Abstract
The mitogen-activated protein kinase-activated protein kinase-5 (MK5) resides predominantly in the nucleus of resting cells, but p38MAPK, extracellular signal-regulated kinases-3 and -4 (ERK3 and ERK4), and protein kinase A (PKA) induce nucleocytoplasmic redistribution of MK5. The mechanism by which PKA causes nuclear export remains unsolved. In the study reported here we demonstrated that Ser-115 is an in vitro PKA phosphoacceptor site, and that PKA, but not p38MAPK, ERK3 or ERK4, is unable to redistribute MK5 S115A to the cytoplasm. However, the phosphomimicking MK5 S115D mutant resides in the cytoplasm in untreated cells. While p38MAPK, ERK3 and ERK4 fail to trigger nuclear export of the kinase dead T182A and K51E MK5 mutants, S115D/T182A and K51E/S115D mutants were able to enter the cytoplasm of resting cells. Finally, we demonstrated that mutations in Ser-115 affect the biological properties of MK5. Taken together, our results suggest that Ser-115 plays an essential role in PKA-regulated nuclear export of MK5, and that it also may regulate the biological functions of MK5.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mitogen-activated protein kinase (MAPK) pathways mediate cellular responses to a wide variety of extracellular signalling molecules. In mammalian cells, they participate in the regulation of processes such as proliferation, differentiation, survival/apoptosis, development, metabolism, motility, and gene expression. Seven distinct mammalian MAPK pathways have been identified. The typical MAPK pathways, represented by the ERK1/2, ERK5, JNK, and p38MAPK modules consist of a cascade of three consecutive phosphorylation events exerted by a MAPK kinase kinase (MAPKKK), a MAPK kinase (MAPKK), and a MAPK. The atypical MAPKs ERK3/4, ERK7 and ERK8 represent the less-characterized MAPK pathways [1–9]. MAPK not only target nonprotein kinase substrates, but they can also phosphorylate other protein kinases designated as MAPK-activated protein kinases (MAPKAPK). The MAPKAPKs include the ribosomal-S6-kinases (RSK1-4), the MAPK-interacting kinases (MNK1 and 2), the mitogen- and stress-activated kinases (MSK1 and 2), and the MAPKAPK (MK2, 3 and 5) subfamilies [10–15].
MK5 and its human homologue p38-regulated/activated protein kinase (PRAK) were originally described as in vitro p38 substrates that could be activated by p38 through phosphorylation of Thr-182 [16–20]. The biological functions of MK5 are poorly understood, as MK5 knockout mice bred onto different backgrounds display either no obvious phenotype or embryonic lethality [10, 20]. Recent studies have demonstrated that MK5 is involved in ras-induced senescence, tumour suppression, inhibition of cell proliferation, rearrangements of the cytoskeleton, and anxiety-related behaviour [18, 21–25], and have identified ERK3, ERK4, p53, Hsp27 and 14-3-3ε as bona fide substrates. [20, 21, 25–29]. MK5-mediated inhibition of proliferation may depend on MK5’s ability to stimulate the transcriptional activity of p53, and subsequent increased expression of the cyclin-dependent protein kinase inhibitor p21Cip1/Waf1, a target gene of p53 [21, 24]. MK5 seems to exert its role in F-actin dynamics through phosphorylation of Hsp27, an event in part controlled by 14-3-3ε [25, 29]. The biological relevance of the other interactions remains incompletely understood.
Several groups have shown that although endogenous and ectopically expressed MK5 predominantly reside in the nucleus of resting cells, MK5 actually shuttles between the nucleus and the cytoplasm [19, 20, 22, 26–28, 30]. The opposing activities of a functional nuclear export signal (NES) and a nuclear localization signal (NLS) may determine the subcellular distribution of MK5. Different studies have demonstrated that the interaction with other proteins influences the accessibility of the NLS/NES motifs to the import/export system and therefore regulates the location of MK5 [19, 30]. MK5 contains a putative p38MAPK docking site that overlaps with the NLS (Fig. 1) and in vitro interaction studies have confirmed that MK5 can interact with p38α and p38β [19, 24, 30–32]. It is still controversial whether p38MAPK and MK5 form direct contact in vivo as they seem to interact very weakly in yeast two-hybrid assays [20, 26], while tandem affinity purification studies have failed to detect MK5-p38MAPK complexes in HEK 293 cells [33]. However, both Tanoue et al. and Li et al. were able to immunoprecipitate MK5-p38MAPK complexes from NIH3T3 cells overexpressing these proteins [24, 32]. Stimulation of endogenous p38MAPK with known activators such as sorbitol, sodium arsenite and TNFα, or overexpression of p38MAPK causes subcellular redistribution of MK5. Overexpression of a p38MAPK mutant unable to bind MK5 fails to redistribute MK5, and similarly mutation in the p38MAPK docking of MK5 abrogates p38MAPK-mediated subcellular redistribution of the protein [19, 24, 30]. These observations suggest that localization of MK5 is affected by physical interaction with p38MAPK [19, 24, 30]. The atypical MAPKs ERK3 and ERK4 can interact with MK5 in vitro and in vivo and overexpression of both proteins has been shown to retain MK5 in the cytoplasm. Binding of ERK3 or ERK4 to MK5 is required for its nuclear export [20, 26–28, 34]. The conformation of MK5 may also influence its subcellular location. In such a scenario, phosphorylation of Thr-182 may induce allosterical changes in MK5, thereby exposing the NES and triggering nuclear export of the phosphoMK5 [10, 19].
Schematic representation of the structure of MK5. The protein consists of 473 amino acids. The catalytic domain and the NES and NLS are shown. The phosphoacceptor site in the activation loop (T182) is indicated, as well as the serine residue (S115) that was identified in this work as a PKA site. The primary sequence of the NES and NLS are given. The p38 docking site, which overlaps with the NLS, is indicated
We have recently shown that activation of the cAMP/PKA signalling pathway or overexpression of the catalytic Cα subunit induces nuclear export of MK5. The kinase activities of both PKA and MK5 are required for nucleocytoplasmic translocation. PKA can phosphorylate MK5, but the target sites have not been mapped, nor has the effect of mutations in the putative phosphorylation sites on the location of the protein been investigated [22]. Here we report that PKA can phosphorylate MK5 at Ser-115 and that mutations in this site interfere with the subcellular distribution of the protein ruled by PKA, but not by p38MAPK, ERK3 or ERK4. Hence, PKA-mediated nuclear export of MK5 differs from the mechanism described for the other interaction partners of MK5.
Materials and methods
Materials
Forskolin and sodium arsenite were purchased from Sigma-Aldrich (St. Louis, MO). Lipofectamine, cell culture medium and serum were obtained from Invitrogen Life Technologies (Carlsbad, CA). Phospho-PKA substrate (RRXS/T) antibody and Myc-tag Alexa Fluor 647 conjugate mouse monoclonal antibody were obtained from Cell Signaling (catalogue number 9624 and 2233, respectively; Danvers, MA). Anti-PRAK A-7 antibodies (catalogue number sc-46667) and anti PKA-Cα C-20 antibodies (catalogue number sc-903) were both from Santa Cruz (Santa Cruz, CA), while anti-actin antibody was purchased from Sigma-Aldrich (catalogue number A2066). The alkaline phosphatase-conjugated secondary antibodies and the oligonucleotides were from Sigma. CDP-star and MagicMark western standard were from Applied Biosystems (CA) and Invitrogen Life Technologies, respectively. Activated PRAK was from Millipore Upstate (Billerica, MA), while purified PKA was from Promega Corporation (Madison, WI). Leptomycin B was from Sigma-Aldrich (St. Louis, MO). The MK5 GGELFHRISQ peptide and the MK5 S115A GGELFHRIAQ peptide were synthesized by Johannes Eksteen (Institute for Pharmacy, University of Tromsø). The F-actin probe Alexa Fluor 594 Phalloidin was purchased from Invitrogen Life Technologies (catalogue number A12381).
Plasmids
The plasmids encoding MKK6 E/E, p38MAPK, and the EGFP fusion proteins with wild-type MK5 and the MK5 mutants K51E, T182A and L337A have all been previously described [19, 26]. Expression plasmids for pEGFP-MK5 S115A and pEGFP-MK5 S115D were generated by site-directed mutagenesis using pEGFP-MK5 DNA as template. Only one strand of the primers used in the mutagenesis reaction is given (the mutated nucleotides are underlined): S115A, 5′-GTC-TGT-GTG-AAG-AAA-GCC-ACT-CAA-GAA-CGG-3′; S115D, 5′-GTC-TGT-GTG-AAG-AAA-GAC-ACT-CAA-GAA-CGG-3′. The pEGFP-MK5 S115D/T182A expression plasmid was generated by site-directed mutagenesis using pEGFP-MK5 T182A as template and the S115D primers mentioned above. Plasmid pEGFP-MK5 S115D/L337A was obtained by site-directed mutagenesis using pEGFP-MK5 L337A as template and the S115D primers. Plasmid pEGFP-MK5 K51E/S115D was generated by site-directed mutagenesis of pEGFP-MK5 K51E [19] with the S115D primers described above. The GST-MK5102–155 expression plasmid was generated by amplifying the coding region of MK5 spanning amino acid residues 102–155 using the primers 5′-CTC GAC TGT TAA ATG GAT CCG AGA TGA TGG AAG-3′ and 5′-CAG AGA GTT ATC GTC GAC AAG CAG ATT TCC AGG-3′. The PCR fragment was thereafter digested with BamHI and SalI and ligated into the BamHI and XhoI sites of the GST vector pGEX-4T-1 (GE Healthcare). All plasmids were verified by sequencing and the expression levels of all EGFP-MK5 mutants were similar to that of wild-type EGFP-MK5 (results not shown). The pEGFP-MK2 plasmid was kindly provided by Dr. Matthias Gaestel [35]. The expression vector pEGFP-MK2insD was obtained by site-directed mutagenesis using the complementary primers (only one primer is given): 5′-GCT-CTT-TAG-TCG-AAT-CGA-TCA-GGA-CCG-AGG-AGA-C-3′. The chimeric EGFP-MK2/mk5, EGFP-MK2/mk5 S115D, and EGFP-MK2/mk5 S115A were created by site-directed mutagenesis. First, EGFP-MK2/mk5 S115D was generated using the complementary oligonucleotides (only one strand is given): 5′-GGT-GGA-GAG-CTC-TTT-CAT-CGA-ATC-GAT-CAG-CAC-CGA-CAC-TTC-ACA-GAA-AGA-GAG-GCG-TCA-GAG-ATC-ATG-AAG-3′. This construct was then used to obtain EGFP-MK2/mk5 and EGFP-MK2/mk5 S115A using the primers (only one strand is shown): 5′-CTC-TTT-CAT-CGA-ATC-AGC-CAG-CAC-CGA-CAC-TTC-3′ and 5′-CTC-TTT-CAT-CGA-ATC-GCT-CAG-CAC-CGA-CAC-TTC-3′, respectively. All plasmid constructs were verified by sequencing. The ERK3 expression plasmid has been previously described [36], while the ERK4 expression plasmid was a generous gift from Dr. Ole Morten Seternes [28].
Cell culture and transfection
PC12 cells were purchased from the European Collection of Cell Cultures (ECACC; Salisbury, UK). The cells were maintained in RPMI 1640 supplemented with 10% horse serum (Gibco) and 5% fetal bovine serum, 2 mM l-glutamine, penicillin (110 U/ml) and streptomycin (100 μg/ml). Cells were transfected using a Nucleofection kit (Amaxa) according to the manufacturer’s instructions. HEK293 cells (ATCC CRL-1573) and HeLa cells (ATCC CCL-2) were purchased from the American Type Culture Collection (LGC Standards, Boras, Sweden) and maintained in Eagle’s minimum essential medium supplemented with 10% fetal calf serum, 2 mM l-glutamine (110 U/ml) and streptomycin (100 μg/ml). HeLa cells also received 1× nonessential amino acids (Invitrogen).
Western blotting
For detection of coimmunoprecipitates and MK5, samples were analysed by SDS-PAGE NuPage 4–12% Bis-Tris SDS-PAGE (Invitrogen) according to the manufacturers protocol and blotted onto a 0.45-μm PVDF membrane (Millipore, Billerica, MA). Immunoblotting was performed by first blocking the membrane with phosphate-buffered saline (PBS) with 0.1% Tween-20 (PBS-T; Sigma-Aldrich) containing 10% (w/v) dried skimmed milk for 1 h and probed with the appropriate primary antibody. After three washes, the membrane was incubated with the appropriate secondary antibody for 1 h. After four washes, antigen–antibody complexes were visualized using CDP Star (Tropix, Bedford, MA) substrate and a Lumi-Imager F1 from Roche (Basel, Switzerland).
Cell staining and microscopy
Cells were rinsed twice with PBS and fixed for 10 min with 4% paraformaldehyde. Next, the cells were washed twice with PBS and then permeabilized for 10 min with 0.1% Triton X-100. The cells were then washed with PBS and examined using confocal laser-scanning Zeiss LSM 510 META and Leica SP5 microscopes. Cell nuclei were visualized by DRAQ5 staining (Biostatus, Shepshed, UK). Routinely 50 cells or more were examined for subcellular localization, and representative images are shown in the Results. The expression plasmid for the HSP27-3A mutant has been previously described [25].
GST-fusion protein purification
GST fusion proteins were purified from Escherichia coli BL21 extracts using glutathione–agarose beads as previously described [19], and the GST moiety was removed from the GST beads using thrombin according to the instructions of the manufacturer. Protein concentrations were measured using a Bio-Rad Protein Assay kit (Bio-Rad Laboratories, Hercules, CA).
In vitro kinase assays
Peptide arrays were synthesized on cellulose paper using a MultiPep automated multiple peptide synthesizer (INTAVIS Bioanalytical Instruments). For phosphorylation, membranes were wetted briefly in ethanol prior to incubation in phosphorylation buffer (20 mM Hepes, pH 7.4, 100 mM NaCl, 5 mM MgCl2, 1 mM DTT and 0.2 mg/ml BSA) for 1 h at room temperature. The array was then incubated at 4°C overnight in blocking buffer (phosphorylation buffer containing 1 mg/ml BSA and 100 μM ATP). Phosphorylation was initiated by incubating the membrane in the presence of 70 U recombinant PKA catalytic subunit (Promega) in 20 ml phosphorylation buffer containing 50 μM ATP and 0.37 MBq [γ-32P]ATP for 30 min at 30°C with agitation. The membrane was washed extensively in 1 M NaCl followed by washes in 5% H3PO4 and dried down [37]. The phosphorylated membrane was then visualized using a BioImaging Analyser (FUJIFILM BAS-5000). MK5102–155 by PKA was phosphorylated in vitro as previously described [22]. For competition, 25-, 2.5, and 0.25-fold molar excesses of peptide were used.
Transient transfection studies and luciferase assay
For transient transfection studies, HEK293 cells were plated at 3 × 105 cells per well in a six-well plate and plasmid DNA was transfected using lipofectamine. The plasmids pGAL4-E1b-LUC and pGAL-p53 have been previously described [38]. Luciferase activity was monitored using a Promega Luciferase 1000 assay system as described previously [22]. Each transfection was performed independently in triplicate and the experiments were repeated four times.
Results
PKA phosphorylates MK5 at Ser-115 in vitro
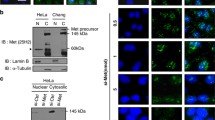
Our previous studies had shown that MK5 specifically interacts with the catalytic subunit of PKA and that PKA can phosphorylate MK5 in vitro. Active PKA also causes MK5 to translocate from the nucleus to the cytoplasm and this process requires the activity of both kinases. The PKA phosphoacceptor site(s) on MK5 were, however, not mapped [22]. Computational prediction revealed that serine residue 115 might be a putative PKA site. This site is encompassed in the HRIS/T motif, which remotely resembles the PKA phosphorylation motif RRXS/T [39]. Ser-115 is not conserved in MK2 and MK3 (Fig. 2a) and may explain why we did not find phosphorylation of MK2 by PKA in vitro [22]. To check whether Ser-115 of MK5 could be phosphorylated by PKA, we first performed an in vitro kinase assay using a peptide array. By this method, we were indeed able to confirm that PKA phosphorylated a peptide encompassing Ser-115 in vitro (Fig. 2a). In addition to phosphorylation of this peptide, we observed weaker phosphorylation of other peptide sequences on the membrane. Next, we examined whether purified PKA could phosphorylate recombinant MK5. As MK5 possesses autophosphorylation activity which would interfere with the interpretation of the data, we generated a MK5 fragment encompassing residues 102–155 as a GST fusion protein and removed the GST moiety by cleavage with thrombin. The purified catalytic subunit of PKA (Cα) clearly phosphorylated the MK5102–155 fragment (Fig. 2b). This phosphorylation was competed out by a MK5 peptide fragment containing Ser-115, but not by a corresponding fragment in which Ser-115 had been replaced by Ala (Fig. 2b). These results indicate that Ser-115 represents an in vitro PKA acceptor site.
PKA phosphorylates MK5 at serine residue 115 in vitro. a In vitro kinase assay of a peptide array membrane that was spotted with three amino acid overlapping oligopeptides 20 amino acids long that spanned the complete MK5 protein. The autoradiographic exposure is shown on the left and the phosphorylated peptide is encircled. The right panel represents the Ponceau-stained membrane used for the peptide kinase assay. The peptide spot corresponding to the phosphorylated peptide is encircled. The sequence of the oligopeptide that was vividly phosphorylated by PKA is given with serine residue 115 indicated in bold. Comparison of the corresponding sequences of MK2 and MK3 reveals that S115 is not conserved in MK2 and MK3. The identical amino acids in the three proteins are underlined. b In vitro kinase assay on recombinant MK5102–155 fragment. GST-MK5102–155 fusion protein was purified from E. coli and the MK5 moiety was released by thrombin. The MK5 fragment was incubated for 30 min at 30°C with Cα in the presence of [γ-32P]ATP. Decreasing amounts (25, 2.5, and 0.25 M excess) of wild-type or S115A mutant competitor peptide encompassing amino acid residues 107–116 were added. Proteins were separated by polyacrylamide gel electrophoresis and phosphorylation was visualized by autoradiography. Bottom panel Coomassie blue staining ensured equal loading of the proteins in each sample. The molecular masses (in kilodaltons) of the protein marker are indicated
Ser-115 is necessary for PKA-induced nucleocytoplasmic distribution of MK5
Because activation of the cAMP/PKA pathway by the cAMP-elevating agent forskolin has been shown to trigger a transient nuclear export of MK5 [22], we wanted to investigate whether Ser-115 is implicated in this process. We therefore generated an EGFP-MK5 S115A mutant and compared its subcellular distribution in resting and forskolin-treated PC12 cells. In resting cells, this mutant resided predominantly in the nucleus (Fig. 3a). In contrast to wild-type MK5, forskolin did not cause nuclear export of EGFP-MK5 S115A (Fig. 3b). Similar results were obtained in HeLa cells (results not shown). To exclude the possibility that activated PKA failed to cause cytoplasmic redistribution of MK5 S115A because the catalytic Cα subunit could no longer interact with this mutant, we performed coimmunoprecipitation studies. HEK293 cells were transfected with expression plasmids for EGFP-MK5 or EGFP-MK5 S115A and the cells were left untreated or stimulated with forskolin for 30 min. EGFP-MK5 fusion proteins were immunoprecipitated with antibodies directed against MK5 and the immunoprecipitates were challenged for the presence of Cα by using anti-Cα antibodies. Our results showed that wild-type MK5 and MK5 S115A interacted with Cα and that no major differences in interaction were observed as judged by the intensity of the coimmunoprecipitated complexes (Fig. 4). Taken together, these findings suggest that Cα can still interact with MK5 S115A but cannot direct nuclear export of this mutant. To further assess whether phosphorylation of Ser-115 is necessary for PKA-induced nuclear exclusion of MK5, we generated the MK5 S115D mutant, which mimics constitutive phosphorylation, and monitored its subcellular location. This phosphomimicking substitution caused EGFP-MK5 S115D to reside also in the cytoplasm of untreated cells (Figs. 3a, d and 6). These results support the assumption that PKA-mediated phosphorylation of MK5 at Ser-115 is implicated in nuclear export of MK5.
Phosphomimicking mutation of Ser-115 affects the subcellular distribution of EGFP-MK5 fusion protein in resting cells. a PC12 cells were transfected with expression plasmids for EGFP-tagged MK5 (top panels), EGFP-tagged MK5 S115A (middle panels), or EGFP-tagged MK5 S115D (bottom panels) and left untreated. b PC12 cells were transfected with expression plasmids for EGFP-MK5 or EGFP-MK5 S115A fusion proteins. Cells were exposed 24 h after transfection to 10 μM forskolin for 30 min and were then fixed. c PC12 cells were transfected with expression plasmids for EGFP-MK5 S115D/T182A and left untreated. d Cells transfected with plasmids encoding EGFP-tagged wild-type MK5, mutant MK5 S115D, MK5 K51E, or MK5 K51E/S115D. The subcellular location of ectopically expressed MK5 variants was monitored by EGFP fluorescence (green channel), while the cell nuclei were visualized by DRAQ5 staining (blue channel). On average 50 cells expressing EGFP-MK5 were analysed and representative images are shown. Similar results were obtained in independent experiments
Substitution of Ser-115 into Ala does not abrogate the interaction between the catalytic subunit of PKA (Cα) and MK5. HEK293 cells were transfected with expression plasmids for EGFP-MK5 or EGFP-MK5 S115A and then either left untreated or stimulated with 10 μM forskolin for 30 min. EGFP-MK5 fusion proteins were immunoprecipitated (IP) from cell lysates using anti-PRAK antibodies. These antibodies are directed against the human homologue of MK5 and crossreact with MK5. The immunoprecipitates were then examined for the presence of Cα by western blot (WB) with anti-Cα antibodies (lanes 5–8). The presence of Cα in the immunocomplexes is indicated by the arrow. Total cell lysates (input control) are shown in lanes 1–4. Bottom panel: to ensure equal loading and expression of EGFP-MK5 and EGFP-MK5 S115A, membranes were stripped and probed with anti-PRAK antibodies. The position of EGFP-MK5 (EGFP-MK5 S115A) is indicated by the arrow. The molecular mass (in kilodaltons) of the protein marker is given
Chimeric MK2 with the S115 motif behave as MK5
Both MK2 and MK5 reside predominantly in the nucleus of untreated cells [19, 22, 27, 35]. However, we have previously shown that PKA does not interact with nor phosphorylate MK2 [22]. Moreover, we found that forskolin did not induce nuclear export of the EGFP-MK2 fusion protein (Fig. 5a). Interestingly, Ser-115 in MK5 is encompassed in the motif RISQ, while the corresponding sequence in MK2 and MK3 is RIQ, lacking an equivalent of Ser-115 (Fig. 5b). To evaluate the importance of this Ser-115 in PKA-induced nuclear export of MK5, we reasoned that a MK2 mutant with an insertion of Asp as a phosphomimicking Ser-115 in the RIQ motif (= MK2insD) may behave identically to MK5 S115D, i.e. with both nuclear and cytoplasmic localizations in untreated cells. Contrary to MK5 S115D, the EGFP-MK2insD mutant remained exclusively in the nucleus of HeLa cells (Fig. 5c) and HEK293 cells (results not shown). Alignment of human and mouse MK5, MK2, and MK3 revealed differences in residues C-terminal to Ser-115 (Fig. 5b). To examine the possible contribution of these residues in PKA-triggered nuclear export, we constructed chimeric MK2 containing the wild-type MK5, and MK5 S115D sequences, to generate the fusion proteins EGFP-MK2/mk5 and EGFP-MK2/mk5 S115D, respectively. EGFP-MK2/mk5 S115D behaved as EGFP-MK5 S115D because it was detected in both the nucleus and cytoplasm of untreated cells (Fig. 5d). Surprisingly, EGFP-MK2/mk5 was found in both the nucleus and the cytoplasm, but was most obvious in the cytoplasm. However, forskolin treatment caused complete nuclear exclusion of EGFP-MK2/mk5 (Fig. 5d). Taken together, these results indicate that Ser-115 is necessary but not sufficient to control subcellular distribution of MK5 in response to an activated PKA pathway. Amino acids C-terminal to Ser-115 also contribute to PKA-regulated subcellular distribution of MK5.
Subcellular localization of MK2 and MK2/mk5 hybrids. a EGFP-MK2 resides predominantly in the nucleus of untreated cells and this subcellular localization is not changed upon activation of the cAMP/PKA pathway by forskolin. Cells were treated with 50 μM forskolin (FSK) for 30 min. b Alignment of the region spanning Ser-115 in MK5 with the corresponding sequence in MK2. The insertion of Asp (d) is highlighted in grey. c Insertion of a phosphomimicking Asp in MK2 that corresponds to Ser-115 in MK5 does not affect the subcellular localization of this MK2insD mutant. d Swapping the MK2 domain with the MK5 motif encompassing Ser-115 changes the subcellular localization of the MK2 from almost exclusively nuclear to both cytoplasmic and nuclear for the MK2/mk5 hybrid protein (top panels). Forskolin triggers nuclear exclusion of MK2/mk5 (middle panels). The MK2/mk5 S115D mutant resides predominantly in the cytoplasm of untreated cells (bottom panels). The EGFP fusion proteins were visualized as described in the legend for Fig. 3. On average 50 cells expressing EGFP-MK5 were analysed and representative images are shown. Similar results were obtained in independent experiments
The PKA phosphomimicking S115D mutation restores cytoplasmic shuttling of kinase dead MK5 mutants
Activated PKA was unable to induce nuclear export of the kinase dead MK5 T182A mutant, which carries a substitution in its activation loop [22]. This may indicate that the kinase activity of MK5 is absolutely required for subcellular translocation by PKA. The finding that another kinase dead MK5 mutant (MK5 K51E) with a mutation in the ATP-binding domain also remained in the nucleus after forskolin treatment supports this assumption [19, 22]. This prompted us to examine whether phosphomimicking of Ser-115 could rescue nuclear export of the kinase dead MK5 T182A and K51E mutants. We generated the MK5 S115D/T182A mutant and subcellular localization studies of this mutant revealed a cytosolic location in untreated cells (Fig. 3c). As could the MK5 S115D/T182A mutant, the double MK5 K51E/S115D mutant was able to enter the cytoplasm in untreated cells (Fig. 3d). Thus activated PKA is unable to translocate kinase dead MK5 out of the nucleus, yet kinase dead MK5 mutants carrying the phosphomimicking S115D substitution can leave the nucleus (see “Discussion”).
PKA-induced nuclear export of MK5 depends on CRM1
Our earlier study showed that stress-induced nuclear export of MK5 relies on a CRM1/NES-dependent mechanism [19]. To test whether PKA-induced nuclear export of MK5 is dependent on CRM1, we treated cells with leptomycin B, a selective inhibitor of CRM-1, prior to the addition of forskolin. While forskolin caused nuclear export of EGFP-MK5, leptomycin B prevented this forskolin-triggered export (Fig. 6a). Leptomycin B exposure also strongly reduced the predominant cytoplasmic localization of the EGFP-MK5 S115D mutant in resting cells (Fig. 6a). Substitution of residue Leu-337 into Ala in the NES generated a MK5 mutant that was unable to shuttle into the cytoplasm [19], while this study demonstrated that the MK5 S115D resides mainly in the cytoplasm (see Figs. 3a and 6). To investigate the involvement of the NES of MK5 in the cytoplasmic distribution of MK5 S115D, we constructed the double EGFP-MK5 S115D/L337A mutant. In contrast to MK5 S115D, this mutant was unable to shuttle to the cytoplasm (Fig. 6b), suggesting that a functional NES is required for PKA-mediated nuclear exclusion of MK5. Taken together, these results indicate that nucleocytoplasmic translocation of MK5 by active PKA requires a functional CRM1/NES mechanism.
Both PKA-induced nucleocytoplasmic redistribution of MK5 and cytoplasmic residence of MK5 S115D are CRM1-dependent processes. a HeLa cells were transfected with EGFP-MK5 (panels A–I) or EGFP-MK5 S115D (panels J–O) and either pretreated with 5 ng/ml leptomycin B (LMB) for 1 h (panels G–I and M–O) or left untreated. Cells were then exposed to 10 μM forskolin (FSK) for 30 min (panels D–I and M–O) or left untreated (panels A–C and J–O). b HeLa cells were transfected with EGFP-MK5 S115D or EGFP-MK5 S115D/L337A expression plasmids. The latter contains a mutation in the NES which renders this motif nonfunctional [19]. The subcellular localization of the EGFP-MK5 fusion proteins was monitored as described in the legend of Fig. 3. On average 50 cells expressing EGFP-MK5 were analysed and representative images are shown. Similar results were obtained in independent experiments
ERK3- and ERK4-mediated nuclear export of MK5 does not require Ser-115
The atypical MAPK ERK3 and ERK4 are two genuine substrates for MK5. ERK4 is predominantly found in the cytoplasm, while ERK3 is distributed in the nucleus and cytoplasm [20, 26–28, 34, 40–42]. Coexpression of ERK3 (or ERK4) and MK5 in HeLa and HEK293 cells results in exclusive cytoplasmic localization of ERK3 (or ERK4) and MK5 [20, 26–28, 34, 40–42]. This ERK3-/ERK4-mediated nuclear export of MK5 does not depend on the kinase activity of MK5 as ectopic expression of both ERK3 and ERK4 causes nuclear exclusion of the kinase dead MK5 T182A mutant [20, 26–28]. To test whether Ser-115 is important for nucleocytoplasmic redistribution of MK5 by ERK3 and ERK4, we cotransfected cells with either wild-type MK5 or MK5 S115A and ERK3 or ERK4 and monitored the subcellular distribution of MK5. ERK3, as well as ERK4, was able to induce nuclear exclusion of both EGFP-MK5 and EGFP-MK5 S115A when coexpressed in HeLa cells (Fig. 7) or PC12 cells (results not shown). Thus ERK3- and ERK4-mediated nuclear export of MK5 is independent of the phosphorylation state of Ser-115.
Overexpression of ERK3 and ERK4 induces nuclear export of wild-type MK5 and the S115A mutant. HeLa cells were cotransfected with either expression plasmids for EGFP-MK5 or EGFP-MK5 S115A alone, or cotransfected with myc-tagged ERK4 (upper panels) or myc-tagged ERK3 expression plasmids (lower panels). The subcellular location of ectopic expressed EGFP-MK5 (or EGFP-MK5 S115A) was monitored by EGFP fluorescence (green channel) and the subcellular distribution of ERK3 (or ERK4) is visualized as the red channel using Alexa Fluor 647-conjugated Myc-tag mouse monoclonal antibody. Merged images show cytoplasmic colocalization of MK5 and ERK3 or ERK4. On average 50 cells expressing EGFP-MK5 were analysed and representative images are shown. Similar results were obtained in independent experiments
p38MAPK-mediated nuclear export of MK5 does not require Ser-115 phosphorylation
Studies by us and others have shown that stimuli that activate the p38MAPK pathway or activated p38MAPK mutants trigger cytoplasmic redistribution of MK5 [19, 24, 30]. This encouraged us to investigate whether the phosphorylation state of Ser-115 can interfere with p38MAPK-mediated subcellular localization of MK5. Exposure of cells to sodium arsenite, a potent activator of the p38MAPK pathway, resulted in nuclear export of EGFP-MK5 S115A (Fig. 8a). Similarly, coexpression of the constitutive active MKK6 E/E mutant and p38MAPK also caused an increase in cytoplasmic residence of EGFP-MK5 S115A (Fig. 8b). These findings strongly suggest that p38MAPK-regulated subcellular localization of MK5 is independent of Ser-115 phosphorylation.
Ser-115 is not implicated in p38MAPK-induced nuclear export of MK5. a HeLa cells transfected with EGFP-MK5 or EGFP-MK5 S115A plasmids were left untreated or exposed to sodium arsenite (250 μM for 120 min). b As in a, but cells were contransfected with plasmids encoding activated MKK6 (MKK6 E/E) and p38MAPK. The subcellular localization of the ectopically expressed MK5 variants was visualized as described in the legend for Fig. 3. On average 50 cells expressing EGFP-MK5 were analysed and representative images are shown. Similar results were obtained in independent experiments
Mutations in Ser-115 affect the biological properties of MK5
Because mutations in Ser-115 control the subcellular localization of MK5, we decided to examine the effect of these mutations on nuclear and cytoplasmic events involving MK5. MK5 has been shown to stimulate the transcriptional activity of p53 by direct phosphorylation of p53 at Ser-37 [21]. As transcription is a nuclear process, we investigated the effect of MK5 S115A, which is located in the nucleus, on p53-mediated transcription. HEK293 cells were therefore cotransfected with a luciferase reporter plasmid under the control of a promoter consisting of the TATA box of the adenovirus E1b gene and five binding motifs for GAL4, an expression plasmids for GAL4-p53 fusion protein, and plasmid DNA encoding EGFP-MK5, EGFP-MK5 S115A, or kinase dead EGFP-MK5 T182A. The last of these mutants also resides exclusively in the nucleus [22]. Substitution of Ser-115 into Ala reduced the ability of MK5 to activate p53-mediated transcription by >50% compared to wild-type MK5 (Fig. 9a). Similarly, MK5 T182A also displayed a reduced potential to stimulate the transcriptional activity of p53 (Fig. 9a).
Mutations in Ser-115 affect the biological properties of MK5. a HEK293 cells were cotransfected with the luciferase plasmid G5-E1b-LUC, a plasmid encoding GAL4-p53 fusion protein, and expression vectors for the fusion proteins EGFP-MK5 wild-type (MK5), EGFP-MK5 S115A (S115A) or kinase dead EGFP-MK5 T182A (T182A). Luciferase activity was determined in three independent parallel experiments and the p53 transcriptional activity in the presence of wild-type MK5 was arbitrarily set as 100% and the activity in the presence of the MK5 S115A or MK5 T182A was related to this. The experiment was repeated three times and similar results were obtained. b HEK293 cells were transfected with an expression plasmid for Hsp27 and with plasmids encoding either EGFP-MK5 (lane 1), EGFP-MK5 S115D (lane 2), EGFP-MK5 S115A (lane 3), or EGFP-MK5 L337A NES (lane 4). The last of these is a constitutive active MK5 that is exclusively localized in the cytoplasm. Phosphorylation of Hsp27 was monitored by western blotting using a specific antibody that recognized the relevant phosphoSer-78 site (top panel). The membrane was stripped and degree of equality in the expression of the EGFP-MK5 variants was assayed by western blotting with an anti-PRAK antibody (bottom panel). c PC12 cells were transfected with plasmids encoding EGFP-MK5 S115D (top panel) or EGFP-MK5 S115A (bottom panel) and F-actin was visualized using Alexa Fluor 594 Phalloidin. The expression of the EGFP-tagged MK5 was visualized by EGFP fluorescence (green channel). The superimposed images demonstrate that the architecture of F-actin is changed in cells expressing EGFP-MK5 S115D, but not in nontransfected cells nor in cells expressing EGFP-MK5 S115A. d PC12 cells were cotransfected with expression plasmids for EGFP-MK5 S115D and Hsp27-3A. Hsp27-3A was visualized using anti-Flag antibody and Alexa 647-coupled secondary antibody (magenta). MK5 S115D and F-actin were stained as described above
To test the effects of S115 mutation on cytoplasmic processes involving MK5, we decided to monitor the ability of MK5 to phosphorylate Hsp27, which is an in vivo substrate for MK5 [25]. HEK293 cells were cotransfected with an expression plasmid for Hsp27 and expression plasmids for EGFP-MK5, EGFP-MK5 S115D, EGFP-MK5 S115A, and EGFP-MK5 L337A NES. The latter encodes a constitutive active MK5, but as the L337A mutation resides in the NES of MK5, the MK5 L337A is exclusively nuclear. To direct this constitutive active MK5 L337A mutant into the cytoplasm, a functional NES was added [22]. Increased Hsp27 phosphorylation at the relevant Ser-78 residue was obtained in the presence of MK5 S115D and MK5 L337A NES (Fig. 9b, lanes 3 and 5, respectively) compared to wild-type and MK5 S115A (Fig. 9b, lanes 2 and 4, respectively). We have previously shown that cytoplasmic MK5 can affect F-actin rearrangement [25]. We therefore compared F-actin architecture in PC12 cells expressing either EGFP-MK5 S115A or EGFP-MK5 S115D. Again, we found that the S115A mutant was predominantly in the nucleus, while the S115D variant was also detected in the cytoplasm (Fig. 9c). Clear changes in F-actin remodelling were detected in cells expressing MK5 S115D, but not in cells expressing MK5 S115A. To address the involvement of HSP27 phosphorylation in MK5 S115D-induced F-actin rearrangement, we overexpressed the HSP27-3A mutant [25]. Overexpression of this mutant ablated MK5 S115D-mediated F-actin remodelling (Fig. 9d). Taken together, these results indicate that mutations in Ser-115 not only affect the subcellular localization of MK5, but also the biological properties of this protein.
Discussion
Several groups, including our own, have shown that MK5 is predominantly nuclear in resting cells, while stimuli that activate the p38MAPK and the PKA pathways, as well as overexpression of MK5 interaction partners cause redistribution of MK5. [19, 20, 22, 24, 26–28, 30, 34, 40]. This subcellular location of MK5 is governed by the opposing action of functional NLS and NES motifs in MK5 [19, 30]. A model was put forward in which the conformation of MK5 regulates the accessibility of either the NLS or the NES motif and therefore controls the subcellular location of the protein. A tertiary structure exposing the NES will promote the cytoplasmic location of MK5, while MK5 taking on a three-dimensional structure with an easy accessible NLS will reside in the nucleus [10, 19]. Structural changes, resulting in altered subcellular distribution of MK5 may be induced through interaction with other proteins or through post-translational modifications. In the case of p38MAPK, ERK3, and ERK4, docking seems to be necessary and sufficient to trigger nucleocytoplasmic translocation of MK5 because the catalytic activity of these proteins was dispensable (Table 1) [19, 20, 26–28, 30, 34, 42]. We have recently identified catalytic subunit Cα of PKA as a genuine MK5 interaction partner and demonstrated that activated PKA can induce a transient nuclear export of MK5 [22]. However, the mechanism by which PKA regulates the subcellular distribution of MK5 remains unsolved. In this work we have shown that Ser-115 is a PKA phosphoacceptor site in vitro and we demonstrated that replacing this residue by Ala resulted in a MK5 mutant that remained nuclear in forskolin-treated cells, while substituting Ser-115 by Asp was sufficient to allow MK5 to enter the cytoplasm of untreated cells. These results suggest that phosphorylation of MK5 at Ser-115 by PKA may regulate its subcellular whereabouts and represents a novel mechanism controlling the location of this protein.
Alignment of the primary sequence of MK2, MK3, and MK5 revealed that, although the region encompassing Ser-115 is somewhat conserved between MK2, MK3 and MK5, there are two major differences. Both MK2 and MK3 lack the corresponding Ser-115 and the residues C-terminal of Ser-115 display higher diversity than the N-terminal ones (Fig. 4b). These differences may explain the dissimilar behaviour of MK2 and MK5 in response to activation of the cAMP/PKA pathway. This prompted us to test whether the addition of this residue and/or replacing the motif encompassing this residue could affect the subcellular distribution of MK2. In contrast to MK5, where the phosphomimicking S115D mutant is present in both the nucleus and the cytoplasm, insertion of an Asp residue at the corresponding site in MK2 did not alter the subcellular localization of the MK2 mutant. This suggests that phosphomicking is not sufficient to trigger nuclear export of MK2. While wild-type MK5 is predominantly in the nucleus, a MK2 hybrid containing the MK5 motif encompassing Ser-115 was unexpectedly detected both in the nucleus and the cytoplasm. However, forskolin treatment triggered complete nuclear exclusion of this MK2 mutant, indicating that phosphorylation of Ser-115 in the MK2 hybrid protein contributes to the regulation of the subcellular distribution of MK2 in response to forskolin. The reason why the hybrid MK2 is predominantly in the cytoplasm of untreated cells remains elusive, but we postulate that the amino acid substitutions may induce a conformational change in the chimeric MK2 protein, resulting in exposure of the NES and subsequent nuclear export. Additional studies are required to confirm this suggestion.
The mechanism of PKA-mediated nucleocytoplasmic distribution of MK5 differs from the other MK5 interaction partners p38MAPK, ERK3, and ERK4 (summarized in Table 1). Nuclear export of MK5 induced by p38MAPK, ERK3, and ERK4 does not rely on Ser-115 and occurs independently of the kinase activity of MK5, as well as phosphorylation of Thr-182 in the activation loop of MK5. This is in contrast with PKA-regulated transport, which required the kinase activity of MK5 and an intact Thr-182. In analogy with p38MAPK and ERK3, we found an importin1/NES-dependency for PKA-triggered nuclear exclusion of MK5.
ERK3, ERK4 and p38MAPK can all phosphorylate MK5 at Thr-182 [16, 19, 20, 26–28], but it is not known whether these enzymes also can phosphorylate Ser-115 and whether this modification contributes to ERK3-, ERK4-, or p38MAPK-regulated subcellular localization of MK5. The findings that ectopic expression of kinase dead mutants caused nuclear export of MK5 [19, 20, 26–28, 30, 34] and that overexpression of ERK3, ERK4 and p38MAPK still resulted in nuclear exclusion of MK5 S115A (Figs. 6 and 7b) points to a phosphoSer-115-independent mechanism. Thr-182 is not essential for ERK3-, ERK4-, and p38MAPK-induced nuclear export of MK5 because the MK5 T182A mutant was still excluded from the nucleus when ERK3, ERK4 or p38MAPK was overexpressed. PKA-mediated transport, however, seems to depend on this residue because no nucleocytoplasmic translocation of MK5 T182A was observed in forskolin-treated cells [22]. Forskolin may theoretically still trigger phosphorylation of Ser-115 in MK5 T182A because PKA can phosphorylate this mutant in vitro and Cα-MK5 T182A complexes can be detected by coimmunoprecipitation (see supplementary Figs. S1 and S2). However, forskolin treatment failed to trigger nuclear export of MK5 T182A, while the MK5 S115D/T182A double mutant could enter the cytoplasm of untreated cells (Fig. 3c). Similarly, the kinase dead MK5 K51E with a mutation in the ATP-binding pocket was not excluded from the nucleus after forskolin treatment [22], while MK5 K51E/S115D was detected in the cytoplasm in resting cells.
Why can MK5 S115D/T182A and MK5 K51E/S115D enter the cytoplasm, while MK5 T182A and MK5 K51E remain in the nucleus upon activation of PKA even though they possess the putative S115 phosphorylation site? This difference in subcellular localization behaviour may be explained by the time point of protein modification. Forskolin treatment of resting cells will trigger posttranslational phosphorylation of MK5, while substitution of Ser-115 into Asp occurs during translation. The MK5 K51E and T182A mutants may possess a conformation that renders Ser-115 inaccessible for posttranslational modification by PKA and/or these mutations may lock MK5 in a conformation that prevents exposure of the NES despite phosphorylation of Ser-115. In both proposed models, forskolin treatment of resting cells will fail to translocate MK5 K51E or MK5 T182A out of the nucleus. At present, we cannot determine which mechanism might be operational, but MK5 T182A could still be phosphorylated by PKA in vitro and still interacts with PKA (see supplementary Figs. S1 and S2), indicating that Ser-115 is still reachable by PKA. MK5 K51E/S115D and MK5 S115D/T182A on the other hand contain amino acid substitutions introduced during protein synthesis. The nascent MK5 S115D polypeptide chain may adopt a structure that exposes the NES or masks the NLS independently of the incorporation of Lys or Glu at position 51 (Thr or Ala at position 182, respectively). The MK5 S115D and MK5 K51E/S115D and MK5 S115D/T182A proteins may therefore be able to enter the cytoplasm.
The involvement of p38MAPK, ERK3 and ERK4 in the subcellular distribution of MK5 has been shown with overexpression of these proteins, but the biological relevance of this redistribution of MK5 remains elusive. We have performed our studies under more physiological conditions by showing that activation of endogenous PKA triggers transient nuclear export of endogenous, as well as ectopically expressed EGFP-MK5 (this study; [22]). We were able to show that MK5 mediates PKA-induced F-actin rearrangements by phosphorylating Hsp27 and that cytoplasmic localization of MK5 is required in this process [25]. Hence, our results indicate that PKA participates in activating MK5, causing it to translocate to the cytoplasm, where it can phosphorylate Hsp27 and modulate F-actin architecture. In accordance with this, we demonstrated that MK5 S115D is a more potent Hsp27 kinase than wild-type MK5. In this study we confirmed the importance of Ser-115 in regulating the biological functions of this protein. Previously, Sun et al. [21] had shown that MK5 stimulates the transcriptional activity of p53 through direct phosphorylation of Ser-37 in the transactivation domain. We found that compared to wild-type MK5, the MK5 S115A mutant is a weaker stimulator of p53-mediated transcription. Whether MK5 S115D could enhance the transcriptional activity of p53 was not addressed. In the luciferase reporter assay we used, it is difficult to compare the effect of MK5 S115D and wild-type MK5 on GAL4-p53-mediated transcription because wild-type MK5 is predominantly in the nucleus, while a large fraction of MK5 S115D resides in the cytoplasm, resulting in unequal nuclear levels of these two proteins. However, because p53 is also implicated in cytoplasmic processes, it is plausible that the MK5 S115D mutant may affect both the nuclear and cytoplasmic functions of p53. Hence, our results indicate that PKA may modulate p53 activity in a MK5-dependent manner.
Several studies have shown that cAMP levels may influence p53 function [43, 44], but the involvement of MK5 has not been addressed. A recent study showed that cAMP inhibits doxorubicin-induced phosphorylation of p53 in pre-B acute lymphoblastic leukaemia NALM-6 cells [45]. Phosphorylation is attenuated at several serines, including Ser-37 which has been shown to be directly phosphorylated by MK5 [21]. The mechanism/phosphatase responsible for cAMP-induced reduction in p53 phosphorylation has not been identified, but it is possible that, at least for Ser-37, cAMP-triggered nuclear export of MK5 places MK5 and p53 in different subcellular compartments resulting in diminished p53 phosphorylation at Ser-37. PKA-induced subcellular localization and activation of MK5 offers a mechanism to differentially phosphorylate nuclear or cytoplasmic substrates of MK5. PKA has also been shown to control the subcellular localization of the MAPKAPK RSK1 and hence to modulate the phosphorylation of nuclear and cytoplasmic substrates for RSK1 [46]. However, the subcellular localization of RSK1 is not governed by PKA-mediated phosphorylation of RSK1, but by binding to the different subunits of PKA [46]. Inactive RSK interacts with the regulatory subunit of PKA, while active RSK1 interacts with the catalytic subunit of PKA. The latter interaction decreases the nuclear accumulation of active RSK1 and increases its cytoplasmic concentration [46].
Whether Ser-115 is an in vivo phosphoacceptor site remains to be established. Because no specific antibody for phospho-Ser 115 is available, we tried to monitor phosphorylation by using phospho-PKA substrate and phosphoSer antibodies to compare the phosphorylation pattern of wild-type and S115A MK5. Unfortunately, in our hands, these antibodies failed to detect phosphorylation of MK5 and CREB in any of the samples tested. We also tried the Phos-tag acrylamide gel electrophoresis method which allows detection of differences in gel electrophoresis mobility between unphosphorylated and phosphorylated proteins [47], but again we could not detect any differences between unphosphorylated and PKA-mediated phosphorylation of MK5 and CREB. Other peptide fragments were weakly phosphorylated by PKA (Fig. 2a). MK5 contains additional putative phosphorylation sites, including Thr-14, Ser-43 and Ser-348. Whether these sites are genuine PKA phosphoacceptor sites is not known, nor has their involvement in PKA-triggered nuclear export of MK5 been examined. However, since mutating Ser-115 into Ala ablates PKA-induced nuclear export, these sites do not seem to play an essential role in the subcellular behaviour of MK5 after activation of PKA.
In conclusion, we provide evidence that Ser-115 plays an essential role in cAMP/PKA-regulated nuclear export of MK5, and that it also contributes to the regulation of the activity of MK5. It has been proposed that nucleocytoplasmic distribution of MK5 depends on the conformational structure of MK5, which will reveal or conceal the NES and NLS (for review see reference [10]). We assume that phosphorylation of MK5 residue Ser-115 alters the three-dimensional structure of the protein, exposing the NES and triggering nuclear export. Our kinetic studies revealed that PKA-induced nuclear export is transient [22], suggesting that phosphoSer-115 MK5 may become dephosphorylated by cytoplasmic protein phosphatase(s) and therefore force MK5 back to the nucleus by changing its conformation and hence exposing its NLS. Identification of these phosphatases and resolving the three-dimensional structure of MK5 will be very useful to fully comprehend the exact molecular mechanism governing the subcellular localization of MK5.
Abbreviations
- ERK:
-
Extracellular signal-regulated kinase
- MAPK:
-
Mitogen-activated protein kinase
- MK5:
-
Mitogen-activated protein kinase-activated protein kinase 5
- MKK6:
-
MAPK kinase 6
- NES:
-
Nuclear export signal
- NLS:
-
Nuclear localization signal
- PC12:
-
Rat phaeochromocytoma cells
- PKA:
-
Protein kinase A or cAMP-dependent protein kinase
References
Johnson GL, Lapadat R (2002) Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 298:1911–1912
Roux PP, Blenis J (2004) ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev 68:320–344
Imajo M, Tsuchiya Y, Nishida E (2006) Regulatory mechanisms and functions of MAP kinase signaling pathways. IUBMB Life 58:312–317
Cuenda A, Rousseau S (2007) p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim Biophys Acta 1773:1358–1375
Cuevas B, Abell A, Johnson G (2007) Role of mitogen-activated protein kinase kinase kinases in signal integration. Oncogene 26:3159–3171
Raman M, Chen W, Cobb MH (2007) Differential regulation and properties of MAPKs. Oncogene 26:3100–3112
Weston CR, Davis RJ (2007) The JNK signal transduction pathway. Curr Opin Cell Biol 19:142–149
Zhang Y, Dong C (2007) Regulatory mechanisms of mitogen-activated kinase signaling. Cell Mol Life Sci 64:2771–2789
Krishna M, Narang H (2008) The complexity of mitogen-activated protein kinases (MAPKs) made simple. Cell Mol Life Sci 65:3525–3544
Gaestel M (2006) MAPKAP kinases – MKs – two’s company, three’s a crowd. Nat Rev Mol Cell Biol 7:120–130
Anjum R, Blenis J (2008) The RSK family of kinases: emerging roles in cellular signalling. Nat Rev Mol Cell Biol 9:747–758
Arthur JS (2008) MSK activation and physiological roles. Front Biosci 13:5866–5879
Buxade M, Parra-Palau JL, Proud CG (2008) The Mnks: MAPK kinase-interacting kinases (MAP kinase signal-integrating kinases). Front Biosci 13:5359–5373
Gaestel M (2008) Specificity of signaling from MAPKs to MAPKAPKs: kinases’ tango nuevo. Front Biosci 13:6050–6059
Perander M, Keyse SM, Seternes OM (2008) Does MK5 reconcile classical and atypical MAP kinases? Front Biosci 13:4617–4624
New L, Jiang Y, Zhao M, Liu K, Zhu W, Flood LJ, Kato Y, Parry GC, Han J (1998) PRAK, a novel protein kinase regulated by the p38 MAP kinase. EMBO J 17:3372–3384
Ni H, Wang XS, Diener K, Yao Z (1998) MAPKAPK5, a novel mitogen-activated protein kinase (MAPK)-activated protein kinase, is a substrate of the extracellular-regulated kinase (ERK) and p38 kinase. Biochem Biophys Res Commun 243:492–496
Chen G, Hitomi M, Han J, Stacey DW (2000) The p38 pathway provides negative feedback for Ras proliferative signaling. J Biol Chem 275:38973–38980
Seternes OM, Johansen B, Hegge B, Johannessen M, Keyse SM, Moens U (2002) Both binding and activation of p38 mitogen-activated protein kinase (MAPK) play essential roles in regulation of the nucleocytoplasmic distribution of MAPK-activated protein kinase 5 by cellular stress. Mol Cell Biol 22:6931–6945
Schumacher S, Laass K, Kant S, Shi Y, Visel A, Gruber AD, Kotlyarov A, Gaestel M (2004) Scaffolding by ERK3 regulates MK5 in development. EMBO J 23:4770–4779
Sun P, Yoshizuka N, New L, Moser BA, Li Y, Liao R, Xie C, Chen J, Deng Q, Yamout M, Dong MQ, Frangou CG, Yates JR III, Wright PE, Han J (2007) PRAK is essential for ras-induced senescence and tumor suppression. Cell 128:295–308
Gerits N, Mikalsen T, Kostenko S, Shiryaev A, Johannessen M, Moens U (2007) Modulation of F-actin rearrangement by the cyclic AMP/cAMP-dependent protein kinase (PKA) pathway is mediated by MAPK-activated protein kinase 5 and requires PKA-induced nuclear export of MK5. J Biol Chem 282:37232–37243
Gerits N, Van Belle W, Moens U (2007) Transgenic mice expressing constitutive active MAPKAPK5 display gender-dependent differences in exploration and activity. Behav Brain Funct 3:58
Li Q, Zhang N, Zhang D, Wang Y, Lin T, Wang Y, Zhou H, Ye Z, Zhang F, Lin SC, Han J (2008) Determinants that control the distinct subcellular localization of p38α-PRAK and p38β-PRAK complexes. J Biol Chem 283:11014–11023
Kostenko S, Johannessen M, Moens U (2009) PKA-induced F-actin rearrangement requires phosphorylation of Hsp27 by the MAPKAP kinase MK5. Cell Signal 21:712–718
Seternes OM, Mikalsen T, Johansen B, Michaelsen E, Armstrong CG, Morrice NA, Turgeon B, Meloche S, Moens U, Keyse SM (2004) Activation of MK5/PRAK by the atypical MAP kinase ERK3 defines a novel signal transduction pathway. EMBO J 23:4780–4791
Kant S, Schumacher S, Singh MK, Kispert A, Kotlyarov A, Gaestel M (2006) Characterization of the atypical MAPK ERK4 and its activation of the MAPK-activated protein kinase MK5. J Biol Chem 281:35511–35519
Åberg E, Perander M, Johansen B, Julien C, Meloche S, Keyse SM, Seterens OM (2006) Regulation of MAPK-activated protein kinase 5 activity and subcellular localization by the atypical MAPK ERK4/MAPK4. J Biol Chem 281:35499–35510
Tak H, Jang E, Kim SB, Park J, Suk J, Yoon YS, Ahn JK, Lee JH, Joe CO (2007) 14-3-3epsilon inhibits MK5-mediated cell migration by disrupting F-actin polymerization. Cell Signal 19:2379–2387
New L, Jiang Y, Han J (2003) Regulation of PRAK subcellular location by p38 MAP kinases. Mol Biol Cell 14:2603–2616
Tanoue T, Adachi M, Moriguchi T, Nishida E (2000) A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat Cell Biol 2:110–116
Tanoue T, Maeda R, Adachi M, Nishida E (2001) Identification of a docking groove on ERK and p38 MAP kinases that regulates the specificity of docking interactions. EMBO J 20:466–479
Shi Y, Kotlyarov A, Laass K, Gruber AD, Butt E, Marcus K, Meyer HE, Friedrich A, Volk HD, Gaestel M (2003) Elimination of protein kinase MK5/PRAK activity by targeted homologous recombination. Mol Cell Biol 23:7732–7741
Déléris P, Rousseau J, Coulombe P, Rodier G, Tanguay PL, Meloche S (2008) Activation loop phosphorylation of the atypical MAP kinase ERK3 and ERK4 is required for binding, activation and cytoplasmic relocalization of MK5. J Cell Physiol 217:778–788
Engel K, Kotlyarov A, Gaestel M (1998) Leptomycin B-sensitive nuclear export of MAPKAP kinase 2 is regulated by phosphorylation. EMBO J 17:3363–3371
Mikalsen T, Johannessen M, Moens U (2005) Sequence- and position-dependent tagging protects extracellular-regulated kinase 3 protein from 26S proteasome-mediated degradation. Int J Biochem Cell Biol 37:2513–2520
Houston E, Lynch MJ, Mohamed A, Collins DM, Hill EV, MacLeod R, Krause E, Baillie GS, Houslay MD (2008) EPAC and PKA allow cAMP dual control over DNA-PK nuclear translocation. Proc Natl Acad Sci U S A 105:12791–12796
Pearson RB, Kemp BE (1991) Protein kinase phosphorylation site sequences and consensus specificity motifs: tabulations. Methods Enzymol 200:62–81
Johannessen M, Olsen PA, Johansen B, Seternes OM, Moens U (2003) Activation of the coactivator four-and-a-half-LIM-only protein FHL2 and the c-fos promoter through inhibition of protein phosphatase 2A. Biochem Pharmacol 65:1317–1328
Coulombe P, Rodier G, Pelletier S, Pellerin J, Meloche S (2003) Rapid turnover of extracellular signal-regulated kinase 3 by the ubiquitin-proteasome pathway defines a novel paradigm of mitogen-activated protein kinase regulation during cellular differentiation. Mol Cell Biol 23:4542–4558
Julien C, Coulombe P, Meloche S (2003) Nuclear export of ERK3 by a CREM1-dependent mechanism regulates its inhibitory action on cell cycle progression. J Biol Chem 278:42615–42624
Perander M, Åberg E, Johansen B, Dreyer B, Guldvik IJ, Outzen H, Keyse SM, Seternes OM (2008) The Ser186 phospho-acceptor site within ERK4 is essential for its ability to interact with and activate PRAK/MK5. Biochem J 411:613–622
Mantovani G, Bondioni S, Lania AG, Rodolfo M, Peverelli E, Polentarutti N, Veliz Rodriguez T, Ferrero S, Bosari S, Beck-Peccoz P, Spada A (2008) High expression of PKA regulatory subunit 1A protein is related to proliferation of human melanoma cells. Oncogene 27:1834–1843
Naderi EH, Findley HW, Ruud E, Blomhoff HK, Naderi S (2009) Activation of cAMP signaling inhibits DNA damage-induced apoptosis in BCP-ALL cells through abrogation of p53 accumulation. Blood 114:608–618
Safa M, Kazemi A, Zand H, Azarkeivan A, Zaker F, Hayat P (2010) Inhibitory role of cAMP on doxorubicin-induced apoptosis in pre-B ALL cells through dephosphorylation of p53 serine residues. Apoptosis 15:196–203
Chaturvedi D, Poppleton HM, Stringfield T, Barbier A, Patel TB (2006) Subcellular localization and biological actions of activated RSK1 are determined by its interactions with subunits of cyclic AMP-dependent protein kinase. Mol Cell Biol 26:4586–4600
Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T (2006) Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics 5:749–757
Acknowledgments
The authors thank Dr. Ole Rumohr Blingsmo and Dr. Elisa Bjørgo (Biotechnology Centre, Oslo, Norway) for synthesis and helpful suggestions with the peptide array, Dr. Ole Morten Seternes (Institute of Pharmacy, University of Tromsø) for the kind gift of the ERK4 expression plasmid, Kenneth Larsen (Bioimaging platform, University of Tromsø) for help with the confocal microscope, and Johannes Eksteen (Institute of Medical Biology, University of Tromsø) for the synthesis of the MK5 peptides. This work was supported by grants from the Norwegian Cancer Society (A01037) and the Mohn Foundation. A.S. is the recipient of a grant from the Norwegian Cancer Society (A01037/015).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
S. Kostenko and A. Shiryaev contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Kostenko, S., Shiryaev, A., Gerits, N. et al. Serine residue 115 of MAPK-activated protein kinase MK5 is crucial for its PKA-regulated nuclear export and biological function. Cell. Mol. Life Sci. 68, 847–862 (2011). https://doi.org/10.1007/s00018-010-0496-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-010-0496-2