Abstract
This review describes the properties of some rare eukaryotic chaperones that each assist in the folding of only one target protein. In particular, we describe (1) the tubulin cofactors, (2) p47, which assists in the folding of collagen, (3) α-hemoglobin stabilizing protein (AHSP), (4) the adenovirus L4-100 K protein, which is a chaperone of the major structural viral protein, hexon, and (5) HYPK, the huntingtin-interacting protein. These various-sized proteins (102–1,190 amino acids long) are all involved in the folding of oligomeric polypeptides but are otherwise functionally unique, as they each assist only one particular client. This raises a question regarding the biosynthetic cost of the high-level production of such chaperones. As the clients of faithful chaperones are all abundant proteins that are essential cellular or viral components, it is conceivable that this necessary metabolic expenditure withstood evolutionary pressure to minimize biosynthetic costs. Nevertheless, the complexity of the folding pathways in which these chaperones are involved results in error-prone processes. Several human disorders associated with these chaperones are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The maintenance of the three-dimensional correct protein structure within the densely packed environment of the cell is a prerequisite for cell survival. Tightly regulated systems have therefore evolved to minimize protein misfolding. Two major mechanisms protecting cells from this threat are molecular chaperones and the ubiquitine–proteasome system. The first facilitates correct folding while the second removes irreversibly damaged or toxic proteins.
Molecular chaperones are a large family of cellular proteins found in prokaryotes and eukaryotes. The term ‘molecular chaperone’ was proposed in 1978 by Laskey and coworkers [1], who showed that misassembly during the formation of nucleosomes in amphibian eggs could be prevented by the inclusion of an abundant acidic nuclear protein called nucleoplasmin. It appeared that nucleoplasmin does not provide steric information required for histones to bind correctly to DNA and that it is not a component of assembled nucleosomes.
Besides being implicated in assisting correct folding of nascent polypeptide chains and preventing protein aggregation after cellular stress, chaperones accomplish important functions in cellular transport and in assembly and disassembly of multiprotein complexes. Also, some viruses encode chaperones while others depend on host chaperones that are often upregulated in response to viral infection. The diverse functions of chaperones are performed through interaction with multiple target proteins whose activity is modulated by changing their conformation and/or their localization. To accomplish these functions, chaperones collaborate with co-chaperone partners, which regulate the enzymatic activity of chaperones and guide them to appropriate partner proteins.
Abundant and ubiquitous chaperones, which are usually called heat-shock proteins, are able to interact with multiple target proteins with diverse functions. Regulation of their activity is quite complex. Another, more specific, category of chaperones are the dedicated chaperones that assist several proteins of a given class, such as proteasome-, histone- and major histocompatibility complex (MHC)-dedicated chaperones or fatty acid-binding proteins (FABPs); the latter are lipid chaperones directing the partitioning of lipids inside cells. Finally, some rare chaperones assist only one target protein. These eukaryotic chaperones, here called faithful chaperones, are the subject of this article. The functional uniqueness of these chaperones raises a question of biosynthetic cost of their production. As the clients of faithful chaperones are all abundant proteins that are essential cellular or viral components, it is conceivable that this necessary metabolic expenditure withstood evolutionary pressure to minimize biosynthetic costs.
Tubulin-specific folding cofactors
Microtubules found in all eukaryotic cells are essential for many cellular functions, such as motility, morphogenesis, intracellular transport, and cell division. They are a major component of the cytoskeleton displaying highly dynamic behavior, controlled primarily by hydrolysis of guanosine triphosphate (GTP). Hence, factors that regulate the microtubule cytoskeleton are critical for determination of cell behavior and its fate. Microtubule synthesis is a dynamic multi-step process that starts with a pool of α- and β-tubulin subunits. The α- and β-tubulins fold in a series of chaperone-assisted steps, where the proper folding and assembly of tubulin α,β-heterodimers involves a stepwise progression mediated by a group of protein cofactors, A through E and ARL2. These cofactors act on tubulin folding intermediates downstream of the cytosolic chaperonin CCT (also known as c-cpn or TriC), through a unique combination of protein interaction domains. These folding steps result in production of a GTP-bound tubulin heterodimer competent for microtubule incorporation. An α,β-tubulin dimer is a barrel-shaped basic microtubule building block assembled in a head-to-tail arrangement, and the asymmetry of the tubulin heterodimer is reflected in the structure and polarity of the microtubule [2].
The preliminary step in this pathway is the binding of newly synthesized tubulin molecules to chaperonin CCT. Following one or more rounds of ATP hydrolysis by CCT, the resulting quasi-native tubulin intermediates interact with five tubulin-specific chaperones named tubulin cofactors A through E (TBCA–TBCE). The native assembly-competent tubulin is released from a supercomplex that contains both α- and β-tubulin and cofactors A–E, and that hydrolyzes GTP as part of this reaction ([3] and references therein); both α- and β-tubulins are GTP-binding proteins [4]. Tubulin subunit exchange can occur only by passage through this supercomplex, thus defining it as a dimer-making machine (Fig. 1). Hydrolysis of GTP by β-tubulin in the supercomplex acts as a switch for the release of native tubulin heterodimer. These data support a model in which the production of tubulin heterodimer via a cofactor complex is coupled to nucleotide hydrolysis by a small GTPase. Assembled tubulin dimers subsequently polymerize into microtubules in a polar fashion, with protofilaments nucleating at the microtuble-organizing center (MTOC) and their extension occurring at the rapidly growing (plus) end. Tubulin cofactors, originally discovered as proteins required for proper tubulin folding and heterodimer formation [5–11], have also been shown to participate in tubulin dissociation [3, 12, 13], transitory tubulin storage [8, 11, 14, 15], and tubulin degradation [16–18].
Schematic illustration of the tubulin heterodimer assembly pathway [5]. CCT denotes cytosolic chaperonin, a ring-like protein complex involved in folding of tubulin subunits. Tubulin-folding cofactors B and E (TBCB and TBCE) are involved in the folding and dimerization pathway of α-tubulin monomers (light blue circle), while cofactors A and D (TBCA and TBCD) fulfil this function for β-tubulin monomers (dark blue circle). Note that TBCE (in addition to TBCD) can also dissociate tubulin dimers into α- and β-tubulin monomers (shown by backward directed arrows), leading to a reduction of the tubulin dimers pool available for microtubules polymerization. Modified from [27]
The first tubulin chaperone identified in the tubulin folding pathway was cofactor A (TBCA, 108 aa) [7, 19]. TBCA binds free native β-tubulin monomers, probably serving as a reservoir for excess β-tubulin [5]. Rbl2p, the TBCA ortholog in Saccharomyces cerevisiae, protects cells from lethal β-tubulin expression by transiently binding free β-tubulin until it forms non-toxic aggregates [15]. The 2.2 Å crystal structure of Rbl2p shows α-helical monomers forming a flat, slightly convex dimer [20]. In contrast, at 1.7 Å resolution, the three-dimensional structure of human TBCA showed not a dimer but a rod-like monomer, consisting of a three-α-helix bundle ending on both ends with a coiled coil, with the second helix kinked by a proline break, offering a convex surface at one face of the protein [21]. The authors observed that the protein–protein interactions of the TBCA are weak, which is consistent with the fact that dimers only form under certain crystallogenesis conditions, at very high protein concentrations. Peptide mapping analysis and competition experiments with peptides showed that TBCA interacts with β-tubulin via the three α-helical regions, but not with the rod-end loops. The crystal structure of Arabidopsis TBCA combined with β-tubulin binding analysis confirmed the monomeric three-helix bundle structure and β-tubulin interaction with the α-helical regions [22].The main interaction occurs with the middle kinked α2 helix, at the convex face of the rod. Strong 3D structural homology is found with the BAG domain of Hsp70 chaperone, confirming that these proteins belong to a family of cofactors with a simple compact architecture. In contrast to Rbl2p that is non-essential in yeast, TBCA is essential for cell viability in plants and humans, and its knockdown produces a decrease in the amount of soluble tubulin, modifications in microtubules, G1 cell cycle arrest and cell death [8, 23].
A second cofactor, TBCB (244 aa), binds partially folded α-tubulin monomers, and a putative tubulin-binding motif within its N-terminal domain has been identified by sequence and structure comparison [24]. TBCB can be nitrated, mainly on Tyr-64 and Tyr-98, and the nitration attenuates the synthesis of new microtubules [25]. It has been demonstrated that TBCB binding to TBCE results in formation of a highly efficient tubulin heterodimer dissociating machine, leading to a complete microtubule loss [18]. After tubulin heterodimer dissociation, TBCB together with TBCE and α-tubulin form a ternary complex, while the free β-tubulin subunit is recovered by TBCA. These complexes may serve to escort α-tubulin towards degradation or recycling. It has been demonstrated that growth factor-induced stimulation of p21-activated kinase 1 (Pak1) participates in the MT dynamics through the phosphorylation of TBCB on Ser-65 and Ser-128 during microtubule regrowth phase [26].
α-tubulin is known to be able to attach to the so called cytoskeleton-associated protein glycine-rich (CAP-Gly) domains that are localized at the C-terminal position in TBCB and in the N-terminal position in TBCE [27]. The structure of the CAP-Gly domain from the Caenorhabditis elegans cytoskeleton-interacting F53F4.3 protein of unknown function has been solved, revealing a novel protein fold containing one α-helix and three β-sheets [28]. The wide distribution of the CAP-Gly motif in cytoskeleton-associated proteins (18 structures of Cap-Gly domains from different proteins have been deposited by S. Yokoyama and coworkers in Protein Data Bank [www.rcsb.org/pdb]), suggests that this motif could be a common domain for attachment to microtubules.
By a combination of modeling and structure predictions, the presence of the ubiquitin-like (Ubl) domain was predicted at the N-terminus of TBCB [29]. This prediction was confirmed by NMR structural analysis of the N-terminal domain of C. elegans TBCB [24]. While its fold closely resembles that of ubiquitin, it is unlikely that it would function as a ubiquitin-like covalent modifier of other target proteins. Instead, the authors hypothesize that the TBCB Ubl domain forms a scaffold for the presentation of residues involved in protein–protein interactions, namely between α-tubulin and TBCB or between TBCB and TBCE.
TBCB binds partially folded α-tubulin monomers, but it is not required for tubulin biogenesis, and is apparently not essential in any of the studied organisms [30]. Nevertheless, it has been shown that TBCB, which in growing neurites localizes at the transition zone of the growth cones, plays a role in microtubule dynamics and in plasticity during neurogenesis; TBCB knockdown enhances axonal growth, while TBCB overexpression leads to microtubule depolymerization, growth cone retraction, and axonal damage followed by neuronal degeneration [31]. The effect of TBCB on microtubule depolymerization was also noted in plants, where overexpression of TBCB analog reduced the number of microtubules [32]. This protein seems to be involved in plant cell division as its knockout caused Arabidopsis embryonic lethality [33].
TBCC (246 aa) together with cofactors D and E stimulates the GTPase activity of native tubulin, a reaction regulated by ADP-ribosylation factor-like 2 protein (ARL2), a 21 kDa GTPase. ARL2 orthologs in yeast are required for MT dynamics and the establishment of growth polarity [34]. Moreover, loss of function of the ARL2 ortholog in C. elegans has been shown to cause defects in the MT cytoskeleton as well as several developmental defects [35]. Finally, in breast cancer cell line, modulations of ARL2 level resulted in changes in the pool of polymerizable soluble tubulin heterodimers, in microtubule dynamic instability and modifications of cell cycle progression [36]. These data confirm earlier findings that an excess of ARL2 caused the loss of microtubules and cell cycle arrest in the M phase [37]. However, since the ARL2 knockdown does not exhibit a distinctive phenotype, it can be concluded that ARL2, one of the regulators of tubulin folding and microtubule integrity, is not essential for life.
TBCD (1,190 aa) interacts with β-tubulin and contributes to the production of polymerizable tubulin heterodimers, but it can also induce the dissociation of soluble α,β-tubulin heterodimers [5, 11]. Overexpression of TBCD in HeLa cells results in complete destruction of the microtubule network [13]. This heterodimer-dissociating activity is lost upon TBCD binding to ARL2, resulting in enhanced α,β heterodimer concentration [12]. Genetic experiments with Schizosaccharomyces pombe and Arabidopsis thaliana have shown that TBCD is essential for life in higher eukaryotes [38, 39]. TBCE (527 aa) is essential for the folding of primary α-tubulin and is required for its heterodimerization with β-tubulin; a stoichiometric excess of purified TBCE dissociates tubulin heterodimers [18]. The TBCE protein, besides the N-terminal cytoskeleton-associated glycine-rich (CAP-Gly) α-tubulin binding domain [40], contains a series of leucine-rich repeats, which can mediate protein–protein interactions [41]. Drosophila devoid of the TBCE gene are embryonic lethal [42], demonstrating that TBCE is an essential gene.
A novel protein has been described, called E-like (El) because of its sequence similarity to the TBCE [16], but which lacks the N-terminal CAP-Gly domain of TBCE implicated in tubulin binding [27]. Despite this, upon overexpression, E-like depolymerizes microtubules in vitro by direct interaction with α,β heterodimers and remains bound to α-tubulin; it commits α-tubulin to proteosomal degradation, thereby regulating microtubule stability. Although El apparently has no direct function in the tubulin folding pathway, suppression of its expression results in an increase in the number of stable microtubules and a tight clustering of endocellular membranes around the MTOC; on the other hand, the properties of dynamic microtubules remain unaffected [27].
The atomic structure of TBCA and partial structure of TBCE are known (see above). Three-dimensional models of other TBCs have been obtained, based on the sequences of yeast homologs of tubulin cofactors and combining modeling and fold prediction tools [29]. These predictions, which suggest the presence of N-terminal spectrin-like and C-terminal RP2/CAP-like domains for TBCC and several Armadillo repeats for TBCD, should be confirmed by atomic structures of TBCs from higher eukaryotes. Nevertheless, all these approaches suggest the presence in TBCs of different types of domains involved in protein–protein binding and allow the expectation of such functions as, for example, the scaffold role of TBCC in super-complex formation, which stems from structure predictions.
In view of their size and high content of protein-binding domains, it is not surprising that some TBCs exert functions that are independent of their role in tubulin heterodimers folding. For example, TBCD is also a centrosomal protein and plays a role in the organization of the mitotic spindle [43]. It has been demonstrated that overexpression of either the full-length protein or one of its two centrosome localization domains leads to the loss of anchoring of the gamma-tubulin ring complex and of nucleation of microtubule growth at centrosomes. In contrast, depletion of cofactor D by short interfering RNA results in mitotic spindle defects [44]. Because none of these changes in cofactor D activity produces a change in the levels of α- or β-tubulins, it seems that these TBCD functions are distinct from its previously described role in tubulin folding. Furthermore, in polarized epithelial MDCK cells, ARL2 and TBCD are involved in disassembly of the apical junctional complex, followed by cell dissociation from the epithelial monolayer, which is not dependent on depolymerization of microtubules [45]. Additionally, in Trypanosoma brucei, ARL2 knockdown by RNA interference results in a severe defect in cytokinesis [46]. Yeast TBCE homolog, Pac2, that displays 30% identity and 54% similarity to human TBCE with conservation of all functional domains, was shown by Voloshin and coworkers [47] to interact with both microtubules and the proteasome, which permitted the authors to hypothesize that Pac2 may have a role in subcellular localization of proteasomes.
The complexity of the folding pathway suggests that tubulin biogenesis might be error-prone, and, indeed, several human disorders are associated with tubulin chaperones. The human giant axonal neuropathy (GAN), an autosomal recessive neurodegenerative disorder, can be linked to faulty degradation of TBCB, resulting in its excess [48]. As described above, TBCB forms a binary complex with TBCE that greatly enhances its efficiency in dissociating tubulin both in vivo and in vitro [49]. An excess of TBCB thus leads to microtubule depolymerization resulting in neuronal degeneration [31]. A missense mutation in the TBCE gene was identified in mice as the cause of progressive motor neuronopathy, associated with irregularly structured β-tubulin, axonal swelling, severe skeletal muscle weakness and early respiratory failure [50]. In humans, mutations/deletions of TBCE cause two important disorders, hypoparathyroidism-retardation-dysmorphism (HRD) and autosomal recessive Kenny-Caffey/Sanjad-Sakati syndrome [51, 52]. Interestingly, it has been observed that cryptic out-of-frame translational initiation of TBCE rescues tubulin formation in heterozygous HRD, which explains the survival of afflicted individuals, who would otherwise lack the capacity to make functional TBCE [53]. A hereditary eye disease, X-linked retinitis pigmentosa, is linked to mutations in the N-terminal domain of the RP2 gene that shares amino acid sequence similarity with the TBCC [54, 55]. Indeed, RP2 is a functional homolog of TBCC since it can replace the β-tubulin GTPase stimulating activity of cofactor C in an in vitro assay [56]. Finally, lissencephaly (agyria-pachygyria), a severe developmental disease characterized by abnormal folds on the surface of the brain and disorganized cortical layering, can be linked to mutations in five different genes, including the α-tubulin gene; this last mutation results in failure of CCT-generated folding intermediates to stably interact with TBCB [57].
The heat shock protein 47 (Hsp47, also known as colligin or J6 protein), collagen chaperone
Collagen is a polymer made up of three polypeptide pro-α chains, coiled together to form a right-handed triple helix, stabilized by a large number of Gly-Pro-X (where X represents any amino acid) repeats. Collagen fibers are the major insoluble, fibrous component of the extracellular matrix, endowed with remarkable tensile strength. Collagen formation is a vital part of wound healing or, under conditions of excessive collagen formation, of pathological fibroses (for review, see [58]).
Collagen assembly begins with the folding and association of C-terminal propeptides into trimeric assemblies within the endoplasmic reticulum (at neutral pH). This initial docking is aided by the formation of disulfides and is modulated by the presence of protein-disulfide isomerase. As a result, the proline-rich regions are close together for efficient and accurate formation of procollagen triple helix, which is the structure able to attach Hsp47. Trimeric procollagen units with bound Hsp47 are then exported to the extracellular space via the Golgi apparatus. Once in the Golgi, at reduced pH the collagen trimers assemble to form procollagen fibrils and dissociate Hsp47, which will recycle back to the endoplasmic reticulum [59, 60]. This chain of events is corroborated by decreased Hsp47 affinity for collagen at lower pH [61]. At the cell surface, collagen N-and C-propeptides are removed by the appropriate peptidases, yielding the triple-helix form of mature collagen fibrils, resistant to digestion with pepsin, trypsin or chymotrypsin.
Throughout the synthesis and processing of collagen, the presence of proteins such as peptidyl prolyl cis–trans-isomerase, prolyl 4-hydroxylase and Hsp47 appears to play a vital role assisting and/or controlling trimer assembly [62, 63]. Prolyl 4-hydroxylase functions as a chaperone interacting with single-stranded procollagens, most probably retaining premature or misfolded procollagen molecules in the endoplasmic reticulum. Peptidyl prolyl cis–trans-isomerases also recognize single-stranded polypeptides of procollagens and accelerate propagation of the triple helix. Hsp47 intervenes later in the collagen folding; it facilitates propagation of collagen triple-helices by inhibiting thermal denaturation of partly folded triple-helical intermediates ([64] and references therein).
Hsp47 was first isolated on a gelatin column and described as a glycoprotein able to bind collagen IV [65]. Early observations suggested that Hsp47 binds specifically to procollagen/collagen, interacting with different kinds of collagen precursors such as nascent procollagen chains and unhydroxylated non-triple helical trimers (review by Nagata [66]). However, it has later been demonstrated that Hsp47 preferentially binds fully formed helical procollagen trimers [67], potentially stabilizing the correctly folded collagen helix against dIssociation, before its transport from the ER. Recently, an intriguing hypothesis has been advanced, based on the observation that procollagen triple helix folds spontaneously into its native conformation at 30–34°C but not at body temperature, and that both procollagen precursor and mature collagen are thermally unstable at body temperature when they acquires random coil structure [68]. Using measured triple helix folding temperature and known binding constants, the authors deduced that procollagen folding at 38°C requires triple helix stabilization by 45–65 kcal/mol. Thermodynamic analysis allowed to predict that ~50 μM Hsp47 will allow procollagen folding at 38°C, where over 20 Hsp47 molecules have to bind to a single triple helix. It is thus conceivable that Hsp47 assists procollagen folding by stabilizing the triple helix. This means that the mechanism of Hsp47 action is different from that of other chaperones, which preferentially bind to unfolded or misfolded polypeptide chains before the native conformation is achieved.
Human Hsp47 is a basic 47-kDa glycoprotein of 418 amino acids with pI close to 9. It has an N-terminal signal sequence, that targets the molecule to the endoplastic reticulum (ER), and a C-terminal ER retention signal (RDEL) [69, 70]. It is induced by heat shock, which is unusual because all other chaperones in ER are induced by stress due to the accumulation of unfolded proteins. Hsp47 is constitutively expressed in collagen-synthesizing cells, but it is not expressed in collagen-non-producing cells [66]. Moreover, an excessive collagen production in pathophysiological conditions is always associated with the upregulation of Hsp47 [71]. Importantly, hsp47 gene disruption caused mortality in embryonic mice, showing that Hsp47, indispensable for normal procollagen biosynthesis, is essential for normal embryogenesis in mice [72, 73]. Collagen I fibrils produced in Hsp47-null cells are abnormally thin and branched, and undergo intracellular aggregation, which can be prevented by transient expression of Hsp47 [74]. Thus, Hsp47 is required for prevention of formation of procollagen aggregates in ER and is indispensable for the formation of thick collagen fibrils. It was demonstrated that addition of Hsp47 prevents self-association of collagen triple helices in vitro [75]. It is possible that Hsp47 not only facilitates triple helix formation but also prevents lateral association of procollagen in ER. In addition, together with several collagen mutations, a missense mutation in the hsp47 gene is involved in the hereditary disease in dogs, osteogenesis imperfecta [76].
Hsp47 belongs to serine protease inhibitors (serpin) family. Serpin family members have a highly conserved secondary structure that consists of a core of three β-sheets surrounded by nine α-helices ([61] and references therein). Hsp47 isolated from the membrane fraction of chick embryos is a monomer, with composition enriched in basic amino acids and glycine [77], while the recombinant mouse Hsp47 was shown to exist as a mixture of two molecular species, a monomer (~45 kDa) and a trimer (~147 kDa) [61]; it seems though that the trimeric form is quite insignificant [78].
The minimal Hsp47 binding site on collagen was the subject of detailed investigations over several years. First, when collagen fragments were used, the stretches of non-hydroxylated Pro-Pro-Gly repeats were identified as the major specific Hsp47 binding site [60]. However, it appears that, in the procollagen biosynthetic pathway, most of the Pro-Pro-Gly sequences are converted before triple helix formation to Pro-Hyp-Gly, to which Hsp47 does not bind. Further search for Hsp47-binding sequences, performed with synthetic collagen peptides, identified Gly-X–Y repeats as the dominant HSP47-binding sequences in the triple-helical procollagens, with a requirement for a certain minimal number of Arg residues at the position Y [79, 80]. In addition, it appears that the arginine residue, also at the position preceding glycine in Gly-X–Y repeats, is an additional important structural determinant for Hsp47 binding [78]. The importance of Arg residues is corroborated by the fact that Hsp47 does not bind to type I and II collagens when side chains of their arginines are modified [79]. When the binding affinities of Hsp47 to triple-helical collagen and a corresponding single-chain peptide were directly compared using conformationally constrained collagen models, Hsp47 showed a strong conformational preference for triple-helical collagens, which confirms that Hsp47 is unlikely to be involved in events preceding the triple helix formation [78].
Misfolded procollagens in Hsp47−/−cells were degraded in the lysosomes via an autophagic pathway [81]. An interesting question is how the accumulation of aggregation-prone misfolded proteins within the ER triggers autophagy, which is an exclusively cytoplasmic phenomenon. It appears that ER stress may be an important inducer of autophagic degradation of ER proteins, including aggregated procollagen. Data provided by Ishida and coworkers [81] suggest that there exists some mechanism to discriminate among misfolded proteins in the ER and determine their processing either by ERAD or autophagy. The molecular conformation of the misfolded protein may determine which degradation pathway is used.
Multiple observations demonstrated that Hsp47 is implicated in molecular maturation of type I and type III collagens, the most prevalent matrix proteins that are involved in formation of fibrotic tissue. Accumulation of fibrotic tissue destroys the normal structure of tissues, resulting, ultimately, in end-stage organ failure. A series of reports have indicated that Hsp47 is identified as an autoantigen in the sera of several rheumatoid arthritis (RA), pulmonary fibroses, systemic sclerosis, and mixed connective tissue disease patients [82] and references therein). Interestingly, Hsp47 has been shown to be upregulated in all studied fibrotic diseases (reference in [83]). Accordingly, the administration of antisense oligonucleotides against Hsp47 was reported to suppress the accumulation of collagen in a renal disease, the tubulointerstitial fibrosis [84], in peritoneal fibrosis [85] and wound-associated scarring [86]. Another fibrotic disease, liver cirrhosis, could be almost completely resolved by delivery of Hsp47 siRNA to cirrhosis rat model [87]. Also, several low molecular weight compounds were found to inhibit the role of Hsp47 in accumulation of fibrotic tissue in vitro and in vivo [88, 89].
Adenovirus 100 K protein, a hexon chaperone
Protein 100 K is a late adenovirus (Ad) polypeptide encoded by the L4 transcription unit and produced in large amounts during virus infection [90]. Depending on the Ad serotype, L4-100 K contains 628–984 amino acid residues, with 805 residues for Ad2 and Ad5, the most frequently studied serotypes. L4-100 K is the first protein translated with the onset of the late phase of infection [91]. However, despite large production, L4-100 K, unlike other late proteins, is not a structural protein––and is not found in mature virions [92]. L4-100 K plays a vital role in the late phase of Ad life cycle, where it is engaged in multiple processes, such as alteration of cellular machinery in favor of translating large amounts of late virus products, involvement in nuclear accumulation necessary for capsid assembly, chaperoning hexon for trimerization and its nuclear transport, acting as a scaffold in the intranuclear assembly of Ad virions and, finally, preventing apoptosis of infected cells.
L4-100 K of Ad2, a native protein from infected human cells as well as the recombinant protein obtained in insect cells, migrates at 100 kDa, with a minor species/band at approximately 98 kDa, resulting from some proteolytic cleavage of the primary gene product [93, 94]. Sucrose density gradient analysis suggested a monomeric status of this chaperone [94, 95]. Nevertheless, some observations suggest that L4-100 K might be able to dimerize [96].
Based on sequence homology among several adenovirus subgenera, the L4-100 K protein can be divided into three regions: an N-terminal region (NTR) of up to 170 amino acids, a central conserved region (CCR) of 600–700 amino acids and a C-terminal region (CTR) of 20–100 amino acids (Fig. 2) [97]. An analysis of structural features of human Ad2 L4-100 K performed by [98] revealed two distinct potential RNA binding motifs: RNA recognition motif (RRM) located between positions 348 and 466, described by Hayes and coworkers [99], and an arginine-glycine-glycine (RGG)-rich RNA binding motif (RGG box) located between positions 727 and 764, containing three RGG repeats and four glycine-arginine-rich (GAR) sequences. RGG box can also function as a nuclear localization signal (NLS), which is more precisely localized within the GAR sequence between positions 749 and 773 [100]. In addition, there is a 10-amino acid sequence in the L4-100 K protein located between residues 383 and 392 and similar to Rev-like nuclear export signal (NES) [101, 102]. Analysis of the L4-100 K amino acid sequences for known protein–protein interaction domains also revealed three potential coiled-coil regions.
Structural regions identified within the adenovirus L4-100 K protein: coiled-coil region (CCR, aa 439–458), RNA recognition motif (RRM, aa 383–458), Rev-like nuclear export signal (NES, aa 383–392), arginine-arginine-glycine (RGG) box (aa 727–743), containing three RGG motifs and overlapping nuclear localization signal (NLS, aa 727–764) (from [98])
L4-100 K chaperone role in Ad hexon trimerization and its nuclear transport has been studied extensively [90, 94, 103, 104]. A client protein of L4-100 K, the hexon, is a major capsid protein of Ad virion present in 240 copies and forming the 20 facets of the viral icosahedron. Each hexon capsomer is trimer of three identical monomers/polypeptides with molecular weights of 110–120 kDa, depending on Ad serotype. These monomers are associated into trimers. The crystallographic structure of Ad2 hexon revealed the trimeric protein as a pseudo-hexagonal base with a triangular top, where the monomers are composed of two eight-stranded β-barrels and four extended loops [105, 106]. Stable complexes of 600–800 kDa containing the L4-100 K protein and the hexon monomer were detected in Ad-infected cells by immunoprecipitation [104, 107, 108] and were isolated by gel-filtration chromatography [104, 109]. In addition, a scaffolding role of L4-100 K protein in the intranuclear assembly pathway of Ad virions has been suggested on the basis of phenotypic analysis of temperature-sensitive mutants of the protein [93].
Initial data on the role of L4-100 K in hexon biogenesis in mammalian cells were confirmed by studies performed in a heterologous eukaryotic system. When expressed alone from recombinant baculovirus, hexon polypeptide was found to be insoluble and accumulated as inclusion bodies in the cytoplasm; a proper folding of hexon into trimers occurred only upon co-expression with L4-100 K protein [94]. EM analysis revealed that recombinant hexons recovered from insect cells have morphological characteristics of native hexon capsomers. The results obtained in the heterologous system implied that no other adenoviral proteins than L4-100 K were required to achieve hexon trimerization and showed that L4-100 K role cannot be fulfilled by insect cell chaperones. Thus, the presence of L4-100 K protein represents the necessary and sufficient condition for hexon trimer formation.
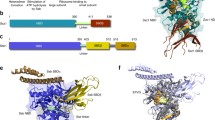
EM observation of purified recombinant Ad2 L4-100 K and native L4-100 K protein from Ad5-infected 293 cells showed a nearly symmetrical, dumbbell-shaped molecule consisting of two globular domains, joined by a rod-like structure (Fig. 3). The overall volume of the molecule allowed calculation of an estimated molecular mass compatible with that of a monomeric L4-100 K protein [94].
Visualisation of Ad L4-100 K protein alone and in the complex with hexon. a Gallery of eight EM images of the recombinant Ad2 L4-100 K protein, the averaged image, and the structural model of the L4-100 K protein (from left to right, upper panel). The L4-100 K protein is a dumbbell-shaped molecule, consisting of two globular domains linked by a rod-like structure. The total length of the molecule is about 120 Å. b Gallery, the averaged image and the model of the recombinant Ad3 hexon-Ad2 L4-100 K protein complex, with the hexon in end-on view (from left to right, lower panel). One of the globular domains of the L4-100 K protein is attached either to the distal domain or to the apical domain of the hexon trimer. c EM analysis of native proteins purified from Ad5-infected 293 cells. Scale bar 30 nm. Most of the visible structures are hexons. Some L4-100 K protein molecules and hexon–100 K protein complexes are marked with black and white arrows, respectively. The column on the right represents five images of hexon–100 K protein complexes and five images of 100 K protein molecules (upper and lower columns, respectively) (from [94])
Results obtained with the L4-100 K ts-mutants maintained at the nonpermissive temperature showed that hexon accumulated in the cytoplasm and failed to be transported to the nucleus [92, 110]. The role of L4-100 K in nuclear transport was therefore postulated. This, combined with the inability to detect trimers in the cytoplasm, led to the hypothesis that hexon protein trimerizes in the nucleus. This constitutes a rather unlikely scenario in the light of results obtained by Hong and coworkers [94]. These novel biochemical data and microscopic observations suggest that within the cytoplasm the L4-100 K interacts with both hexon monomers and trimers, whereas in the nucleus it is found exclusively in complexes with hexon trimers. The authors postulated that within the cytoplasm hexon monomers are assisted by the L4-100 K chaperone protein in their folding into trimers and, subsequently, only the complexes of L4-100 K protein with hexon trimer (but not monomer) are transferred to the nucleus. In these experiments, only hexon and L4-100 K genes were delivered to the insect cells by the recombinant baculovirus, and these data together with earlier studies prove that L4-100 K is a chaperone for hexon trimerization and nuclear transport. It was proposed that another viral protein, pVI, that shuttles between the nucleus and the cytoplasm, is implicated in the nuclear import of hexons [111, 112]. It is conceivable that at different stages of Ad life-cycle nuclear import of hexon could be mediated by two different partners.
Interaction domains in L4-100 K protein and on hexon have not yet been precisely identified. EM images of 100 K protein–hexon trimer complexes suggests that the chaperon–hexon interaction occurred via one of the globular domains of the L4-100 K molecule with either the distal or the apical domain of the hexon capsomer (Fig. 3) [94]. Since the atomic structure of the chaperone is not available, it is difficult to identify the domains/regions of L4-100 K polypeptide involved in the contacts. Our data from biochemical analyses of hexon intaraction with L4-100 K deletion mutants suggest that both N- and C-terminal domains of L4-100 K are involved in hexon binding (unpublished results). Furthermore, L4-100 K protein–hexon interaction is not serotype-specific since the L4-100 K from Ad2 (subgroup C) assisted not only in the trimerization of hexons from serotypes belonging to the same subgroup (e.g., Ad5) but also to a different one, subgroup B, (e.g., Ad3) [94]. This suggests that the 100 K protein regions involved in hexon folding are functionally conserved, at least between chaperones from subgroups B and C adenoviruses.
Another important function of L4-100 K is prevention of cellular mRNA translation through elimination of the cap (m7G)-dependent translation pathway and promoting viral mRNA translation [96, 98, 99, 113–117]. Underlying these processes is the interaction of L4-100 K with the scaffolding element of the translation initiation complex, namely, eukaryotic initiation factor G (eIF4G). The L4-100 K binds by its N-terminal part to the C-terminus of eIF4G, near or at the place occupied by Mnk1 protein kinase, which is responsible for phosphorylation of the cap-binding protein eIF4E. Competitive displacement of Mnk1 results in dephosphorylation of eIF4E leading to inhibition of cap-dependent cellular mRNA translation [98, 114]. An eIF4G-binding element, which is critical for inhibition of host-cell protein synthesis, is localized in the N-terminal region of the L4-100 K protein (positions 280–345) (Fig. 2).
Interestingly, late Ad messenger RNAs are capped, but are translated despite the inhibition of cap-dependent cellular protein synthesis by L4-100 K (see above). This is due to the presence in the viral mRNAs of 5′ noncoding region (5′NCR), also called the tripartite leader. During late viral infection, L4-100 K, through its RNA-binding element located in the middle region (but not through RGG RNA-binding domain, present in the C-terminal part) together with the cap initiation complex recruits with higher specificity mRNAs that contain the tripartite leader 5′NCR [96, 115]. The L4-100 K-tripartite leader complex enhances association with the initiation factor 4G (eIF4G) and poly(A) binding protein, which is accompanied by increased translation through ribosome shunting. The ribosome shunting is a form of translation initiation, in which 40S ribosome subunits are directed by the tripartite leader to the downstream initiating codon in a nonlinear manner, bypassing intervening RNA regions [96, 115–118]. These data emphasize the multifunctionality of L4-100 K protein, which, by N-terminal part interaction with eIF4G, prevents eIF4E phosphorylation thus inhibiting cap-dependent translation of host messengers, while using its RNA-binding middle region it promotes recruitment of tripartite leader-containing viral messengers to ribosomes, thereby promoting cap-dependent translation of viral messengers.
The L4-100 K protein is also involved in modulation of host immune response to viral infection, in which cytotoxic lymphocytes are the main players. Andrade and coworkers have shown that L4-100 K is a substrate of Granzymes––B (GrB) and H (GrH)––cellular proteases present in granules of cytotoxic lymphocytes, while at the same time it is a potent and specific inhibitor of human GrB [97, 119, 120]. This inhibition prevents GrB-induced apoptosis of Ad-infected cells so that the virus can complete its life cycle. Differently from GrB, GrH is not inhibited by L4-100 K [120]. Instead, it cleaves L4-100 K which decreases the inhibitory efficacy of L4-100 K and allows the recovery of GrB activity.
Biochemical analysis demonstrated that the specificity of the Ad5 L4-100 K protein for human GrB resides in two distinct regions of L4-100 K. It is proposed that Ad5 L4-100 K first attaches to the GrB active site through its IEQD48↓P49 sequence. The initial complex between the protease and L4-100 K is stabilized by interaction of the C-terminal 688–781 (CT688-781) fragment of L4-100 K with the GrB sequence outside the enzyme catalytic site (exosite), which has not been precisely localized [97]. Ad5 L4-100 K inhibitory activity is uniquely directed against the human protease and does not affect the mouse and rat orthologs, despite the degree of sequence homology among GrBs. Although the L4-100 K protein sequence is well conserved among different adenovirus subgenera, no homology is observed at the amino- and carboxy-terminal extremes. Therefore, the N-terminal region containing the motif essential for human GrB inhibition is unique for the Ad5 L4-100 K. Additionally, CT688-781 in L4-100 K interacts with human GrB exosite, but not with that of mouse or rat. Stable binding and efficient inhibition of GrB is dependent on Asp-48 in L4-100 K. The D48A L4-100 K mutant that is not active in GrB inhibition is nevertheless functional in other viral pathways, which suggests that GrB inhibitory effect evolved independently from other L4-100 K functions [119].
Multiple functions and interactions of L4-100 K are regulated by its posttranslational modifications including phosphorylation and methylation. The tyrosine phosphorylation, which has no influence on interaction with the initiation factor 4G, importantly enhances preferential binding of L4-100 K protein to tripartite leader mRNAs; mutations of tyrosine 365 and/or 682 were sufficient to abolish L4-100 K protein preferential binding to mRNA tripartite leader essential for promoting ribosome shunting [96]. It was recently shown that methylation of arginine located in the RGG domain regulates the L4-100 K binding to hexon and promotes the capsid assembly in the nucleus [100].
The importance of L4-100 K for the Ad life cycle was confirmed by the phenotype of L4-100 K deleted Ad5 vectors, produced in the L4-100 K-complementing K-16 cell line. Such recombinant adenoviruses were prepared for gene therapy applications with the aim to overcome the unwanted toxicity associated with Ad late gene expression. After infection of non-complementing cells, those vectors, as could be expected, had a significantly decreased ability to express several Ad late genes and were blocked in their ability to produce infectious virions. Such properties and decreased hepatotoxicity observed after in vivo administration suggest that the L4-100 K deletion strategy may be useful in preparation of safer Ad vectors for gene therapy application [121].
The α-hemoglobin-stabilizing protein (AHSP), also known as EDRF (erythroid differentiation related factor) and ERAF (erythroid associated factor)
Mammalian hemoglobin A (HbA) is a tetramer of two α- and two β-globin chains. It is crucial to erythrocyte formation that the two globin chains be produced at balanced levels since any disruptions resulting in an excess of either chain have significant, deleterious effects on red cell survival, a situation most clearly illustrated by important human diseases, such as thalassemias [122]. In addition, free α-Hb (α-globin plus heme, or holo-α-globin) is a potent oxidant, catalyzing the production of reactive oxygen species (ROSs), which damage erythroid precursors and mature erythrocytes [123].
Using a screen for genes induced by the essential erythroid transcription factor GATA-1, Kihm and coworkers [124] identified the AHSP protein (102 aa) that stabilizes free α-hemoglobin. AHSP mRNA is expressed exclusively in hematopoietic tissues including bone marrow, spleen and fetal liver. AHSP is an abundant, erythroid-specific protein that forms a stable heterodimeric complex with free α-hemoglobin. Although AHSP is able to interact with multiple forms of α-globin, including apo-, ferrous and ferric states bound to a variety of ligands, it does not interact with β-hemoglobin or HbA (α(2)β(2)). Moreover, AHSP specifically protects free α-hemoglobin from precipitation both in vitro and in live cells, and reduces oxidant-induced precipitation of α-globin in solution [124, 125]. Finally, AHSP promotes refolding of denatured α-globin and HbA assembly from newly synthesized α-globin in vitro [126].
Gene ablation studies in mice demonstrated that AHSP is required for normal erythropoiesis. AHSP (−/−) homozygous knockout mice exhibit mild hemolytic anemia, shortened erythrocyte lifespan and high levels of reactive oxygen species, consistent with the presence of unstable α-globin [124]. AHSP-null erythrocytes are short-lived, contain Hb precipitates, and exhibit signs of oxidative damage. Predictably, loss of AHSP exacerbates β-thalassemia in mice and results in increased α-globin precipitation, suggesting that altered AHSP expression or function could modify thalassemia phenotypes in humans [124, 127]. Recently, the first mutant of AHSP, AHSP(V56G), was discovered associated with clinical symptoms of mild thalassemia [128]. The kinetics analysis showed that the AHSP(V56G) apparently does not bind long enough (0.5 vs. 2 s for the WT) to stabilize α-globin.
Biophysical characterization demonstrated that monomeric AHSP in solution forms a moderate affinity complex with α-globin with 1:1 stoichiometry and with an association constant of 1 × 107 M−1 [129]. A far-UV circular dichroism spectrum showed that slightly elongated AHSP is primarily α-helical in conformation, probably with an extended C-terminal region. The crystal structure of AHSP bound to Fe(II)-αHb revealed that AHSP binds α-globin at the α1β1 dimer interface, opposite the heme binding pocket, where it is easily displaced by attachment of β-globin [125]. The structure of AHSP bound to ferrous αHb is thought to represent a transitional complex through which αHb is converted to a non-reactive, hexacoordinate ferric form. The crystal structure of this ferric α-Hb-AHSP complex at 2.4 Å resolution revealed a striking bis-histidine configuration, in which both the proximal and the distal histidines of α-Hb coordinate the heme iron atom. To attain this unusual conformation, segments of α-Hb undergo drastic structural rearrangements, including the repositioning of several α-helices. Moreover, conversion to the ferric bis-histidine configuration strongly and specifically inhibits redox chemistry catalysis and heme loss from α-Hb. The observed structural changes, which impair the chemical reactivity of heme iron, explain how AHSP stabilizes α-Hb and prevents its damaging effects in cells [130]. Taken together, X-ray crystallography, NMR spectroscopy, and mutagenesis data indicate the importance of an evolutionarily conserved proline, Pro-30, in loop 1 of AHSP. In complex with αHb, AHSP Pro-30 adopts a cis-peptidyl conformation and establishes contact with the N-terminus of helix G in α-Hb. As described above, complex formation suppresses the heme-catalyzed evolution of reactive oxygen species by converting α-Hb to a conformation in which the heme is coordinated at both axial positions by histidine side chains (bis-histidine coordination). Mutations that stabilize the cis-peptidyl conformation of free AHSP also enhance the conversion of α-Hb. These findings suggest that AHSP loop 1 can transmit structural changes to the heme pocket of α-Hb [131].
Since AHSP and β-Hb have overlapping binding sites on αHb, the addition of β-Hb to either Fe(II)- or Fe(III) αHb.AHSP displaces AHSP to generate tetrameric (α(2)β(2)) HbA species. These findings suggest a biochemical pathway through which AHSP might participate in normal Hb synthesis and modulate the severity of thalassemias. Indeed, some thalassemia phenotypes could be explained by a decreased expression of the Ahsp gene [132], or by an α-Hb variant displaying an impaired interaction with the AHSP or partner β chain [133], thereby hampering formation of a α1β1 dimer [134, 135]. Recently, in a murine model of β(IVS-2-654)-thalassemia, the expression of wt AHSP was able to improve the anemia phenotype and partially relieve this thalassemia syndrome [48], suggesting a possible role for AHSP in the treatments of thalassemia.
HYPK, the Huntingtin-interacting protein
HYPK is one of 13 Huntingtin-interacting proteins isolated by double hybrid technique [136], but possibly the only one with chaperone activity. HYPK (NP_057484) shares no significant sequence homology with any protein of known sequence/structure in the databases. It is not found in yeast. It is a small protein with theoretical molecular weight of 14,665 and theoretical pI of 4.9 [137]. HYPK is an intrinsically unstructured protein with an endoplasmic reticulum retention signal (Fig. 4). Indeed, the protein is significantly enriched in glutamate (15.89%), lysine (6.62%), arginine (8.61%) and other charged amino acids, and depleted in tryptophan, phenylalanine, and cysteine, order-promoting residues.
Schematic model of the predicted domains of HYPK chaperone, a Huntingtin-interacting protein. Domains are shown from left to right: acidic, putative SH3-binding and coiled coil (after [142])
The partner of HYPK, Huntingtin protein (Htt), an approximately 350-kDa protein of unknown function, is implicated in Huntington disease (HD), an autosomal dominant neurodegenerative disease characterized by loss of striatal neurons. The neuropathology of HD is thought to be due to elongation of a polyglutamine segment (poly Q) in Htt N-terminal extremity. HYPK was found to reduce Htt polyglutamine aggregation upon overexpression; it interacts physically with the N-terminal extremity of Htt and alters the numbers and distribution of aggregates [138]. The aggregates formed by the mutated N-terminal Htt lead to enhanced apoptosis [139], and overexpression of HYPK reduces apoptosis based on poly Q-mediated activation of caspase-2, caspase-3 and caspase-8.
The seminal study of Raychaudhuri and collaborators [138] demonstrated that HYPK is a chaperone. Recombinant HYPK reduced temperature-induced aggregation of alcohol dehydrogenase and malate dehydrogenase in a dose (concentration of HYPK)-dependent manner. Also, renaturation kinetics of unfolded bovine carbonic anhydrase (BCA) was accelerated significantly in the presence of HYPK. Furthermore, when HeLa cells expressing heat-sensitive luciferase protein and transfected with GFP-HYPK underwent heat shock at 45°C for 30 min, luciferase activity was ~15-fold higher in the presence of HYPK, indicating a significant increase in properly folded luciferase. These results showed unequivocally that HYPK exhibits chaperone-like activity. HYPK does not have any sequence homology to known chaperones but it has been found linked with ribosome-associated chaperone complex containing MPP11 and Hsp70L1 (homologues of Hsp40 and Hsp70) [139]. The structural basis of recognition by HYPK of oligomeric structures in the form of poly Q is not yet known. In addition, it was recently shown that oxidation-dependent oligomers of Htt form spontaneously in cell and mouse HD models [140, 141]. The role of these oligomers in HT is not clear, and nothing is as yet known on the eventual role of HYPK in their life cycle.
It seems that HYPK might have roles beyond prevention of Htt aggregation. Endogenous HeLa cells HYPK was found to be a cytosolic partner of the N-terminal-acetyltransferase complex responsible for Nα-terminal acetylation, the most common protein modifications in eukaryotes [142]. HYPK appeared to be required for N-terminal acetylation of some substrates and for cell survival. Indeed, HYPK knockdown resulted in HeLa cell death and accumulation of cells in the G1/G0 phase.
Concluding remarks
The mechanism of protein folding in the cell remains one of the central problems in biology. It is an extremely active field of research, including some aspects of biology, chemistry, biochemistry, computer science and physics. Folding of newly synthesized polypeptides in the crowded cellular environment requires the assistance of so-called molecular chaperones. Chaperones are defined as a group of proteins that assist in non-covalent folding and unfolding, and assembly and disassembly of macromolecular structures, but are not permanent components of these structures during their performance of normal biological functions [143]. Through their remarkable ability to distinguish between native and non-native protein structures, molecular chaperones participate in a variety of cellular processes that depend critically on the conformation of the proteins involved.
The vast majority of chaperone research concerns their role in protein folding, whereas much less is known about their function in the assembly of oligomeric complexes. We were interested in a subgroup consisting of five eukaryotic molecular chaperones that assists not only in folding but also in assembly of oligomeric polypeptides. As each of these chaperones assists only one client, it raises the question regarding the biosynthetic cost of the high-level production of such chaperones. The clients of faithful chaperones are all abundant proteins that are essential cellular or viral components; it is thus conceivable that this necessary metabolic expenditure withstood evolutionary pressure to minimize biosynthetic costs. Future work will be necessary to provide a better understanding of unexplored facets of these and potentially other chaperone activities, such as their thermodynamics and the mechanism of recognition of a particular client, all based on observation of protein movement and interactions in individual living cells. This knowledge is important for understanding the efficiency of chaperone action and is also necessary for productive therapeutic interventions.
Abbreviations
- aa:
-
Amino acid
- Ad:
-
Adenovirus
- ER:
-
Endoplasmic reticulum
- ERAD:
-
ER-associated degradation
- Hb:
-
Hemoglobin
- Pro-Pro-Gly and Pro-Hyp-Gly:
-
Tripeptides proline-proline-glycine and proline-hydroxyproline-glycine
- Arg, Ser:
-
Arginine, serine
References
Laskey RA, Honda BM, Mills AD, Finch JT (1978) Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature 275:416–420. doi:10.1038/275416a0
Wade RH (2007) Microtubules: an overview. Methods Mol Med 137:11–16 Review
Tian G, Bhamidipati A, Cowan NC, Lewis SA (1999) Tubulin folding cofactors as GTPase-activating proteins. GTP hydrolysis and the assembly of the alpha/beta-tubulin heterodimer. J Biol Chem 274:24054–24058. doi:10.1074/jbc.274.34.24054
Spiegelman BM, Penningroth SM, Kirschner MW (1977) Turnover of tubulin and the N site GTP in Chinese hamster ovary cells. Cell 12:587–600. doi:10.1152/ajpheart.01252.2008
Lopez-Fanarraga M, Avila J, Guasch A, Coll M, Zabala JC (2001) Review: postchaperonin tubulin folding cofactors and their role in microtubule dynamics. J Struct Biol 135(2):219–229. doi:10.1074/jbc.274.34.24054
Szymanski D (2002) Tubulin folding cofactors: half a dozen for a dimer. Curr Biol 12:R767–R769. doi:10.1371/journal.pone.0008846 Review
Campo R, Fontalba A, Sanchez LM, Zabala JC (1994) A 14 kDa release factor is involved in GTP-dependent beta-tubulin folding. FEBS Lett 353(2):162–166
Archer JE, Vega LR, Solomon F (1995) Rbl2p, a yeast protein that binds to beta-tubulin and participates in microtubule function in vivo. Cell 82:425–434. doi:10.1186/1471-2105-4-46
Melki R, Rommelaere H, Leguy R, Vandekerckhove J, Ampe C (1996) Cofactor A is a molecular chaperone required for beta-tubulin folding: functional and structural characterization. Biochemistry 35(32):10422–10435. doi:10.1021/bi960788r
Tian G, Huang Y, Rommelaere H, Vandekerckhove J, Ampe C, Cowan NJ (1996) Pathway leading to correctly folded beta-tubulin. Cell 86:287–296. doi:10.1016/S0092-8674(00)80100-2
Tian G, Lewis SA, Feierbach B, Stearns T, Rommelaere H, Ampe C, Cowan NJ (1997) Tubulin subunits exist in an activated conformational state generated and maintained by protein cofactors. J Cell Biol 138:821–832. doi:10.1083/jcb.138.4.821
Bhamidipati A, Lewis SA, Cowan NJ (2000) ADP ribosylation factor-like protein 2 (Arl2) regulates the interaction of tubulin-folding cofactor D with native tubulin. J Cell Biol 149:1087–1096. doi:10.1083/jcb.149.5.1087
Martín L, Fanarraga ML, Aloria K, Zabala JC (2000) Tubulin folding cofactor D is a microtubule destabilizing protein. FEBS Lett 470(1):93–95
Archer JE, Magendantz M, Vega LR, Solomon F (1998) Formation and function of the Rbl2p-beta-tubulin complex. Mol Cell Biol 18(3):1757–1762
Abruzzi KC, Smith A, Chen W, Solomon F (2002) Protection from free beta-tubulin by the beta-tubulin binding protein Rbl2p. Mol Cell Biol 22:138–147. doi:10.1128/MCB.22.1.138-147.2002
Bartolini F, Tian G, Piehl M, Cassimeris L, Lewis SA, Cowan NJ (2005) Identification of a novel tubulin-destabilizing protein related to the chaperone cofactor. J Cell Sci 118:1197–1207. doi:10.1242/jcs.01719
Keller CE, Lauring BP (2005) Possible regulation of microtubules through destabilization of tubulin. Trends Cell Biol 15(11):571–573. doi:10.1016/j.tcb.2005.09.008
Kortazar D, Carranza G, Bellido J, Villegas JC, Fanarraga ML, Zabala JC (2006) Native tubulin-folding cofactor E purified from baculovirus-infected Sf9 cells dissociates tubulin dimers. Protein Expr Purif 49:196–202. doi:10.1016/j.pep.2006.03.005
Gao Y, Melki R, Walden PD, Lewis SA, Ampe C, Rommelaere H, Vandekerckhove J, Cowan NJ (1994) A novel cochaperonin that modulates the ATPase activity of cytoplasmic chaperonin. J Cell Biol 125(5):989–996
Steinbacher S (1999) Crystal structure of the post-chaperonin β-tubulin binding cofactor Rbl2p. Nat Struct Biol 6:1029–1032. doi:10.1038/14912
Guasch A, Aloria K, Pérez R, Avila J, Zabala JC, Coll M (2002) Three-dimensional structure of human tubulin chaperone cofactor A. J Mol Biol 318:1139–1149. doi:10.1016/S0022-2836(02)00185-7
Lu L, Nan J, Mi W, Li LF, Wei CH, Su XD, Li Y (2010) Crystal structure of tubulin folding cofactor A from Arabidopsis thaliana and its beta-tubulin binding. FEBS Lett 584(16):3533–3539. doi:10.1016/j.febslet.2010.07.017
Nolasco S, Bellido J, Gonçalves J, Zabala JC, Soares H (2005) Tubulin cofactor A gene silencing in mammalian cells induces changes in microtubule cytoskeleton, cell cycle arrest and cell death. FEBS Lett 579(17):3515–3524. doi:10.1016/j.febslet.2005.05.022
Lytle BL, Peterson FC, Qiu SH, Luo M, Zhao Q, Markley JL, Volkman BF (2004) Solution structure of a ubiquitin-like domain from tubulin-binding cofactor B. J Biol Chem 279(45):46787–46793. doi:10.1074/jbc.M409422200
Rayala SK, Martin E, Sharina IG, Molli PR, Wang X, Jacobson R, Murad F, Kumar R (2007) Dynamic interplay between nitration and phosphorylation of tubulin cofactor B in the control of microtubule, dynamics. Proc Natl Acad Sci USA 104:19470–19475. doi:10.1073/pnas.0705149104 Erratum in: Proc Natl Acad Sci USA 2008 105(1):399
Vadlamudi RK, Barnes CJ, Rayala S, Li F, Balasenthil S, Marcus S, Goodson HV, Sahin AA, Kumar R (2005) p21-activated kinase 1 regulates microtubule dynamics by phosphorylating tubulin cofactor B. Mol Cell Biol 25:3726–3736. doi:10.1128/MCB.25.9.3726-3736.2005
Steinmetz MO, Akhmanova A (2008) Capturing protein tails by CAP-Gly domains. Trends Biochem Sci 33(11):535–545. doi:10.1016/j.tibs.2008.08.006
Li S, Finley J, Liu ZJ, Qiu SH, Chen H, Luan CH, Carson M, Tsao J, Johnson D, Lin G, Zhao J, Thomas W, Nagy LA, Sha B, DeLucas LJ, Wang BC, Luo M (2002) Crystal structure of the cytoskeleton-associated protein glycine-rich (CAP-Gly) domain. J Biol Chem 277:48596–48601. doi:10.1074/jbc.M208512200
Grynberg M, Jaroszewski L, Godzik A (2003) Domain analysis of the tubulin cofactor system: a model for tubulin folding and dimerization. BMC Bioinformatics 4:46–56. doi:10.1186/1471-2105-4-46
Srayko M, Kaya A, Stamford J, Hyman AA (2005) Identification and characterization of factors required for microtubule growth and nucleation in the early C. elegans embryo. Dev Cell 9(2):223–236. doi:10.1016/j.devcel.2005.07.003
Lopez-Fanarraga M, Carranza G, Bellido J, Kortazar D, Villegas JC, Zabala JC (2007) Tubulin cofactor B plays a role in the neuronal growth cone. J Neurochem 100(6):1680–1687. doi:10.1111/j.1471-4159.2006.04328.x
Dhonukshe P, Bargmann BO, Gadella TW Jr (2006) Arabidopsis tubulin folding cofactor B interacts with α-tubulin in vivo. Plant Cell Physiol 47:1406–1411. doi:10.1093/pcp/pcl001
Du Y, Cui M, Qian D, Zhu L, Wei C, Yuan M, Zhang Z, Li Y (2010) AtTFC B is involved in control of cell division. Front Biosci (Elite Ed) 2:752–763
Radcliffe PA, Vardy L, Toda T (2000) A conserved small GTP-binding protein Alp41 is essential for the cofactor-dependent biogenesis of microtubules in fission yeast. FEBS Lett 468:84–88
Antoshechkin I, Han M (2002) The C. elegans evl-20 gene is a homolog of the small GTPase ARL2 and regulates cytoskeleton dynamics during cytokinesis and morphogenesis. Dev Cell 2:579–591. doi:10.1016/S1534-5807(02)00146-6
Beghin A, Honore S, Messana C, Matera EL, Aim J, Burlinchon S, Braguer D, Dumontet C (2007) ADP ribosylation factor like 2 (Arl2) protein influences microtubule dynamics in breast cancer cells. Exp Cell Res 313:473–485. doi:10.1016/j.yexcr.2006.10.024
Zhou C, Cunningham L, Marcus AI, Li Y, Kahn RA (2006) Arl2 and Arl3 regulate different microtubule-dependent processes. Mol Biol Cell 17:2476–2487. doi:10.1091/mbc.E05-10-0929
Hirata D, Masuda H, Eddison M, Toda T (1998) Essential role of tubulin-folding cofactor D in microtubule assembly and its association with microtubules in fission yeast. EMBO J 17(3):658–666. doi:10.1093/emboj/17.3.658
Steinborn K, Maulbetsch C, Priester B, Trautmann S, Pacher T, Geiges B, Küttner F, Lepiniec L, Stierhof YD, Schwarz H, Jürgens G, Mayer U (2002) The Arabidopsis PILZ group genes encode tubulin-folding cofactor orthologs required for cell division but not cell growth. Genes Dev 16(8):571–959. doi:10.1101/gad.221702
Pierre P, Pepperkok R, Kreis TE (1994) Molecular characterization of two functional domains of CLIP-170 in vivo. J Cell Sci 107:1909–1920
Kobe B, Deisenhofer J (1994) The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci 19:415–421
Jin S, Pan L, Liu Z, Wang Q, Xu Z, Zhang YQ (2009) Drosophila tubulin-specific chaperone E functions at neuromuscular synapses and is required for microtubule network formation. Development 136:1571–1581. doi:10.1242/dev.029983
Fanarraga ML, Carranza G, Castaño R, Jiménez V, Villegas JC, Zabala JC (2010) Emerging roles for tubulin folding cofactors at the centrosome. Commun Integr Biol 3(4):306–308. doi:10.4161/cib.3.4.11976
Cunningham LA, Kahn RA (2008) Cofactor D functions as a centrosomal protein and is required for the recruitment of the gamma-tubulin ring complex at centrosomes and organization of the mitotic spindle. J Biol Chem 283(11):7155–7165. doi:10.1074/jbc.M706753200
Schultz T, Shmuel M, Hyman T, Altschuler Y (2008) Β-tubulin cofactor D and ARL2 take part in apical junctional complex disassembly and abrogate epithelial structure. FASEB J 22:168–182. doi:10.1096/fj.06-7786com
Price HP, Peltan A, Stark M, Smith DF (2010) The small GTPase ARL2 is required for cytokinesis in Trypanosoma brucei. Mol Biochem Parasitol May 31.[Epub ahead of print] doi:10.1016/j.molbiopara.2010.05.016
Voloshin O, Gocheva Y, Gutnick M, Movshovich N, Bakhrat A, Baranes-Bachar K, Bar-Zvi D, Parvari R, Gheber L, Raveh D (2010) Tubulin chaperone E binds microtubules and proteasomes and protects against misfolded protein stress. Cell Mol Life Sci 67(12):2025–2038. doi:10.1007/s00018-010-0308-8
Wang W, Ding J, Allen E, Zhu P, Zhang L, Vogel H, Yang Y (2005) Gigaxonin interacts with tubulin folding cofactor B and controls its degradation through the ubiquitin-proteasome pathway. Curr Biol 15:2050–2055. doi:10.1016/j.cub.2005.10.052
Kortazar D, Fanarraga ML, Carranza G, Bellido J, Villegas JC, Avila J, Zabala JC (2007) Role of cofactors B (TBCB) and E (TBCE) in tubulin heterodimer dissociation. Exp Cell Res 313:425–436. doi:10.1016/j.yexcr.2006.09.002
Bommel H, Xie G, Rossoll W, Wiese S, Jablonka S, Boehm T, Sendtner M (2002) Missense mutation in the tubulin-specific chaperone E (Tbce) gene in the mouse mutant progressive motor neuronopathy, a model of human motoneuron disease. J Cell Biol 159:563–569. doi:10.1083/jcb.200208001
Parvari R, Hershkovitz E, Grossman N, Gorodischer R, Loeys B, Zecic A, Mortier G, Gregory S, Sharony R, Kambouris M, Sakati N, Meyer BF, Al Aqeel AI, Al Humaidan AK, Al Zanhrani F, Al Swaid A, Al Othman J, Diaz GA, Weiner R, Khan KT, Gordon R, Gelb BD (2002) HRD/Autosomal Recessive Kenny-Caffey Syndrome Consortium. Mutation of TBCE causes hypoparathyroidism-retardation-dysmorphism and autosomal recessive Kenny-Caffey syndrome. Nat Genet 32(3):448–452. doi:10.1038/ng1012
Hershkovitz E, Parvari R, Diaz GA, Gorodischer R (2004) Hypoparathyroidism-retardation-dysmorphism (HRD) syndrome-a review. J Pediatr Endocrinol Metab 17:1583–1590
Tian G, Huang MC, Parvari R, Diaz GA, Cowan NJ (2006) Cryptic out-of-frame translational initiation of TBCE rescues tubulin formation in compound heterozygous HRD. Proc Natl Acad Sci USA 103:13491–13496. doi:10.1073/pnas.0602798103
Evans RJ, Hardcastle AJ, Cheetham ME (2006) Focus on molecules: X-linked Retinitis Pigmentosa 2 protein, RP2. Exp Eye Res 82:543–544. doi:10.1016/j.exer.2005.10.023
Kühnel K, Veltel S, Schlichting I, Wittinghofer A (2006) Crystal structure of the human retinitis pigmentosa 2 protein and its interaction with Arl3. Structure 14:367–478. doi:10.1016/j.str.2005.11.008
Bartolini F, Bhamidipati A, Thomas S, Schwahn U, Lewis SA, Cowan NJ (2002) Functional overlap between retinitis pigmentosa 2 protein and the tubulin-specific chaperone cofactor C. J Biol Chem 277:14629–14634. doi:10.1074/jbc.M200128200
Tian G, Kong XP, Jaglin XH, Chelly J, Keays D, Cowan NJ (2008) A Pachygyria-causing α-tubulin mutation results in inefficient cycling with cct and a deficient interaction with TBCB. Mol Biol Cell 19:1152–1161. doi:/10.1091/mbc.E07-09-0861
Heino J (2007) The collagen family members as cell adhesion proteins. Bioessays 29(10):1001–1010. doi:10.1002/bies.20636
Bonfanti L, Mironov AA Jr, Martínez-Menárguez JA, Martella O, Fusella A, Baldassarre M, Buccione R, Geuze HJ, Mironov AA, Luini A (1998) Procollagen traverses the Golgi stack without leaving the lumen of cisternae: evidence for cisternal maturation. Cell 95(7):993–1003. doi:10.1016/S0092-8674(00)81723-7
Koide T, Asada S, Nagata K (1999) Substrate recognition of collagen-specific molecular chaperone HSP47. Structural requirements and binding regulation. J Biol Chem 274:34523–34526. doi:10.1074/jbc.274.49.34523
Dafforn TR, Della M, Miller AD (2001) The molecular interactions of heat shock protein 47 (Hsp47) and their implications for collagen biosynthesis. J Biol Chem 276(52):49310–49319. doi:10.1074/jbc.M108896200
Engel J, Prockop DJ (1991) The zipper-like folding of collagen triple helices and the effects of mutations that disrupt the zipper. Annu Rev Biophys Biophys Chem 20:137–152
Bulleid NJ, Wilson R, Lees JF (1996) Type-III procollagen assembly in semi-intact cells: chain association, nucleation and triple-helix folding do not require formation of inter-chain disulphide bonds but triple-helix nucleation does require hydroxylation. Biochem J 317:195–202
Nagata K (2003) HSP47 as a collagen-specific molecular chaperone: function and expression in normal mouse development. Semin Cell Dev Biol 14(5):275–282 Review
Kurkinen M, Taylor A, Garrels JI, Hogan BL (1984) Cell surface-associated proteins which bind native type IV collagen or gelatin. J Biol Chem 259:5915–5922
Nagata K (1996) Hsp47: a collagen-specific molecular chaperone. Trends Biochem Sci 21(1):22–26
Tasab M, Batten MR, Bulleid NJ (2000) Hsp47: a molecular chaperone that interacts with and stabilizes correctly-folded procollagen. EMBO J 19(10):2204–2211. doi:10.1093/emboj/19.10.2204
Makareeva E, Leikin S (2007) Procollagen triple helix assembly: an unconventional chaperone-assisted folding paradigm. PLoS One 2(10):e1029. doi:10.1371/journal.pone.0001029
Satoh M, Hirayoshi K, Yokota S, Hosokawa N, Nagata K (1996) Intracellular interaction of collagen-specific stress protein HSP47 with newly synthesized procollagen. J Cell Biol 133(2):469–483
Saga S, Nagata K, Chen WT, Yamada KM (1987) pH-dependent function, purification, and intracellular location of a major collagen-binding glycoprotein. J Cell Biol 105:517–527
Razzaque MS, Nazneen A, Taguchi T (1998) Immunolocalization of collagen and collagen-binding heat shock protein 47 in fibrotic lung diseases. Mod Pathol 11(12):1183–1188
Nagai N, Hosokawa M, Itohara S, Adach E, Matsushita T, Hosokawa N, Nagata K (2000) J. Cell Biol 150:1499–1506. doi:10.1083/jcb.150.6.1499
Marutani T, Yamamoto A, Nagai N, Kubota H, Nagata K (2004) Accumulation of type IV collagen in dilated ER leads to apoptosis in Hsp47-knockout mouse embryos via induction of CHOP. J Cell Sci 117:5913–5922. doi:10.1242/jcs.01514
Ishida Y, Kubota H, Yamamoto A, Kitamura A, Bächinger HP, Nagata K (2006) Type I collagen in Hsp47-null cells is aggregated in endoplasmic reticulum and deficient in N-propeptide processing and fibrillogenesis. Mol Biol Cell 17:2346–2355. doi:10.1091/mbc.E05-11-1065
Thomson CA, Ananthanarayanan VS (2000) Structure-function studies on hsp47: pH-dependent inhibition of collagen fibril formation in vitro. Biochem J 349(Pt 3):877–883
Drögemüller C, Becker D, Brunner A, Haase B, Kircher P, Seeliger F, Fehr M, Baumann U, Lindblad-Toh K, Leeb T (2009) A missense mutation in the SERPINH1 gene in Dachshunds with osteogenesis imperfecta. PLoS Genet 5(7):e1000579. doi:10.1371/journal.pgen.1000579
Nagata K, Saga S, Yamada KM (1988) Characterization of a novel transformation-sensitive heat-shock protein (HSP47) that binds to collagen. Biochem Biophys Res Commun 153(1):428–434. doi:10.1016/S0006-291X(88)81242-7
Koide T, Nishikawa Y, Asada S, Yamazaki CM, Takahara Y, Homma DL, Otaka A, Ohtani K, Wakamiya N, Nagata K, Kitagawa K (2006) Specific recognition of the collagen triple helix by chaperone HSP47. II. The HSP47-binding structural motif in collagens and related proteins. J Biol Chem 281:11177–11185. doi:10.1074/jbc.M601369200
Koide T, Takahara Y, Asada S, Nagata K (2002) Xaa-Arg-Gly triplets in the collagen triple helix are dominant binding sites for the molecular chaperone HSP47. J Biol Chem 277(8):6178–6182. doi:10.1074/jbc.M106497200
Tasab M, Jenkinson L, Bulleid NJ (2002) Sequence-specific recognition of collagen triple helices by the collagen-specific molecular chaperone HSP47. J Biol Chem 277(38):35007–35012. doi:10.1074/jbc.M202782200
Ishida Y, Yamamoto A, Kitamura A, Lamandé SR, Yoshimori T, Bateman JF, Kubota H, Nagata K (2009) Autophagic elimination of misfolded procollagen aggregates in the endoplasmic reticulum as a means of cell protection. Mol Biol Cell 20:2744–2754. doi:10.1091/mbc.E08-11-1092
Yokota S, Kubota H, Matsuoka Y, Naitoh M, Hirata D, Minota S, Takahashi H, Fujii N, Nagata K (2003) Prevalence of HSP47 antigen and autoantibodies to HSP47 in the sera of patients with mixed connective tissue disease. Biochem Biophys Res Commun 303(2):413–418. doi:10.1016/S0006-291X(03)00352-8
Taguchi T, Razzaque MS (2007) The collagen-specific molecular chaperone HSP47: is there a role in fibrosis? Trends Mol Med 13(2):45–53. doi:10.1016/j.molmed.2006.12.001
Xia Z, Abe K, Furusu A, Miyazaki M, Obata Y, Tabata Y, Koji T, Kohno S (2008) Suppression of renal tubulointerstitial fibrosis by small interfering RNA targeting heat shock protein 47. Am J Nephrol 28(1):34–46. doi:10.1159/000108759
Miyazaki M, Nishino T, Abe K, Furusu A, Koji T, Kohno S (2003) Contrib Nephrol 139:141–155
Ohba S, Wang ZL, Baba TT, Nemoto TK, Inokuchi T (2003) Antisense oligonucleotide against 47-kDa heat shock protein (Hsp47) inhibits wound-induced enhancement of collagen production. Arch Oral Biol 48(9):627–633. doi:10.1016/S0003-9969(03)00117-1
Sato Y, Murase K, Kato J, Kobune M, Sato T, Kawano Y, Takimoto R, Takada K, Miyanishi K, Matsunaga T, Takayama T, Niitsu Y (2008) Resolution of liver cirrhosis using vitamin A-coupled liposomes to deliver siRNA against a collagen-specific chaperone. Nat Biotechnol 26(4):431–442. doi:10.1038/nbt1396
Thomson CA, Atkinson HM, Ananthanarayanan VS (2005) Identification of small molecule chemical inhibitors of the collagen-specific chaperone Hsp47. J Med Chem 48:1680–1684. doi:10.1021/jm049148+
Gelosa P, Banfi C, Gianella A, Brioschi M, Pignieri A, Nobili E, Castiglioni L, Cimino M, Tremoli E, Sironi L (2010) Peroxisome proliferator-activated receptor alpha agonism prevents renal damage and the oxidative stress and inflammatory processes affecting the brains of stroke-prone rats. J Pharmacol Exp Ther 335(2):324–331. doi:10.1124/jpet.110.171090
Oosterom-Dragon EA, Ginsberg HS (1980) Purification and preliminary immunological characterization of the type 5 adenovirus, nonstructural 100,000-Da protein. J Virol 33:1203–1207
Bablanian R, Russell WC (1974) Adenovirus polypeptide synthesisin the presence of non replicatigpoliovirus. J Gen Virol 24:261–279. doi:10.1099/0022-1317-24-2-261
Russell WC, Skehel JJ (1972) The polypeptides of adenovirus-infected cells. J Gen Virol 15:45–57. doi:10.1099/0022-1317-15-1-45
Morin N, Boulanger P (1986) Hexon trimerization occurring in an assembly-defective, 100 K temperature-sensitive mutant of adenovirus 2. Virology 152:11–31
Hong SS, Szolajska E, Schoehn G, Franqueville L, Myhre S, Lindholm L, Ruigrok RW, Boulanger P, Chroboczek J (2005) The 100 K-chaperone protein from adenovirus serotype 2 (Subgroup C) assists in trimerization and nuclear localization of hexons from subgroups C and B adenoviruses. J Mol Biol 352(1):125–138. doi:10.1016/j.jmb.2005.06.070
Lemay P, Boulanger P (1980) Physicochemical characteristics of structural and non-structural proteins of human adenovirus 2. Ann Virol (Inst Past) 131E:259–275
Xi Q, Cuesta R, Schneider RJ (2005) Regulation of translation by ribosome shunting through phosphotyrosine-dependent coupling of adenovirus protein 100 K to viral mRNAs. J Virol 79(9):5676–5683. doi:10.1128/JVI.79.9.5676-5683.2005
Andrade F, Casciola-Rosen LA, Rosen A (2003) A novel domain in adenovirus L4–100 K is required for stable binding and efficient inhibition of human granzyme B: possible interaction with a species-specific exosite. Mol Cell Biol 23(17):6315–6326. doi:10.1128/MCB.23.17.6315-6326.2003
Cuesta R, Xi Q, Schneider RJ (2004) Structural basis for competitive inhibition of eIF4G-Mnk1 interaction by the adenovirus 100-kDa protein. J Virol 78:7707–7716. doi:10.1128/JVI.78.14.7707-7716.2004
Hayes BW, Telling GC, Myat MM, Williams JF, Flint SJ (1990) The adenovirus L4 100-kDa protein is necessary for efficient translation of viral late mRNA species. J Virol 64:2732–2734
Koyuncu OO, Dobner T (2009) Arginine methylation of human adenovirus type 5 L4 100-kDa protein is required for efficient virus production. J Virol 83(10):4778–4790. doi:10.1128/JVI.02493-08
Görlich D, Mattaj IW (1996) Nucleocytoplasmic transport. Science 271(5255):1513–1518
Murphy R, Wente SR (1996) An RNA-export mediator with an essential nuclear export signal. Nature 383(6598):357–360. doi:10.1038/383357a0
Leibowitz J, Horwitz MS (1975) Synthesis and assembly of adenovirus peptide III, reversible inhibition of hexon assmebly in adenovirus type 5 temperature-sensitive mutants. Virology 66:10–24. doi:10.1016/0042-6822(75)90175-0
Cepko CL, Sharp PA (1982) Assembly of adenovirus major capsid protein is mediated by a nonvirion protein. Cell 31:407–415. doi:10.1016/0092-8674(82)90134-9
Athappilly FK, Murali R, Rux JJ, Cai Z, Burnett RM (1994) The refined crystal structure of hexon, the major coat protein of adenovirus type 2, at 2.9 Å resolution. J Mol Biol 242:430–455. doi:10.1006/jmbi.1994.1593
Rux JJ, Burnett RM (2000) Type-specific epitope locations revealed by X-ray crystallographic study of adenovirus type 5 hexon. Mol Ther 1:18–30. doi:10.1006/mthe.1999.0001
Cepko CL, Sharp PA (1983) Analysis of Ad5 hexonand 100 K ts mutants using conformation specific monoclonal antibodies. Virology 129:137–154
Gambke C, Deppert W (1983) Specific complex of the late nonstructural 100,000-Da protein with newly synthesized hexon in adenovirus type 2-infected cells. Virology 124:1–12
Morin N, Boulanger P (1984) Morphogenesis of human adenovirus type 2, sequence of entry of proteins into previral and viral particles. Virology 136:153–167
Stinski MF, Ginsberg HS (1974) Antibody to the type 5 adenovirus hexon polypeptide, detection of nascent polypeptides in the cytoplasm of infected KB cells. Intervirology 4:226–236
Kauffman RS, Ginsberg HS (1976) Characterization of a temperature-sensitive, hexon transport mutant of type 5 adenovirus. J Virol 19(2):643–658
Wodrich H, Guan T, Cingolani G, Von Seggern D, Nemerow G, Gerace L (2003) Switch from capsid protein import to adenovirus assembly by cleavage of nuclear transport signals. EMBO J 22:6245–6255. doi:10.1093/emboj/cdg614
Dolph PJ, Racaniello V, Villamarin A, Palladino F, Schneider RJ (1988) The adenovirus tripartite leader may eliminate the requirement for cap-binding protein complex during translation initiation. J Virol 62(6):2059–2066
Cuesta R, Xi Q, Schneider RJ (2000) Adenovirus specific translation by selective disassembly of cap-initiation complex eIF4F. EMBO J 19:3465–3474. doi:10.1093/emboj/19.13.3465
Xi Q, Cuesta R, Schneider RJ (2004) Tethering of eIF4G to adenoviral mRNAs by viral 100 k protein drives ribosome shunting. Genes Dev 18(16):1997–2009. doi:10.1101/gad.1212504
Yueh A, Schneider RJ (1996) Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev 10(12):1557–1567. doi:10.1101/gad.10.12.1557
Yueh A, Schneider RJ (2000) Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes Dev 14(4):414–421. doi:10.1101/gad.14.4.414
Cuesta R, Xi Q, Schneider RJ (2001) Preferential translation of adenovirus mRNAs in infected cells. Cold Spring Harb Symp Quant Biol 66:259–267 Review
Andrade F, Bull HG, Thornberry NA, Ketner GW, Casciola-Rosen LA, Rosen A (2001) Adenovirus L4–100 K assembly protein is a granzyme B substrate that potently inhibits granzyme B-mediated cell death. Immunity 14:751–761. doi:10.1016/S1074-7613(01)00149-2
Andrade F, Fellows E, Jenne DE, Rosen A, Young CS (2007) Granzyme H destroys the function of critical adenoviral proteins required for viral DNA replication and granzyme B inhibition. EMBO J 26(8):2148–2157. doi:10.1038/sj.emboj.7601650
Hodges B, Evans HK, Everett RS, Ding EY, Serra D, Amalfitano A (2001) Adenovirus vectors with the 100 K gene deleted and their potential for multiple gene therapy applications. J Virol 75:5913–5920. doi:10.1128/JVI.75.13.5913-5920.2001
Nathan DG, Gunn RB (1966) Thalassemia: the consequences of unbalanced hemoglobin synthesis. Am J Med 41:815–830. doi:10.1016/0002-9343(66)90039-8
Brunori M, Falcino G, Fioreti E, Giardina B, Rotilio G (1975) Formation of superoxide in the autoxidation of the isolated α and β chains of human hemoglobin and its involvement in hemichrome precipitation. Eur J Biochem 53:99–104. doi:10.1111/j.1432-1033.1975.tb04046.x
Kihm AJ, Kong Y, Hong W, Russell JE, Rouda S, Adachi K, Simon MC, Blobel GA, Weiss MJ (2002) An abundant erythroid protein that stabilizes free α-haemoglobin. Nature 417:758–763. doi:10.1038/nature00803
Feng LL, Gell DA, Zhou S, Gu LQ, Kong YC, Li J, Hu MJ, Yan N, Lee C, Rich AM, Armstrong RS, Lay PA, Gow AJ, Weiss MJ, Mackay JP, Shi YP (2004) Molecular mechanism of AHSP-mediated stabilization of α-hemoglobin. Cell 119(5):629–640. doi:10.1016/j.cell.2004.11.025
Yu X, Kong Y, Doré LC, Abdulmalik O, Katein AM, Zhou S, Cho JK, Gell DA, Mackay JP, Gow AJ, Weiss MJ (2007) An erythroid chaperone that facilitates folding of α-globin subunits for hemoglobin synthesis. J Clin Invest 117:1856–1865. doi:10.1172/JCI31664
Kong Y, Zhou S, Kihm AJ, Katein AM, Yue X, Gell DA, Mackay JP, Adachi K, Foster-Brown L, Louden CS, Gow AJ, Weiss MJ (2004) Loss of α-hemoglobin-stabilizing protein impairs erythropoiesis and exacerbates β-thalassemia. J Clin Invest 114(10):1457–1466. doi:10.1172/JCI21982
Brillet T, Baudin-Creuza V, Vasseur C, Domingues-Hamdi E, Kiger L, Wajcman H, Pissard S, Marden MC (2010) Alpha-hemoglobin stabilizing protein (AHSP), a kinetic scheme of the action of a human mutant, AHSPV56G. J Biol Chem 285(23):17986–17992. doi:10.1074/jbc.M109.098491
Gell DA, Kong Y, Eaton SA, Weiss MJ, Mackay JP (2002) Biophysical characterization of the α-globin binding protein α-hemoglobin stabilizing protein. J Biol Chem 277(43):40602–40609. doi:10.1074/jbc.M206084200
Feng L, Zhou S, Gu L, Gell DA, Mackay JP, Weiss MJ, Gow AJ, Shi Y (2005) Structure of oxidized alpha-haemoglobin bound to AHSP reveals a protective mechanism for haem. Nature 435(7042):697–701. doi:10.1038/nature03609
Gell DA, Feng L, Zhou S, Jeffrey PD, Bendak K, Gow A, Weiss MJ, Shi Y, Mackay JP (2009) A cis-proline in alpha-hemoglobin stabilizing protein directs the structural reorganization of alpha-hemoglobin. J Biol Chem 284(43):29462–29469. doi:10.1074/jbc.M109.027045
Galanello R, Perseu L, Giagu N, Sole G, Perra C (2003) AHSP expression in β-thalassemia carriers with thalassemia intermedia phenotype. Blood 102 Abstract 1881
Lacerra G, Scarano C, Musollino G, Flagiello A, Pucci P, Carestia C (2008) Hb Foggia or alpha 117(GH5)Phe → Ser: a new alpha 2 globin allele affecting the alpha Hb-AHSP interaction. Haematologica 93(1):141–142. doi:10.3324/haematol.11789
Turbpaiboon C, Limjindaporn T, Wongwiwat W, U-Pratya Y, Siritanaratkul N, Yenchitsomanus PT, Jitrapakdee S, Wilairat P (2006) Impaired interaction of α-haemoglobin-stabilising protein with α-globin termination mutant in a yeast two-hybrid system. Br J Haematol 132:370–373. doi:10.1111/j.1365-2141.2005.05865.x
Yu X, Mollan TL, Butler A, Gow AJ, Olson JS, Weiss MJ (2009) Analysis of human alpha globin gene mutations that impair binding to the alpha hemoglobin stabilizing protein. Blood 113(23):5961–5969. doi:10.1182/blood-2008-12-196030
Faber PW, Barnes GT, Srinidhi J, Chen J, Gusella JF, MacDonald ME (1998) Huntingtin interacts with a family of WW domain proteins. Hum Mol Genet 7(9):1463–1474. doi:10.1093/hmg/7.9.1463
Raychaudhuri S, Majumder P, Sarkar S, Giri K, Mukhopadhyay D, Bhattacharyya NP (2008) Huntingtin interacting protein HYPK is intrinsically unstructured. Proteins 71(4):1686–1698. doi:10.1002/prot.21856
Raychaudhuri S, Sinha M, Mukhopadhyay D, Bhattacharyya NP (2008) HYPK, a Huntingtin interacting protein, reduces aggregates and apoptosis induced by N-terminal Huntingtin with 40 glutamines in Neuro2a cells and exhibits chaperone-like activity. Hum Mol Genet 17(2):240–255. doi:10.1093/hmg/ddm301
Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, Bates GP (1996) Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87:493–506. doi:10.1016/S0092-8674(00)81369-0
Otto H, Conz C, Maier P, Wölfle T, Suzuki CK, Jenö P, Rücknagel P, Stahl J, Rospert S (2005) The chaperones MPP11 and Hsp70L1 form the mammalian ribosome-associated complex. Proc Natl Acad Sci USA 102:10064–10069. doi:10.1073/pnas.0504400102
Fox J, Connor T, Stiles M, Kama J, Lu Z, Dorsey K, Liebermann G, Sapp E, Cherny R, Banks M, Volitakis I, Difiglia M, Berezovska O, Bush A, Hersch S (2011) Cysteine oxidation within N-terminal mutant huntingtin promotes oligomerization and delays clearance of soluble protein. J Biol Chem 286:18320–18330. doi:10.1074/jbc.M110.199448
Arnesen T, Starheim KK, Van Damme P, Evjenth R, Dinh H, Betts MJ, Ryningen A, Vandekerckhove J, Gevaert K, Anderson D (2010) The chaperone-like protein HYPK acts together with NatA in cotranslational N-terminal acetylation and prevention of Huntingtin aggregation. Mol Cell Biol 30(8):1898–1909. doi:10.1128/MCB.01199-09
Ellis RJ (2005) Molecular chaperones: the orthodox view. In: Pockley AG, Henderson B (eds) Molecular chaperones and cell signalling. Cambridge University Press, London, pp 3–21
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Szolajska, E., Chroboczek, J. Faithful chaperones. Cell. Mol. Life Sci. 68, 3307–3322 (2011). https://doi.org/10.1007/s00018-011-0740-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-011-0740-4