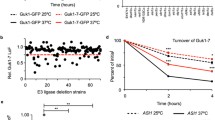
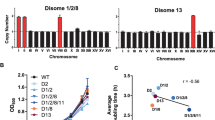
Abstract
The proteostasis network (PN) comprises a plethora of proteins that are dedicated to aid in protein folding and maintenance; some with overlapping functions. Despite this, there are multiple pathophysiological states associated with depletion of chaperones. This is counter-intuitive, assuming cells have the ability to re-program transcriptional outputs in accordance with its proteostasic limitations. Here, we have used S. cerevisiae to understand how cells respond to different types of proteostasis impairments. We monitored the proteostasis status and transcriptome of single deletions of fourteen different Protein Quality Control (PQC) genes. In most cases, cellular response did not activate proteostasis components or pathways that could either complement the function of the missing PQC gene or restore proteostasis. Over-expression of alternate machineries could restore part of the proteostasis defect in two representative PQC gene deletion strains. We posit that S. cerevisiae inherently lacks the ability to sense and respond optimally to defects in proteostasis caused due to deletion of specific PQC components.






Similar content being viewed by others
References
Daughdrill GW, Pielak GJ, Uversky VN, Cortese MS, Dunker AK (2005) Natively disordered proteins. Protein folding handbook, pp 275–357. https://doi.org/10.1002/9783527619498.ch41
Oldfield CJ, Dunker AK (2014) Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem 83:553–584
Kramer G, Boehringer D, Ban N, Bukau B (2009) The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol 16(6):589–597
Zhang G, Ignatova Z (2011) Folding at the birth of the nascent chain: coordinating translation with co-translational folding. Curr Opin Struct Biol 21(1):25–31
Hartl FU, Bracher A, Hayer-Hartl M (2011) Molecular chaperones in protein folding and proteostasis. Nature 475(7356):324–332 Epub 2011/07/22
Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU (2013) Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem 82:323–355 Epub 2013/06/12
Frydman J (2001) Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem 70:603–647 Epub 2001/06/08
Klaips CL, Jayaraj GG, Hartl FU (2018) Pathways of cellular proteostasis in aging and disease. J Cell Biol 217(1):51–63 (Epub 2017/11/12)
Wolff S, Weissman JS, Dillin A (2014) Differential scales of protein quality control. Cell 157(1):52–64
Macario AJ, de Macario EC (2005) Sick chaperones, cellular stress, and disease. N Engl J Med 353(14):1489–1501
Park S-H, Kukushkin Y, Gupta R, Chen T, Konagai A, Hipp MS et al (2013) PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell 154(1):134–145
Wang X, Venable J, LaPointe P, Hutt DM, Koulov AV, Coppinger J et al (2006) Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell 127(4):803–815
Koulov AV, LaPointe P, Lu B, Razvi A, Coppinger J, Dong M-Q et al (2010) Biological and structural basis for Aha1 regulation of Hsp90 ATPase activity in maintaining proteostasis in the human disease cystic fibrosis. Mol Biol Cell 21(6):871–884
Gong Y, Kakihara Y, Krogan N, Greenblatt J, Emili A, Zhang Z et al (2009) An atlas of chaperone-protein interactions in Saccharomyces cerevisiae: implications to protein folding pathways in the cell. Mol Syst Biol 5:275 (Epub 2009/06/19)
David DC, Ollikainen N, Trinidad JC, Cary MP, Burlingame AL, Kenyon C (2010) Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol 8(8):e1000450 (Epub 2010/08/17)
Labbadia J, Cunliffe H, Weiss A, Katsyuba E, Sathasivam K, Seredenina T et al (2011) Altered chromatin architecture underlies progressive impairment of the heat shock response in mouse models of Huntington disease. J Clin Investig 121(8):3306–3319
Katsuno M, Sang C, Adachi H, Minamiyama M, Waza M, Tanaka F et al (2005) Pharmacological induction of heat-shock proteins alleviates polyglutamine-mediated motor neuron disease. Proc Natl Acad Sci 102(46):16801–16806
Zhou H, Li S-H, Li X-J (2001) Chaperone suppression of cellular toxicity of huntingtin is independent of polyglutamine aggregation. J Biol Chem 276:48417–48424. https://doi.org/10.1074/jbc.M104140200
Stenoien DL, Cummings CJ, Adams HP, Mancini MG, Patel K, DeMartino GN et al (1999) Polyglutamine-expanded androgen receptors form aggregates that sequester heat shock proteins, proteasome components and SRC-1, and are suppressed by the HDJ-2 chaperone. Hum Mol Genet 8(5):731–741
Kabani M, Martineau CN (2008) Multiple hsp70 isoforms in the eukaryotic cytosol: mere redundancy or functional specificity? Curr Genom 9(5):338–348 (Epub 2009/05/28)
Werner-Washburne M, Stone DE, Craig EA (1987) Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol Cell Biol 7(7):2568–2577
Meyer AE, Hung NJ, Yang P, Johnson AW, Craig EA (2007) The specialized cytosolic J-protein, Jjj1, functions in 60S ribosomal subunit biogenesis. Proc Natl Acad Sci USA 104(5):1558–1563 (Epub 2007/01/24)
Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N et al (2003) Global analysis of protein expression in yeast. Nature 425(6959):737–741 (Epub 2003/10/17)
Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li G-W et al (2012) A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151(5):1042–1054
Sharma D, Masison DC (2008) Functionally redundant isoforms of a yeast Hsp70 chaperone subfamily have different antiprion effects. Genetics 179(3):1301–1311
Blatch GL, Edkins AL (2014) The networking of chaperones by co-chaperones: control of cellular protein homeostasis. Springer, Berlin
Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, Pons C, Tan G et al (2016) A global genetic interaction network maps a wiring diagram of cellular function. Science 353(6306):1420 (Epub 2016/10/07)
Kemmeren P, Sameith K, van de Pasch LA, Benschop JJ, Lenstra TL, Margaritis T et al (2014) Large-scale genetic perturbations reveal regulatory networks and an abundance of gene-specific repressors. Cell 157(3):740–752 (Epub 2014/04/29)
Usaj M, Tan Y, Wang W, VanderSluis B, Zou A, Myers CL et al (2017) TheCellMap.org: a web-accessible database for visualizing and mining the global yeast genetic interaction network. G3 (Bethesda) 7(5):1539–1549 (Epub 2017/03/23)
Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G et al (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11(12):4241–4257
Davidson JF, Whyte B, Bissinger PH, Schiestl RH (1996) Oxidative stress is involved in heat-induced cell death in Saccharomyces cerevisiae. Proc Natl Acad Sci 93(10):5116–5121
Taylor RC, Dillin A (2011) Aging as an event of proteostasis collapse. Cold Spring Harbor Perspect Biol. https://doi.org/10.1101/cshperspect.a004440
Kaushik S, Cuervo AM (2015) Proteostasis and aging. Nat Med 21(12):1406–1415 (Epub 2015/12/10)
Magrané J, Smith RC, Walsh K, Querfurth HW (2004) Heat shock protein 70 participates in the neuroprotective response to intracellularly expressed β-amyloid in neurons. J Neurosci 24(7):1700–1706
Sõti C, Nagy E, Giricz Z, Vígh L, Csermely P, Ferdinandy P (2005) Heat shock proteins as emerging therapeutic targets. Br J Pharmacol 146(6):769–780
Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ et al (2005) Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev 19(23):2816–2826
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120 (Epub 2014/04/04)
Bray NL, Pimentel H, Melsted P, Pachter L (2016) Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34(5):525–527 (Epub 2016/04/05)
Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BM et al (2013) EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29(8):1035–1043 (Epub 2013/02/23)
Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27(3):431–432 (Epub 2010/12/15)
Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P et al (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 39(Database issue):561–568 (Epub 2010/11/04)
Oughtred R, Chatr-aryamontri A, Breitkreutz BJ, Chang CS, Rust JM, Theesfeld CL et al (2016) BioGRID: a resource for studying biological interactions in yeast. Cold Spring Harbor Protocols 2016(1):pdb top080754 (Epub 2016/01/06)
Team RC (2014) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, p 2014
Wei T, Simko V (2013) corrplot: Visualization of a correlation matrix. R Package Version 073 230(231):11
Acknowledgements
We are grateful to Dr. Mohammed Faruq for aiding us with the Illumina sequencing platform. We thank Dr. Deepak Sharma (affiliated to CSIR-IMTECH) for assistance with reagents. This work was primarily funded by OLP1104 grant by CSIR to KC and partially by the grant YSS/2015/000532 from SERB to KM along with SNU core funding. We thank the HPC facility of CSIR-IGIB, for aiding us with computing resources. AG1 (Asmita), AG2 (Abhilash) and LM thank UGC for their fellowship. MV is grateful to CSIR, SD to SNU-core funding, and DPD to DBT for their fellowships.
Author information
Authors and Affiliations
Contributions
AG1 and KC designed the work. KC, KM and DD supervised the work and analysis. Sequencing was done by AG1. LM made the TS mutants of Nat-R. DPD made Mutants of yeGFP. AG1, SD, MV did the yeast experiments. The transcriptomics experiments were done by AG1. Analysis was primarily done by AG2 along with AG1 and KC. AG1 and KC wrote the manuscript with input from all authors. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
18_2019_3013_MOESM1_ESM.pdf
Supplementary Figure S1 (A) Representative blots for cycloheximide chase of TS 22 and WT Nat are shown. Ponceau was used for loading normalisation. (B) Growth curve of Wt Nat and TS22 at 30 °C done on Bioscreen. Average of 3 biological replicates is plotted. (C) Steady state amounts of Wt Nat and TS22 upon 20 µM celastrol treatment at 30 °C were determined by western blot analysis (n = 3). Representative blot shown. (D) Representative blots for steady state amounts of Wt Nat at 30 °C in different chaperone deletion strains were determined by western blot analysis. Ponceau was used for loading normalisation. (E) Quantification from 3 biological replicates are plotted for WT Nat in different PQC gene deletion strains. (F) Amounts of TS22 normalized to the amount of WT Nat in the respective chaperone deletion strains are quantitated (n = 3). (G) Growth (at 30 °C) corresponding to the activity of TS22 in PQC deletion strains when grown in different concentrations of cloNat in shown. The error bars are the standard deviation from three biological replicates. The left panel is for rqc2Δ, the middle panel is for sse1Δ and the right panel is for hsp82Δ. Wt Nat in each corresponding strain acts as the control (PDF 3686 kb)
18_2019_3013_MOESM2_ESM.pdf
Supplementary Figure S2 A) HSE-GFP reporter levels in these PQC deletions at 25 °C and 37 °C as obtained fromBrandman et al. [24]. B) Expression levels of HSF1-inducible ssa4 and PKA-inducible hsp12 mRNA are shown for the different PQC deletions. C) The panel shows the expression of canonical chronic HSR (stressed for 6 h at 37 °C) genes in different PQC gene deletions. The 37 °C HSR column is the positive control which shows these genes are highly upregulated upon chronic HSR. The fold change in the expression of these genes in PQC deletion strains w.r.t BY4741 (parental strain) was compared to the global alteration in transcripts using Mann–Whitney test. The p values obtained were corrected for multiple comparisons using FDR and the genes in any of these strains that did not cross the cut-off significance for p value 0.05 are without any colour (blank boxes). The colour scheme is graded from red (down-regulated) to blue (upregulated) showing median fold change values. The size of the circle represents the median fold change shift in either direction (PDF 150 kb)
18_2019_3013_MOESM3_ESM.pdf
Supplementary Figure S3 A) For each pair of the PQC deletions transcriptome alteration was quantified w.r.t WT strain. Pairwise comparison was then done between the different PQC deletions using transcriptome alterations. The upper triangular matrix shows the Pearson correlation between the PQC deletions. Lower triangular part of the graph is scatter plots of the transcriptomes of the deletion pairs. The trend lines are LOESS fits and are guides for visualization. B) Transcriptional alteration of each gene (fold change in a PQC gene deletion with respect to WT strain from this study) is plotted against their Boone epistasis score (ε) as reported (27) (PDF 4477 kb)
18_2019_3013_MOESM4_ESM.pdf
Supplementary Figure S4 A) BY4741 and sse1Δ bearing either empty vector or galactose-inducible over expression of Sse1 or Sse2 were spotted on YPD, YP + Raffinose or YP + Galactose and grown at 30 °C. B) BY4741 and jjj1Δ bearing either empty vector or galactose-inducible over expression of Jjj1 or Asc1 or Vam7 or Vid22 were spotted on YPD, YP + Raffinose or YP + Galactose and grown at 30 °C. C) Representative blot showing steady state amounts of TS22 at 30 °C in By4741 and jjj1Δ with or without 2.5 mM paraquat treatment. D) Upper graph corresponds to activity, measured by growth in different concentrations of Clonat, of Wt Nat in jjj1Δ in the presence and absence of paraquat. The lower graph corresponds to activity of Wt Nat in BY4741 in the presence and absence of paraquat. Error bars are from standard deviation of three biological replicates (PDF 6298 kb)
Rights and permissions
About this article
Cite this article
Ghosh, A., Gangadharan, A., Verma, M. et al. Cellular responses to proteostasis perturbations reveal non-optimal feedback in chaperone networks. Cell. Mol. Life Sci. 76, 1605–1621 (2019). https://doi.org/10.1007/s00018-019-03013-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-019-03013-8