Abstract
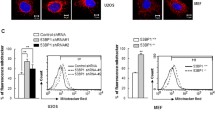
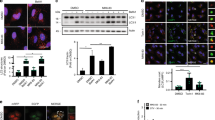
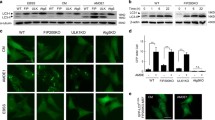
Etoposide-induced protein 2.4 (EI24), located on the endoplasmic reticulum (ER) membrane, has been proposed to be an essential autophagy protein. Specific ablation of EI24 in neuronal and liver tissues causes deficiency of autophagy flux. However, the molecular mechanism of the EI24-mediated autophagy process is still poorly understood. Like neurons and hepatic cells, pancreatic β cells are also secretory cells. Pancreatic β cells contain large amounts of ER and continuously synthesize and secrete insulin to maintain blood glucose homeostasis. Yet, the effect of EI24 on autophagy of pancreatic β cells has not been reported. Here, we show that the autophagy process is inhibited in EI24-deficient primary pancreatic β cells. Further mechanistic studies demonstrate that EI24 is enriched at the ER–mitochondria interface and that the C-terminal domain of EI24 is important for the integrity of the mitochondria-associated membrane (MAM) and autophagy flux. Overexpression of EI24, but not the EI24-ΔC mutant, can rescue MAM integrity and decrease the aggregation of p62 and LC3II in the EI24-deficient group. By mass spectrometry-based proteomics following immunoprecipitation, EI24 was found to interact with voltage-dependent anion channel 1 (VDAC1), inositol 1,4,5-trisphosphate receptor (IP3R), and the outer mitochondrial membrane chaperone GRP75. Knockout of EI24 impairs the interaction of IP3R with VDAC1, indicating that these proteins may form a quaternary complex to regulate MAM integrity and the autophagy process.







Similar content being viewed by others
Change history
21 November 2019
In the published article, few errors were noticed and this has been corrected with this erratum publication.
References
Yu L, Chen Y, Tooze SA (2018) Autophagy pathway: cellular and molecular mechanisms. Autophagy 14(2):207–215
Farre JC, Subramani S (2016) Mechanistic insights into selective autophagy pathways: lessons from yeast. Nat Rev Mol Cell Biol 17(9):537–552
Eskelinen EL, Saftig P (2009) Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim Biophys Acta 1793(4):664–673
Levine B, Kroemer G (2008) Autophagy in the pathogenesis of disease. Cell 132(1):27–42
Chen CG et al (2017) Human beta cell mass and function in diabetes: recent advances in knowledge and technologies to understand disease pathogenesis. Mol Metab 6(9):943–957
Jung HS et al (2008) Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab 8(4):318–324
Sheng QF et al (2017) Autophagy protects pancreatic beta cell mass and function in the setting of a high-fat and high-glucose diet. Sci Rep 7:16348
Diakopoulos KN et al (2015) Impaired autophagy induces chronic atrophic pancreatitis in mice via sex- and nutrition-dependent processes. Gastroenterology 148(3):626
Yuan L et al (2018) Etoposide-induced protein 2.4 functions as a regulator of the calcium ATPase and protects pancreatic beta-cell survival. J Biol Chem 293(26):10128–10140
Zhao YG et al (2012) The p53-induced gene Ei24 is an essential component of the basal autophagy pathway. J Biol Chem 287(50):42053–42063
Tian Y et al (2010) C elegans screen identifies autophagy genes specific to multicellular organisms. Cell 141(6):1042–1055
Xie ZP, Klionsky DJ (2007) Autophagosome formation: core machinery and adaptations. Nat Cell Biol 9(10):1102–1109
Hayashi-Nishino M et al (2009) A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol 11(12):1433–1437
Yla-Anttila P et al (2009) 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 5(8):1180–1185
Ravikumar B et al (2010) Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol 12(8):747–757
Bodemann BO et al (2011) RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell 144(2):253–267
Hamasaki M et al (2013) Autophagosomes form at ER–mitochondria contact sites. Nature 495(7441):389–393
Stoica R et al (2014) ER–mitochondria associations are regulated by the VAPB–PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat Commun 5:3996
Gomez-Suaga P et al (2017) The ER–mitochondria tethering complex VAPB–PTPIP51 regulates autophagy. Curr Biol 27(3):371–385
Arasaki K, Tagaya M (2017) Legionella blocks autophagy by cleaving STX17 (syntaxin 17). Autophagy 13(11):2008–2009
Lieu K et al (2014) The p53-induced factor Ei24 inhibits nuclear import through an importin beta-binding-like domain. J Cell Biol 205(3):301–312
Mauthe M et al (2018) Chloroquine inhibits autophagic flux by decreasing autophagosome–lysosome fusion. Autophagy 14(8):1435–1455
Szabadkai G et al (2006) Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J Cell Biol 175(6):901–911
Flis VV, Daum G (2013) Lipid transport between the endoplasmic reticulum and mitochondria. Cold Spring Harb Perspect Biol 5(6):a013235
Mejia EM, Hatch GM (2016) Mitochondrial phospholipids: role in mitochondrial function. J Bioenerg Biomembr 48(2):99–112
Gelmetti V et al (2017) PINK1 and BECN1 relocalize at mitochondria-associated membranes during mitophagy and promote ER–mitochondria tethering and autophagosome formation. Autophagy 13(4):654–669
Li Y et al (2011) P32 regulates mitochondrial morphology and dynamics through parkin. Neuroscience 199:346–358
Suzuki J et al (2014) Imaging intraorganellar Ca2+ at subcellular resolution using CEPIA. Nat Commun 5:4153
Luo C et al (2019) A genetically encoded ratiometric calcium sensor enables quantitative measurement of the local calcium microdomain in the endoplasmic reticulum. Biophys Rep 5(1):31–42
Levy JMM, Thorburn A (2011) Targeting autophagy during cancer therapy to improve clinical outcomes. Pharmacol Ther 131(1):130–141
Papadakis M et al (2013) Tsc1 (hamartin) confers neuroprotection against ischemia by inducing autophagy. Nat Med 19(3):351–357
Chong CR et al (2006) A clinical drug library screen identifies astemizole as an antimalarial agent. Nat Chem Biol 2(8):415–416
Sarparanta J, Garcia-Macia M, Singh R (2017) Autophagy and mitochondria in obesity and type 2 diabetes. Curr Diabetes Rev 13(4):352–369
Lim H et al (2018) A novel autophagy enhancer as a therapeutic agent against metabolic syndrome and diabetes. Nat Commun 9:1438
Yang L et al (2010) Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab 11(6):467–478
Rowland AA, Voeltz GK (2012) Endoplasmic reticulum-mitochondria contacts: function of the junction. Nat Rev Mol Cell Biol 13(10):607–625
de Brito OM, Scorrano L (2008) Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456(7222):605–610
Csordas G et al (2006) Structural and functional features and significance of the physical linkage between ER and mitochondria. J Cell Biol 174(7):915–921
Filadi R et al (2015) Mitofusin 2 ablation increases endoplasmic reticulum–mitochondria coupling. Proc Natl Acad Sci USA 112(17):E2174–E2181
Zheng P et al (2018) DNA damage triggers tubular endoplasmic reticulum extension to promote apoptosis by facilitating ER–mitochondria signaling. Cell Res 28(8):833–854
Betz C et al (2013) mTOR complex 2-Akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology. Proc Natl Acad Sci USA 110(31):12526–12534
Yuan L et al (2013) Low-level shear stress induces human mesenchymal stem cell migration through the SDF-1/CXCR42 axis via MAPK signaling pathways. Stem Cells Dev 22(17):2384–2393
Acknowledgements
This project was supported by the National Key R&D Program of China (2017YFA0505300 and 2016YFA0501500), the National Natural Science Foundation of China (31421002, 21778069 and 31401174), Project of the Chinese Academy of Sciences (XDB08030203) and Project of Chinese Academy of Sciences-Peking University Leading Cooperation Team.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest regarding the contents of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yuan, L., Liu, Q., Wang, Z. et al. EI24 tethers endoplasmic reticulum and mitochondria to regulate autophagy flux. Cell. Mol. Life Sci. 77, 1591–1606 (2020). https://doi.org/10.1007/s00018-019-03236-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-019-03236-9