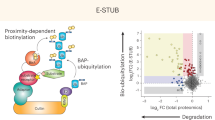
Abstract
Ubiquitin ligases play an integral role in fine-tuning signaling cascades necessary for normal cell function. Aberrant regulation of ubiquitin ligases has been implicated in several neurodegenerative diseases, generally, due to mutations within the E3 ligase itself. Several proteomic-based methods have recently emerged to facilitate the rapid identification of ligase–substrate pairs—a previously challenging feat due to the transient nature of ligase–substrate interactions. These novel methods complement standard immunoprecipitations (IPs) and include proximity-dependent biotin identification (BioID), ubiquitin ligase–substrate trapping, tandem ubiquitin-binding entities (TUBEs), and a molecular trapping unit known as the NEDDylator. The implementation of these techniques is expected to facilitate the rapid identification of novel substrates of E3 ubiquitin ligases, a process that is likely to enhance our understanding of neurodegenerative diseases and highlight novel therapeutic targets for the treatment of neurodegenerative diseases.
Similar content being viewed by others
Abbreviations
- AD:
-
Alzheimer’s disease
- ALS:
-
Amyotrophic lateral sclerosis
- AP–MS:
-
Affinity purification–mass spectrometry
- BioID:
-
Proximity-dependent biotin identification
- CHX:
-
Cycloheximide
- CRAPome:
-
Contaminant repository for affinity purification
- DUBs:
-
Deubiquitylases
- E1:
-
Ubiquitin-activating enzyme
- E2:
-
Ubiquitin-conjugating enzyme
- E3:
-
Ubiquitin ligase
- FTD:
-
Frontotemporal dementia
- His6 :
-
Hexahistidine
- HB-NEDD8:
-
Histidine–biotin-tagged NEDD8
- HD:
-
Huntington’s disease
- IP:
-
Immunoprecipitation
- IP-LC/MS:
-
Immunoprecipitation followed by mass spectrometry
- MS:
-
Mass spectrometry
- NiNTA:
-
Nickel–nitrilotriacetic acid
- PD:
-
Parkinson’s disease
- TR-TUBE:
-
Trypsin-resistant tandem ubiquitin-binding entities
- TUBE:
-
Tandem ubiquitin-binding entity
- UBA:
-
Ubiquitin-associated domain
- UPS:
-
Ubiquitin-proteasome system
References
Ross CA, Poirier MA (2004) Protein aggregation and neurodegenerative disease. Nat Med 10(Suppl):S10–S17
Iwatsubo T, Odaka A, Suzuki N, Mizusawa H et al (1994) Visualization of A beta 42(43) and A beta 40 in senile plaques with end-specific A beta monoclonals: evidence that an initially deposited species is A beta 42(43). Neuron 13:45–53
Spillantini MG, Crowther RA, Jakes R, Cairns NJ et al (1998) Filamentous alpha-synuclein inclusions link multiple system atrophy with Parkinson’s disease and dementia with Lewy bodies. Neurosci Lett 251:205–208
Spillantini MG, Crowther RA, Jakes R, Hasegawa M et al (1998) alpha-Synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with lewy bodies. Proc Natl Acad Sci USA 95:6469–6473
Arai T, Hasegawa M, Akiyama H, Ikeda K et al (2006) TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun 351:602–611
Neumann M, Sampathu DM, Kwong LK, Truax AC et al (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314:130–133
Behrouzi R, Liu X, Wu D, Robinson AC et al (2016) Pathological tau deposition in motor neurone disease and frontotemporal lobar degeneration associated with TDP-43 proteinopathy. Acta Neuropathol Commun 4:33
Davies SW, Turmaine M, Cozens BA, DiFiglia M et al (1997) Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell 90:537–548
Becher MW, Kotzuk JA, Sharp AH, Davies SW et al (1998) Intranuclear neuronal inclusions in Huntington’s disease and dentatorubral and pallidoluysian atrophy: correlation between the density of inclusions and IT15 CAG triplet repeat length. Neurobiol Dis 4:387–397
DiFiglia M, Sapp E, Chase KO, Davies SW et al (1997) Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277:1990–1993
Gutekunst CA, Li SH, Yi H, Mulroy JS et al (1999) Nuclear and neuropil aggregates in Huntington’s disease: relationship to neuropathology. J Neurosci 19:2522–2534
Hasegawa M, Fujiwara H, Nonaka T, Wakabayashi K et al (2002) Phosphorylated alpha-synuclein is ubiquitinated in alpha-synucleinopathy lesions. J Biol Chem 277:49071–49076
Lowe J, Lennox G, Jefferson D, Morrell K et al (1988) A filamentous inclusion body within anterior horn neurones in motor neurone disease defined by immunocytochemical localisation of ubiquitin. Neurosci Lett 94:203–210
Lowe J, Blanchard A, Morrell K, Lennox G et al (1988) Ubiquitin is a common factor in intermediate filament inclusion bodies of diverse type in man, including those of Parkinson’s disease, Pick’s disease, and Alzheimer’s disease, as well as Rosenthal fibres in cerebellar astrocytomas, cytoplasmic bodies in muscle, and mallory bodies in alcoholic liver disease. J Pathol 155:9–15
Lecker SH, Goldberg AL, Mitch WE (2006) Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol JASN 17:1807–1819
Saeki Y (2017) Ubiquitin recognition by the proteasome. J Biochem 161:113–124
Cuervo AM, Wong ES, Martinez-Vicente M (2010) Protein degradation, aggregation, and misfolding. Mov Disord 25(Suppl 1):S49–S54
Ciechanover A, Kwon YT (2015) Degradation of misfolded proteins in neurodegenerative diseases: therapeutic targets and strategies. Exp Mol Med 47:e147
Tsakiri EN, Trougakos IP (2015) The amazing ubiquitin-proteasome system: structural components and implication in aging. Int Rev Cell Mol Biol 314:171–237
Ye Y, Rape M (2009) Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol 10:755–764
Sluimer J, Distel B (2018) Regulating the human HECT E3 ligases. Cell Mol Life Sci CMLS 75:3121–3141
Eletr ZM, Huang DT, Duda DM, Schulman BA et al (2005) E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat Struct Mol Biol 12:933–934
Akutsu M, Dikic I, Bremm A (2016) Ubiquitin chain diversity at a glance. J Cell Sci 129:875–880
Tan JM, Wong ES, Kirkpatrick DS, Pletnikova O et al (2008) Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum Mol Genet 17:431–439
d’Azzo A, Bongiovanni A, Nastasi T (2005) E3 ubiquitin ligases as regulators of membrane protein trafficking and degradation. Traffic 6:429–441
Morris JR, Solomon E (2004) BRCA1: BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum Mol Genet 13:807–817
McDowell GS, Philpott A (2013) Non-canonical ubiquitylation: mechanisms and consequences. Int J Biochem Cell Biol 45:1833–1842
Ardley HC, Robinson PA (2004) The role of ubiquitin-protein ligases in neurodegenerative disease. Neuro-degener Dis 1:71–87
Klein C, Westenberger A (2012) Genetics of Parkinson’s disease. Cold Spring Harbor Perspect Med 2:a008888
Kitada T, Asakawa S, Hattori N, Matsumine H et al (1998) Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392:605–608
Williams KL, Topp S, Yang S, Smith B et al (2016) CCNF mutations in amyotrophic lateral sclerosis and frontotemporal dementia. Nature Commun 7:11253
Tsai PC, Liao YC, Chen PL, Guo YC et al (2018) Investigating CCNF mutations in a Taiwanese cohort with amyotrophic lateral sclerosis. Neurobiol Aging 62:243 e241–243 e246
Greer PL, Hanayama R, Bloodgood BL, Mardinly AR et al (2010) The Angelman syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell 140:704–716
Petrucelli L, Dickson D, Kehoe K, Taylor J et al (2004) CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum Mol Genet 13:703–714
Lee VM, Trojanowski JQ (1999) Neurodegenerative tauopathies: human disease and transgenic mouse models. Neuron 24:507–510
Lin JS, Lai EM (2017) Protein-protein interactions: co-immunoprecipitation. Methods Mol Biol 1615:211–219
Pierce NW, Kleiger G, Shan SO, Deshaies RJ (2009) Detection of sequential polyubiquitylation on a millisecond timescale. Nature 462:615–619
Clague MJ, Barsukov I, Coulson JM, Liu H et al (2013) Deubiquitylases from genes to organism. Physiol Rev 93:1289–1315
Abbas T, Sivaprasad U, Terai K, Amador V et al (2008) PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev 22:2496–2506
Morishima Y, Wang AM, Yu Z, Pratt WB et al (2008) CHIP deletion reveals functional redundancy of E3 ligases in promoting degradation of both signaling proteins and expanded glutamine proteins. Hum Mol Genet 17:3942–3952
Smith A, Simanski S, Fallahi M, Ayad NG (2007) Redundant ubiquitin ligase activities regulate wee1 degradation and mitotic entry. Cell Cycle 6:2795–2799
Li Y, Xie P, Lu L, Wang J et al (2017) An integrated bioinformatics platform for investigating the human E3 ubiquitin ligase–substrate interaction network. Nature Commun 8:347
Harper JW, Tan MK (2012) Understanding cullin-RING E3 biology through proteomics-based substrate identification. Mol Cell Proteom MCP 11:1541–1550
Coyaud E, Mis M, Laurent EM, Dunham WH et al (2015) BioID-based identification of Skp cullin F-box (SCF)beta-TrCP1/2 E3 ligase substrates. Mol Cell Proteom MCP 14:1781–1795
Roux KJ, Kim DI, Raida M, Burke B (2012) A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196:801–810
Mark KG, Simonetta M, Maiolica A, Seller CA et al (2014) Ubiquitin ligase trapping identifies an SCF(Saf1) pathway targeting unprocessed vacuolar/lysosomal proteins. Mol Cell 53:148–161
Mark KG, Loveless TB, Toczyski DP (2016) Isolation of ubiquitinated substrates by tandem affinity purification of E3 ligase-polyubiquitin-binding domain fusions (ligase traps). Nat Protoc 11:291–301
Loveless TB, Topacio BR, Vashisht AA, Galaang S et al (2015) DNA damage regulates translation through beta-TRCP targeting of CReP. PLoS Genet 11:e1005292
Yoshida Y, Saeki Y, Murakami A, Kawawaki J et al (2015) A comprehensive method for detecting ubiquitinated substrates using TR-TUBE. Proc Natl Acad Sci USA 112:4630–4635
Zhuang M, Guan S, Wang H, Burlingame AL et al (2013) Substrates of IAP ubiquitin ligases identified with a designed orthogonal E3 ligase, the NEDDylator. Mol Cell 49:273–282
D’Angiolella V, Donato V, Forrester FM, Jeong YT et al (2012) Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell 149:1023–1034
D’Angiolella V, Donato V, Vijayakumar S, Saraf A et al (2010) SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature 466:138–142
Peschiaroli A, Dorrello NV, Guardavaccaro D, Venere M et al (2006) SCFbetaTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol Cell 23:319–329
Fuchs SY, Spiegelman VS, Kumar KG (2004) The many faces of beta-TrCP E3 ubiquitin ligases: reflections in the magic mirror of cancer. Oncogene 23:2028–2036
Kim TY, Siesser PF, Rossman KL, Goldfarb D et al (2015) Substrate trapping proteomics reveals targets of the betaTrCP2/FBXW11 ubiquitin ligase. Mol Cell Biol 35:167–181
Mavrommati I, Faedda R, Galasso G, Li J et al (2018) beta-TrCP- and casein kinase II-mediated degradation of cyclin F controls timely mitotic progression. Cell reports 24:3404–3412
Guardavaccaro D, Frescas D, Dorrello NV, Peschiaroli A et al (2008) Control of chromosome stability by the beta-TrCP-REST-Mad2 axis. Nature 452:365–369
Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH et al (2006) S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science 314:467–471
Wu G, Xu G, Schulman BA, Jeffrey PD et al (2003) Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell 11:1445–1456
Kruiswijk F, Yuniati L, Magliozzi R, Low TY et al (2012) Coupled activation and degradation of eEF2K regulates protein synthesis in response to genotoxic stress. Sci Signal 5:ra40
Low TY, Peng M, Magliozzi R, Mohammed S et al (2014) A systems-wide screen identifies substrates of the SCFbetaTrCP ubiquitin ligase. Sci Signal 7:rs8
Roux KJ, Kim DI, Burke B (2013) BioID: a screen for protein-protein interactions. Curr Protocols Protein Sci 74:19–23
Li Y, Sousa R (2012) Expression and purification of E. coli BirA biotin ligase for in vitro biotinylation. Protein Expr Purif 82:162–167
Cronan JE Jr (1990) Biotination of proteins in vivo. A post-translational modification to label, purify, and study proteins. J Biol Chem 265:10327–10333
Kwon K, Streaker ED, Ruparelia S, Beckett D (2000) Multiple disordered loops function in corepressor-induced dimerization of the biotin repressor. J Mol Biol 304:821–833
Frescas D, Pagano M (2008) Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer 8:438–449
Rhee HW, Zou P, Udeshi ND, Martell JD et al (2013) Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339:1328–1331
Varnaite R, MacNeill SA (2016) Meet the neighbors: mapping local protein interactomes by proximity-dependent labeling with BioID. Proteomics 16:2503–2518
Hofmann K, Bucher P (1996) The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem Sci 21:172–173
Su V, Lau AF (2009) Ubiquitin-like and ubiquitin-associated domain proteins: significance in proteasomal degradation. Cell Mol Life Sci CMLS 66:2819–2833
Wilkinson CR, Seeger M, Hartmann-Petersen R, Stone M et al (2001) Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat Cell Biol 3:939–943
Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V et al (2009) Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep 10:1250–1258
Heride C, Urbe S, Clague MJ (2014) Ubiquitin code assembly and disassembly. Curr Biol CB 24:R215–R220
Hebron ML, Lonskaya I, Sharpe K, Weerasinghe PP et al (2013) Parkin ubiquitinates Tar-DNA binding protein-43 (TDP-43) and promotes its cytosolic accumulation via interaction with histone deacetylase 6 (HDAC6). J Biol Chem 288:4103–4115
Ling SC, Polymenidou M, Cleveland DW (2013) Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79:416–438
Dickey CA, Patterson C, Dickson D, Petrucelli L (2007) Brain CHIP: removing the culprits in neurodegenerative disease. Trends Mol Med 13:32–38
Dickey CA, Koren J, Zhang YJ, Xu YF et al (2008) Akt and CHIP coregulate tau degradation through coordinated interactions. Proc Natl Acad Sci USA 105:3622–3627
Sahara N, Murayama M, Mizoroki T, Urushitani M et al (2005) In vivo evidence of CHIP up-regulation attenuating tau aggregation. J Neurochem 94:1254–1263
Kubo S, Hatano T, Takanashi M, Hattori N (2013) Can parkin be a target for future treatment of Parkinson’s disease? Expert Opin Therapeutic Targets 17:1133–1144
Wauer T, Simicek M, Schubert A, Komander D (2015) Mechanism of phospho-ubiquitin-induced PARKIN activation. Nature 524:370–374
Trempe JF, Sauve V, Grenier K, Seirafi M et al (2013) Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science 340:1451–1455
Gladkova C, Maslen SL, Skehel JM, Komander D (2018) Mechanism of parkin activation by PINK1. Nature 559:410–414
Lee A, Rayner SL, DeLuca A, Gwee SSL et al (2017) Casein kinase II phosphorylation of cyclin F at serine 621 regulates the Lys48-ubiquitylation E3 ligase activity of the SCF((cyclin F)) complex. Open Biol 7:170058
Lee A, Rayner SL, Gwee SSL, De Luca A et al (2018) Pathogenic mutation in the ALS/FTD gene, CCNF, causes elevated Lys48-linked ubiquitylation and defective autophagy. Cell Mol life Sci CMLS 75:335–354
Galper J, Rayner SL, Hogan AL, Fifita JA et al (2017) Cyclin F: a component of an E3 ubiquitin ligase complex with roles in neurodegeneration and cancer. Int J Biochem Cell Biol 89:216–220
Acknowledgements
SLR holds a Macquarie University Research Training Program Scholarship. This research was supported by Grant-in-Aid funding from the Motor Neurone Disease Research Institute of Australia (GIA1628, GIA1638, and GIA1715), National Health & Medical Research Council (APP1095215 and APP1107644), and donations made towards MND research at Macquarie University.
Author information
Authors and Affiliations
Contributions
SLR, MPM, AL, and RC conceptualised the article content. SLR wrote the manuscript. MM, MPM, BS, MM, AL, and RC assisted in revising and editing the manuscript.
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Rayner, S.L., Morsch, M., Molloy, M.P. et al. Using proteomics to identify ubiquitin ligase–substrate pairs: how novel methods may unveil therapeutic targets for neurodegenerative diseases. Cell. Mol. Life Sci. 76, 2499–2510 (2019). https://doi.org/10.1007/s00018-019-03082-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-019-03082-9