Abstract
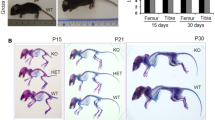
Fibrillin microfibrils are ubiquitous elements of extracellular matrix assemblies that play crucial roles in regulating the bioavailability of growth factors of the transforming growth factor beta superfamily. Recently, several “a disintegrin and metalloproteinase with thrombospondin motifs” (ADAMTS) proteins were shown to regulate fibrillin microfibril function. Among them, ADAMTS17 is the causative gene of Weill-Marchesani syndrome (WMS) and Weill-Marchesani-like syndrome, of which common symptoms are ectopia lentis and short stature. ADAMTS17 has also been linked to height variation in humans; however, the molecular mechanisms whereby ADAMTS17 regulates skeletal growth remain unknown. Here, we generated Adamts17-/- mice to examine the role of Adamts17 in skeletogenesis. Adamts17-/- mice recapitulated WMS, showing shorter long bones, brachydactyly, and thick skin. The hypertrophic zone of the growth plate in Adamts17-/- mice was shortened, with enhanced fibrillin-2 deposition, suggesting increased incorporation of fibrillin-2 into microfibrils. Comprehensive gene expression analysis of growth plates using laser microdissection and RNA sequencing indicated alteration of the bone morphogenetic protein (BMP) signaling pathway after Adamts17 knockout. Consistent with this, phospho-Smad1 levels were downregulated in the hypertrophic zone of the growth plate and in Adamts17-/- primary chondrocytes. Delayed terminal differentiation of Adamts17-/- chondrocytes, observed both in primary chondrocyte and primordial metatarsal cultures, and was prevented by BMP treatment. Our data indicated that Adamts17 is involved in skeletal formation by modulating BMP-Smad1/5/8 pathway, possibly through inhibiting the incorporation of fibrillin-2 into microfibrils. Our findings will contribute to further understanding of disease mechanisms and will facilitate the development of therapeutic interventions for WMS.






Similar content being viewed by others
Change history
16 October 2019
In the published article, the Fig. 4a was published incorrectly.
References
Kronenberg HM (2003) Developmental regulation of the growth plate. Nature 423(6937):332–336. https://doi.org/10.1038/nature01657
Melrose J, Shu C, Whitelock JM, Lord MS (2016) The cartilage extracellular matrix as a transient developmental scaffold for growth plate maturation. Matrix Biol 52–54:363–383. https://doi.org/10.1016/j.matbio.2016.01.008
Hubmacher D, Apte SS (2015) ADAMTS proteins as modulators of microfibril formation and function. Matrix Biol 47:34–43. https://doi.org/10.1016/j.matbio.2015.05.004
Le Goff C, Mahaut C, Wang LW, Allali S, Abhyankar A, Jensen S, Zylberberg L, Collod-Beroud G, Bonnet D, Alanay Y, Brady AF, Cordier MP, Devriendt K, Genevieve D, Kiper PO, Kitoh H, Krakow D, Lynch SA, Le Merrer M, Megarbane A, Mortier G, Odent S, Polak M, Rohrbach M, Sillence D, Stolte-Dijkstra I, Superti-Furga A, Rimoin DL, Topouchian V, Unger S, Zabel B, Bole-Feysot C, Nitschke P, Handford P, Casanova JL, Boileau C, Apte SS, Munnich A, Cormier-Daire V (2011) Mutations in the TGFbeta binding-protein-like domain 5 of FBN1 are responsible for acromicric and geleophysic dysplasias. Am J Hum Genet 89(1):7–14. https://doi.org/10.1016/j.ajhg.2011.05.012
Faivre L, Gorlin RJ, Wirtz MK, Godfrey M, Dagoneau N, Samples JR, Le Merrer M, Collod-Beroud G, Boileau C, Munnich A, Cormier-Daire V (2003) In frame fibrillin-1 gene deletion in autosomal dominant Weill-Marchesani syndrome. J Med Genet 40(1):34–36
Dagoneau N, Benoist-Lasselin C, Huber C, Faivre L, Megarbane A, Alswaid A, Dollfus H, Alembik Y, Munnich A, Legeai-Mallet L, Cormier-Daire V (2004) ADAMTS10 mutations in autosomal recessive Weill-Marchesani syndrome. Am J Hum Genet 75(5):801–806. https://doi.org/10.1086/425231
Mularczyk EJ, Singh M, Godwin ARF, Galli F, Humphreys N, Adamson AD, Mironov A, Cain SA, Sengle G, Boot-Handford RP, Cossu G, Kielty CM, Baldock C (2018) ADAMTS10-mediated tissue disruption in Weill-Marchesani Syndrome. Hum Mol Genet 27:3657–3687. https://doi.org/10.1093/hmg/ddy276
Morales J, Al-Sharif L, Khalil DS, Shinwari JM, Bavi P, Al-Mahrouqi RA, Al-Rajhi A, Alkuraya FS, Meyer BF, Al Tassan N (2009) Homozygous mutations in ADAMTS10 and ADAMTS17 cause lenticular myopia, ectopia lentis, glaucoma, spherophakia, and short stature. Am J Hum Genet 85(5):558–568. https://doi.org/10.1016/j.ajhg.2009.09.011
Shah MH, Bhat V, Shetty JS, Kumar A (2014) Whole exome sequencing identifies a novel splice-site mutation in ADAMTS17 in an Indian family with Weill-Marchesani syndrome. Mol Vis 20:790–796
van Duyvenvoorde HA, Lui JC, Kant SG, Oostdijk W, Gijsbers AC, Hoffer MJ, Karperien M, Walenkamp MJ, Noordam C, Voorhoeve PG, Mericq V, Pereira AM, Claahsen-van de Grinten HL, van Gool SA, Breuning MH, Losekoot M, Baron J, Ruivenkamp CA, Wit JM (2014) Copy number variants in patients with short stature. Eur J Hum Genet 22(5):602–609. https://doi.org/10.1038/ejhg.2013.203
Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Willer CJ, Jackson AU, Vedantam S, Raychaudhuri S, Ferreira T, Wood AR, Weyant RJ, Segre AV, Speliotes EK, Wheeler E, Soranzo N, Park JH, Yang J, Gudbjartsson D, Heard-Costa NL, Randall JC, Qi L, Vernon Smith A, Magi R, Pastinen T, Liang L, Heid IM, Luan J, Thorleifsson G, Winkler TW, Goddard ME, Sin Lo K, Palmer C, Workalemahu T, Aulchenko YS, Johansson A, Zillikens MC, Feitosa MF, Esko T, Johnson T, Ketkar S, Kraft P, Mangino M, Prokopenko I, Absher D, Albrecht E, Ernst F, Glazer NL, Hayward C, Hottenga JJ, Jacobs KB, Knowles JW, Kutalik Z, Monda KL, Polasek O, Preuss M, Rayner NW, Robertson NR, Steinthorsdottir V, Tyrer JP, Voight BF, Wiklund F, Xu J, Zhao JH, Nyholt DR, Pellikka N, Perola M, Perry JR, Surakka I, Tammesoo ML, Altmaier EL, Amin N, Aspelund T, Bhangale T, Boucher G, Chasman DI, Chen C, Coin L, Cooper MN, Dixon AL, Gibson Q, Grundberg E, Hao K, Juhani Junttila M, Kaplan LM, Kettunen J, Konig IR, Kwan T, Lawrence RW, Levinson DF, Lorentzon M, McKnight B, Morris AP, Muller M, Suh Ngwa J, Purcell S, Rafelt S, Salem RM, Salvi E, Sanna S, Shi J, Sovio U, Thompson JR, Turchin MC, Vandenput L, Verlaan DJ, Vitart V, White CC, Ziegler A, Almgren P, Balmforth AJ, Campbell H, Citterio L, De Grandi A, Dominiczak A, Duan J, Elliott P, Elosua R, Eriksson JG, Freimer NB, Geus EJ, Glorioso N, Haiqing S, Hartikainen AL, Havulinna AS, Hicks AA, Hui J, Igl W, Illig T, Jula A, Kajantie E, Kilpelainen TO, Koiranen M, Kolcic I, Koskinen S, Kovacs P, Laitinen J, Liu J, Lokki ML, Marusic A, Maschio A, Meitinger T, Mulas A, Pare G, Parker AN, Peden JF, Petersmann A, Pichler I, Pietilainen KH, Pouta A, Ridderstrale M, Rotter JI, Sambrook JG, Sanders AR, Schmidt CO, Sinisalo J, Smit JH, Stringham HM, Bragi Walters G, Widen E, Wild SH, Willemsen G, Zagato L, Zgaga L, Zitting P, Alavere H, Farrall M, McArdle WL, Nelis M, Peters MJ, Ripatti S, van Meurs JB, Aben KK, Ardlie KG, Beckmann JS, Beilby JP, Bergman RN, Bergmann S, Collins FS, Cusi D, den Heijer M, Eiriksdottir G, Gejman PV, Hall AS, Hamsten A, Huikuri HV, Iribarren C, Kahonen M, Kaprio J, Kathiresan S, Kiemeney L, Kocher T, Launer LJ, Lehtimaki T, Melander O, Mosley TH Jr, Musk AW, Nieminen MS, O'Donnell CJ, Ohlsson C, Oostra B, Palmer LJ, Raitakari O, Ridker PM, Rioux JD, Rissanen A, Rivolta C, Schunkert H, Shuldiner AR, Siscovick DS, Stumvoll M, Tonjes A, Tuomilehto J, van Ommen GJ, Viikari J, Heath AC, Martin NG, Montgomery GW, Province MA, Kayser M, Arnold AM, Atwood LD, Boerwinkle E, Chanock SJ, Deloukas P, Gieger C, Gronberg H, Hall P, Hattersley AT, Hengstenberg C, Hoffman W, Lathrop GM, Salomaa V, Schreiber S, Uda M, Waterworth D, Wright AF, Assimes TL, Barroso I, Hofman A, Mohlke KL, Boomsma DI, Caulfield MJ, Cupples LA, Erdmann J, Fox CS, Gudnason V, Gyllensten U, Harris TB, Hayes RB, Jarvelin MR, Mooser V, Munroe PB, Ouwehand WH, Penninx BW, Pramstaller PP, Quertermous T, Rudan I, Samani NJ, Spector TD, Volzke H, Watkins H, Wilson JF, Groop LC, Haritunians T, Hu FB, Kaplan RC, Metspalu A, North KE, Schlessinger D, Wareham NJ, Hunter DJ, O'Connell JR, Strachan DP, Wichmann HE, Borecki IB, van Duijn CM, Schadt EE, Thorsteinsdottir U, Peltonen L, Uitterlinden AG, Visscher PM, Chatterjee N, Loos RJ, Boehnke M, McCarthy MI, Ingelsson E, Lindgren CM, Abecasis GR, Stefansson K, Frayling TM, Hirschhorn JN (2010) Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467(7317):832–838. https://doi.org/10.1038/nature09410
Matsumura H, Hasuwa H, Inoue N, Ikawa M, Okabe M (2004) Lineage-specific cell disruption in living mice by Cre-mediated expression of diphtheria toxin A chain. Biochem Biophys Res Commun 321(2):275–279. https://doi.org/10.1016/j.bbrc.2004.06.139
Ovchinnikov DA, Deng JM, Ogunrinu G, Behringer RR (2000) Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis 26(2):145–146
Mack SA, Maltby KM, Hilton MJ (2014) Demineralized murine skeletal histology. Methods Mol Biol 1130:165–183. https://doi.org/10.1007/978-1-62703-989-5_12
Chehrehasa F, Meedeniya AC, Dwyer P, Abrahamsen G, Mackay-Sim A (2009) EdU, a new thymidine analogue for labelling proliferating cells in the nervous system. J Neurosci Methods 177(1):122–130. https://doi.org/10.1016/j.jneumeth.2008.10.006
Kawamoto T, Kawamoto K (2014) Preparation of thin frozen sections from nonfixed and undecalcified hard tissues using Kawamot's film method (2012). Methods Mol Biol 1130:149–164. https://doi.org/10.1007/978-1-62703-989-5_11
Mori Y, Chung UI, Tanaka S, Saito T (2014) Determination of differential gene expression profiles in superficial and deeper zones of mature rat articular cartilage using RNA sequencing of laser microdissected tissue specimens. Biomed Res 35(4):263–270
Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11(3):R25. https://doi.org/10.1186/gb-2010-11-3-r25
Mirando AJ, Dong Y, Kim J, Hilton MJ (2014) Isolation and culture of murine primary chondrocytes. Methods Mol Biol 1130:267–277. https://doi.org/10.1007/978-1-62703-989-5_20
Yahara Y, Takemori H, Okada M, Kosai A, Yamashita A, Kobayashi T, Fujita K, Itoh Y, Nakamura M, Fuchino H, Kawahara N, Fukui N, Watanabe A, Kimura T, Tsumaki N (2016) Pterosin B prevents chondrocyte hypertrophy and osteoarthritis in mice by inhibiting Sik3. Nat Commun 7:10959. https://doi.org/10.1038/ncomms10959
Martin I, Jakob M, Schafer D, Dick W, Spagnoli G, Heberer M (2001) Quantitative analysis of gene expression in human articular cartilage from normal and osteoarthritic joints. Osteoarthr Cartil 9(2):112–118. https://doi.org/10.1053/joca.2000.0366
Houston DA, Staines KA, MacRae VE, Farquharson C (2016) Culture of murine embryonic metatarsals: a physiological model of endochondral ossification. J Vis Exp 118:54978. https://doi.org/10.3791/54978
Wang LW, Kutz WE, Mead TJ, Beene LC, Singh S, Jenkins MW, Reinhardt DP, Apte SS (2018) Adamts10 inactivation in mice leads to persistence of ocular microfibrils subsequent to reduced fibrillin-2 cleavage. Matrix Biol 77:117–128. https://doi.org/10.1016/j.matbio.2018.09.004
Charbonneau NL, Jordan CD, Keene DR, Lee-Arteaga S, Dietz HC, Rifkin DB, Ramirez F, Sakai LY (2010) Microfibril structure masks fibrillin-2 in postnatal tissues. J Biol Chem 285(26):20242–20251. https://doi.org/10.1074/jbc.M109.087031
Hubmacher D, Wang LW, Mecham RP, Reinhardt DP, Apte SS (2015) Adamtsl2 deletion results in bronchial fibrillin microfibril accumulation and bronchial epithelial dysplasia—a novel mouse model providing insights into geleophysic dysplasia. Dis Model Mech 8(5):487–499. https://doi.org/10.1242/dmm.017046
Kutz WE, Wang LW, Bader HL, Majors AK, Iwata K, Traboulsi EI, Sakai LY, Keene DR, Apte SS (2011) ADAMTS10 protein interacts with fibrillin-1 and promotes its deposition in extracellular matrix of cultured fibroblasts. J Biol Chem 286(19):17156–17167. https://doi.org/10.1074/jbc.M111.231571
Gabriel LA, Wang LW, Bader H, Ho JC, Majors AK, Hollyfield JG, Traboulsi EI, Apte SS (2012) ADAMTSL4, a secreted glycoprotein widely distributed in the eye, binds fibrillin-1 microfibrils and accelerates microfibril biogenesis. Invest Ophthalmol Vis Sci 53(1):461–469. https://doi.org/10.1167/iovs.10-5955
Saito M, Kurokawa M, Oda M, Oshima M, Tsutsui K, Kosaka K, Nakao K, Ogawa M, Manabe R, Suda N, Ganjargal G, Hada Y, Noguchi T, Teranaka T, Sekiguchi K, Yoneda T, Tsuji T (2011) ADAMTSL6beta protein rescues fibrillin-1 microfibril disorder in a Marfan syndrome mouse model through the promotion of fibrillin-1 assembly. J Biol Chem 286(44):38602–38613. https://doi.org/10.1074/jbc.M111.243451
Tsutsui K, Manabe R, Yamada T, Nakano I, Oguri Y, Keene DR, Sengle G, Sakai LY, Sekiguchi K (2010) ADAMTSL-6 is a novel extracellular matrix protein that binds to fibrillin-1 and promotes fibrillin-1 fibril formation. J Biol Chem 285(7):4870–4882. https://doi.org/10.1074/jbc.M109.076919
Hubmacher D, Schneider M, Berardinelli SJ, Takeuchi H, Willard B, Reinhardt DP, Haltiwanger RS, Apte SS (2017) Unusual life cycle and impact on microfibril assembly of ADAMTS17, a secreted metalloprotease mutated in genetic eye disease. Sci Rep 7:41871. https://doi.org/10.1038/srep41871
Sengle G, Carlberg V, Tufa SF, Charbonneau NL, Smaldone S, Carlson EJ, Ramirez F, Keene DR, Sakai LY (2015) Abnormal activation of BMP signaling causes myopathy in FBN2 null mice. PLoS Genet 11(6):e1005340. https://doi.org/10.1371/journal.pgen.1005340
Sakai LY, Keene DR (2018) Fibrillin protein pleiotropy: Acromelic dysplasias. Matrix Biol. doi:10.1016/j.matbio.2018.09.005
Salazar VS, Capelo LP, Cantu C, Zimmerli D, Gosalia N, Pregizer S, Cox K, Ohte S, Feigenson M, Gamer L, Nyman JS, Carey DJ, Economides A, Basler K, Rosen V (2019) Reactivation of a developmental Bmp2 signaling center is required for therapeutic control of the murine periosteal niche. eLife 8:e42386. doi:10.7554/eLife.42386.
Le Goff C, Morice-Picard F, Dagoneau N, Wang LW, Perrot C, Crow YJ, Bauer F, Flori E, Prost-Squarcioni C, Krakow D, Ge G, Greenspan DS, Bonnet D, Le Merrer M, Munnich A, Apte SS, Cormier-Daire V (2008) ADAMTSL2 mutations in geleophysic dysplasia demonstrate a role for ADAMTS-like proteins in TGF-beta bioavailability regulation. Nat Genet 40(9):1119–1123. https://doi.org/10.1038/ng.199
Le Goff C, Mahaut C, Abhyankar A, Le Goff W, Serre V, Afenjar A, Destree A, di Rocco M, Heron D, Jacquemont S, Marlin S, Simon M, Tolmie J, Verloes A, Casanova JL, Munnich A, Cormier-Daire V (2011) Mutations at a single codon in Mad homology 2 domain of SMAD4 cause Myhre syndrome. Nat Genet 44(1):85–88. https://doi.org/10.1038/ng.1016
Sengle G, Charbonneau NL, Ono RN, Sasaki T, Alvarez J, Keene DR, Bachinger HP, Sakai LY (2008) Targeting of bone morphogenetic protein growth factor complexes to fibrillin. J Biol Chem 283(20):13874–13888. https://doi.org/10.1074/jbc.M707820200
Wohl AP, Troilo H, Collins RF, Baldock C, Sengle G (2016) Extracellular regulation of bone morphogenetic protein activity by the microfibril component fibrillin-1. J Biol Chem 291(24):12732–12746. https://doi.org/10.1074/jbc.M115.704734
Zilberberg L, Todorovic V, Dabovic B, Horiguchi M, Courousse T, Sakai LY, Rifkin DB (2012) Specificity of latent TGF-beta binding protein (LTBP) incorporation into matrix: role of fibrillins and fibronectin. J Cell Physiol 227(12):3828–3836. https://doi.org/10.1002/jcp.24094
Wu M, Chen G, Li YP (2016) TGF-beta and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res 4:16009. https://doi.org/10.1038/boneres.2016.9
Acknowledgements
We thank J. Sugita for technical assistance. This study was supported by Grants-in-Aid for Scientific Research from the Japanese Ministry of Education, Culture, Sports, Science and Technology (16K10810, 17K10924, 17H04311).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
All experiments performed in this study comply with the current laws of Japan.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Oichi, T., Taniguchi, Y., Soma, K. et al. Adamts17 is involved in skeletogenesis through modulation of BMP-Smad1/5/8 pathway. Cell. Mol. Life Sci. 76, 4795–4809 (2019). https://doi.org/10.1007/s00018-019-03188-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-019-03188-0