Golgi membrane. Plant J 64:1028–1037
Tamura K, Shimada T, Kondo M, Nishimura M, Hara-Nishimura I (2005) KATAMARI1/MURUS3 is a novel Golgi membrane protein that is required for endomembrane organization in Arabidopsis. Plant Cell 17:1764–1776
Chevalier L, Bernard S, Ramdani Y, Lamour R, Bardor M, Lerouge P, Follet-Gueye ML, Driouich A (2010) Subcompartment localization of the side chain xyloglucan-synthesizing enzymes within Golgi stacks of tobacco suspension-cultured cells. Plant J 64:977–989
Lee C, Teng Q, Zhong R, Ye ZH (2011) The Four Arabidopsis REDUCED WALL ACETYLATION genes are expressed in secondary wall-containing cells and required for the acetylation of Xylan. Plant Cell Physiol 52:1289–1301
Ebert B, Birdseye D, Liwanag AJM, Laursen T, Rennie EA, Guo X, Catena M, Rautengarten C, Stonebloom SH, Gluza P, Pidatala VR, Andersen MCF, Cheetamun R, Mortimer JC, Heazlewood JL, Bacic A, Clausen MH, Willats WGT, Scheller HV (2018) The three members of the Arabidopsis glycosyltransferase family 92 are functional ß-1,4-galactan synthases. Plant Cell Physiol 59:2624–2636
Li S, Bashline L, Zheng Y, Xin X, Huang S, Kong Z, Kim SH, Cosgrove DJ, Gu Y (2016) Cellulose synthase complexes act in a concerted fashion to synthesize highly aggregated cellulose in secondary cell walls of plants. Proc Natl Acad Sci U S A 113:11348–11353
McNair G (2015) COBRA-LIKE4: a GPI-anchored protein functioning as a mediator of cellulose ultrastructure in herbaceous and woody plants. University of British Columbia, Vancouver
Peña MJ, Zhong R, Zhou GK, Richardson EA, O’Neill MA, Darvill AG, York WS, Yeb ZH (2007) Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19:549–563
Lee C, Teng Q, Huang W, Zhong R, Ye ZH (2010) The Arabidopsis family GT43 glycosyltransferases form two functionally nonredundant groups essential for the elongation of glucuronoxylan backbone. Plant Physiol 153:526–541
Chiniquy D, Sharma V, Schultink A, Baidoo EE, Rautengarten C, Cheng K, Carroll A, Ulvskov P, Harholt J, Keasling JD, Pauly M, Scheller HV, Ronald PC (2012) XAX1 from glycosyltransferase family 61 mediates xylosyltransfer to rice xylan. Proc Natl Acad Sci U S A 109:17117–17122
Nguyen HP, Jeong HY, Kim H, Kim YC, Lee C (2016) Molecular and biochemical characterization of rice pectin methylesterase inhibitors (OsPMEIs). Plant Physiol Biochem 101:105–112
Williamson RE, Burn JE, Birch R, Baskin TI, Arioli T, Betzner AS, Cork A (2001) Morphology of rsw1, a cellulose-deficient mutant of Arabidopsis thaliana. Protoplasma 215:116–127
Fujita M, Himmelspach R, Ward J, Whittington A, Hasenbein N, Liu C, Truong TT, Galway ME, Mansfield SD, Hocart CH, Wasteneys GO (2013) The anisotropy1 D604N mutation in the Arabidopsis cellulose synthase1 catalytic domain reduces cell wall crystallinity and the velocity of cellulose synthase complexes. Plant Physiol 162:74–85
Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci U S A 100:1450–1455
Desprez T, Vernhettes S, Fagard M, Refrégier G, Desnos T, Aletti E, Py N, Pelletier S, Höfte H (2002) Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol 128:482–490
Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Höfte H (2000) Procuste1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of arabidopsis. Plant Cell 12:2409–2423
Stork J, Harris D, Griffiths J, Williams B, Beisson F, Li-Beisson Y, Mendu V, Haughn G, DeBolt S (2010) CELLULOSE SYNTHASE9 serves a nonredundant role in secondary cell wall synthesis in Arabidopsis epidermal testa cells. Plant Physiol 153:580–589
Robert S, Bichet A, Grandjean O, Kierzkowski D, Satiat-Jeunemaître B, Pelletier S, Hauser MT, Höfte H, Vernhettes S (2005) An Arabidopsis endo-1,4-β-d-glucanase involved in cellulose synthesis undergoes regulated intracellular cycling. Plant Cell 17:3378–3389
Bringmann M, Li E, Sampathkumar A, Kocabek T, Hauser MT, Perssona S (2012) POM-POM2/CELLULOSE SYNTHASE INTERACTING1 is essential for the functional association of cellulose synthase and microtubules in Arabidopsis. Plant Cell 24:163–177
Hashimoto-Sugimoto M, Higaki T, Yaeno T, Nagami A, Irie M, Fujimi M, Miyamoto M, Akita K, Negi J, Shirasu K, Hasezawa S, Iba K (2013) A Munc13-like protein in Arabidopsis mediates H+-ATPase translocation that is essential for stomatal responses. Nat Commun 4:2215
Gu F, Bringmann M, Combs JR, Yang J, Bergmann DC, Nielsen E (2016) Arabidopsis CSLD5 functions in cell plate formation in a cell cycle-dependent manner. Plant Cell 28:1722–1737
Xiao C, Zhang T, Zheng Y, Cosgrove DJ, Anderson CT (2016) Xyloglucan deficiency disrupts microtubule stability and cellulose biosynthesis in Arabidopsis, altering cell growth and morphogenesis. Plant Physiol 170:234–249
Reiter WD, Chapple C, Somerville CR (1997) Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. Plant J 12:335–345
Vanzin GF, Madson M, Carpita NC, Raikhel NV, Keegstra K, Reiter WD (2002) The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc Natl Acad Sci U S A 99:3340–3345
Faria-Blanc N, Mortimer JC, Dupree P (2018) A transcriptomic analysis of Xylan mutants does not support the existence of a secondary cell wall integrity system in Arabidopsis. Front Plant Sci 9:384
Verger S, Chabout S, Gineau E, Mouille G (2016) Cell adhesion in plants is under the control of putative O-fucosyltransferases. Development 143:2536–2540
Peaucelle A, Louvet R, Johansen JN, Salsac F, Morin H, Fournet F, Belcram K, Gillet F, Hofte H, Laufs P, Mouille G, Pelloux J (2011) The transcription factor BELLRINGER modulates phyllotaxis by regulating the expression of a pectin methylesterase in Arabidopsis. Development 138:4733–4741
Peaucelle A, Louvet R, Johansen JN, Höfte H, Laufs P, Pelloux J, Mouille G (2008) Arabidopsis phyllotaxis is controlled by the methyl-esterification status of cell-wall pectins. Curr Biol 18:1943–1948
Gramegna G, Modesti V, Savatin DV, Sicilia F, Cervone F, De Lorenzo G (2016) GRP-3 and KAPP, encoding interactors of WAK1, negatively affect defense responses induced by oligogalacturonides and local response to wounding. J Exp Bot 67:1715–1729
Yan C, Yan S, Zeng X, Zhang Z, Gu M (2007) Fine mapping and isolation of Bc7(t), allelic to OsCesA4. J Genet Genomics 34:1019–1027
Petti C, Hirano K, Stork J, DeBolt S (2015) Mapping of a cellulose-deficient mutant named dwarf1-1 in Sorghum bicolor to the green revolution gene gibberellin20-oxidase reveals a positive regulatory association between gibberellin and cellulose biosynthesis. Plant Physiol 169:705–716
Hématy K, Sado PE, Van Tuinen A, Rochange S, Desnos T, Balzergue S, Pelletier S, Renou JP, Höfte H (2007) A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr Biol 17:922–931
Guo H, Li L, Ye H, Yu X, Algreen A, Yin Y (2009) Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc Natl Acad Sci 106:7648–7653
Schoenaers S, Balcerowicz D, Breen G, Hill K, Zdanio M, Mouille G, Holman TJ, Oh J, Wilson MH, Nikonorova N, Vu LD, De Smet I, Swarup R, De Vos WH, Pintelon I, Adriaensen D, Grierson C, Bennett MJ, Vissenberg K (2018) The auxin-regulated CrRLK1L kinase ERULUS controls cell wall composition during root hair tip growth. Curr Biol 28:722–732.e6
Xu SL, Rahman A, Baskin TI, Kieber JJ (2008) Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell 20:3065–3079
Mori K, Renhu N, Naito M, Nakamura A, Shiba H, Yamamoto T, Suzaki T, Iida H, Miura K (2018) Ca2+ -permeable mechanosensitive channels MCA1 and MCA2 mediate cold-induced cytosolic Ca 2+ increase and cold tolerance in Arabidopsis. Sci Rep 8:550
Jensen GS, Haswell ES (2012) Functional analysis of conserved motifs in the mechanosensitive channel homolog MscS-Like2 from Arabidopsis thaliana. PLoS One 7:e40336
Wilson ME, Jensen GS, Haswell ES (2011) Two mechanosensitive channel homologs influence division ring placement in Arabidopsis chloroplasts. Plant Cell 23:2939–2949
Tocquin P, Corbesier L, Havelange A, Pieltain A, Kurtem E, Bernier G, Périlleux C (2003) A novel high efficiency, low maintenance, hydroponic system for synchronous growth and flowering of Arabidopsis thaliana. BMC Plant Biol 3:2
Acknowledgements
We would like to acknowledge financial support from: EEA Grant (7F14155 CYTOWALL to NG-B/TH) and Horizon 2020 through a Marie Skłodowska-Curie fellowship ‘SugarOsmoSignaling’ (to TE).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Gigli-Bisceglia, N., Engelsdorf, T. & Hamann, T. Plant cell wall integrity maintenance in model plants and crop species-relevant cell wall components and underlying guiding principles. Cell. Mol. Life Sci. 77, 2049–2077 (2020). https://doi.org/10.1007/s00018-019-03388-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-019-03388-8