Abstract
The peroxisome proliferator-activated receptor (PPAR) family includes three transcription factors: PPARα, PPARβ/δ, and PPARγ. PPAR are nuclear receptors activated by oxidised and nitrated fatty acid derivatives as well as by cyclopentenone prostaglandins (PGA2 and 15d-PGJ2) during the inflammatory response. This results in the modulation of the pro-inflammatory response, preventing it from being excessively activated. Other activators of these receptors are nonsteroidal anti-inflammatory drug (NSAID) and fatty acids, especially polyunsaturated fatty acid (PUFA) (arachidonic acid, ALA, EPA, and DHA). The main function of PPAR during the inflammatory reaction is to promote the inactivation of NF-κB. Possible mechanisms of inactivation include direct binding and thus inactivation of p65 NF-κB or ubiquitination leading to proteolytic degradation of p65 NF-κB. PPAR also exert indirect effects on NF-κB. They promote the expression of antioxidant enzymes, such as catalase, superoxide dismutase, or heme oxygenase-1, resulting in a reduction in the concentration of reactive oxygen species (ROS), i.e., secondary transmitters in inflammatory reactions. PPAR also cause an increase in the expression of IκBα, SIRT1, and PTEN, which interferes with the activation and function of NF-κB in inflammatory reactions.
Similar content being viewed by others
Background
The prostaglandin synthesis pathway is an important element of inflammatory responses. The pathway is induced by cytokines [1, 2], LPS [3, 4], or xenobiotics, including metal compounds [5,6,7] and fluoride [8,9,10]. At the beginning of the pathway, PLA2 releases arachidonic acid (AA) from the cell membranes [11]. Next, AA is enzymatically converted, at first by lipoxygenases (LOX) or cytochromes P450 (CYP), or by the best known COX, into a large group of compounds called eicosanoids [12, 13]. Like any biochemical response, increasing synthesis of various eicosanoids in the COX pathway is subject to strict regulation. There are numerous regulatory mechanisms in this pathway which cause both an increase and decrease in synthesis and in activity of particular eicosanoids [14, 15]. Regulation relating to modification of activity of prostaglandins and thromboxanes receptors is an example of the above [16]. The activity of COX-2 is also enhanced by the product of its pathway, i.e., by PGE2 in an autocrine manner [17, 18]. Activation of prostaglandin E2 receptors (EPs) causes an increase in the level of cAMP and activation of the cAMP response element-binding protein (CREB) which leads to increased COX-2 expression [19, 20]. The increase in COX-2 expression may also depend on the activation of mitogen-activated protein kinase (MAPK) cascades [21] and phosphoinositide 3-kinase (PI3K) [18, 22]. Apart from the effect of individual products of the COX pathway, LOX products also increase COX-2 expression [23]. Despite this, AA, regardless of COX, LOX, and CYP activity, is itself capable of causing oxidative stress and, in particular, of activating NADPH oxidase [24, 25]. This results in an increase in the level of reactive oxygen species (ROS) and activation of c-Jun N-terminal kinase (JNK) MAPK and NF-κB, which increases COX-2 expression.
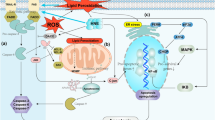
Nonetheless, self-regulation of the COX pathway does not consist solely of positive feedback. The pathway also involves mechanisms which inhibit a too strong inflammatory response. In the center of the COX auto-inhibition pathway, peroxisome proliferator-activated receptors (PPAR) are found (Fig. 1). Activation of inflammatory responses by LPS or other pro-inflammatory particles causes an increase in the expression of PPARα and a decrease in the expression of PPARγ [26,27,28,29]. Blocking the mechanism with a knock-out gene for PPARα causes an increase in the intensity of inflammatory responses to IL-1 or LPS [28, 30].
Self-regulation of NF-κB activity and COX-2 expression. In inflammatory reactions, NF-κB is activated and, partly as a result of this, an increase in expression and activity of enzymes of the prostaglandin synthesis pathway takes place. Released AA is converted into PGD2 or PGE2. In inflammatory reactions, the production and concentration of NO also increase. With time, all of the compounds react together or undergo further non-enzymatic transformation. AA in reaction with NO is subject to nitration. PGD2 and PGE2 convert to 15d-PGJ2 and PGA2, respectively. Compounds with anti-inflammatory properties are formed, which activate PPARα and PPARγ. Activated PPARα and PPARγ inhibit the activity of NF-κB, which leads to inhibition of inflammatory reactions by the products of these reactions
Ligands of peroxisome proliferator-activated receptors
PPAR are nuclear receptors and transcription factors. They include PPARα, PPARβ/δ, and PPARγ. These transcription factors control genes responsible for the oxidation of lipids [31, 32]. PPARα is a transcription factor that increases gene expression of acyl-CoA oxidase and carnitine palmitoyltransferase I, i.e., enzymes involved in β-oxidation. PPARγ increases adiponectin concentration and expression of transporters GLUT1 and GLUT4. PPARβ/δ increases the expression of pyruvate dehydrogenase kinase-4 and carnitine palmitoyltransferase 1A, which increases the intensity of fatty acid oxidation.
All of PPAR have a similar structure—ligand-binding domain (LBD). LBD is in the shape of the letter Y and is of polar character [33,34,35,36]. In the first arm, hydrophilic amino acid residue can be found, which is responsible for ligand binding. This part also contains helix12, which is stabilized during ligand binding and PPAR activation. The remaining two arms of the ligand-binding domain consist of hydrophobic amino acid residue with far less hydrophilic residue. These two parts of LBD are responsible for the specificity of ligand binding when activating PPAR.
This LBD structure enables the activation of PPAR by polar structure ligands, and, in particular, by a fatty acids and derivatives of fatty acids (Table 1). Nonetheless, not all fatty acids equally activate PPAR. The arms with hydrophobic residue in LBD stabilize ligand [33]. Therefore, only fatty acids with 14 and more carbon atoms are capable of activating PPAR [33, 37, 38]. However, saturated fatty acids with 20 and more carbon atoms do not fit in the LBD and thus are not activators of PPAR. Double bonds also play an important role in the structure of a fatty acid as a potential ligand. Monounsaturated fatty acids in cis configuration have a favorable conformation to better match LBD than saturated fatty acids and fatty acids in trans configuration of the same length. Fatty acids in trans configuration have similar conformation as unsaturated fatty acids [33]. Long-chain polyunsaturated fatty acid (PUFA) is also ligands for PPAR [33, 37, 39, 40]. For example, AA connects with LBD at a concentration of IC50 equal to 1.2 ± 0.2 μM and 1.6 ± 0.2 μM for PPARα and PPARγ, respectively [33, 41]. Eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) have similar properties. However, at a much higher concentration, 100 μM, inhibition of PPARα activity by AA, EPA, and DHA takes place [38].
Hydrophilic residue is also present in LBD, which enables activation of PPAR by derivatives of fatty acids arising from enzymatic and non-enzymatic oxidation [35, 42]. Owing to self-regulation of inflammatory responses, AA metabolites fulfill special roles as activators. An example of the above is the products of the 5-LOX pathway, 5(S)-hydroxyeicosatetraenoic acid (5(S)-HETE), and leukotriene B4 (LTB4), which also activate PPARα [37, 40, 43]. Nonetheless, this effect is less significant compared to the impact of free AA—fatty acids from which they arise. Other important activators of PPAR are 8(S)-HETE and hydroperoxyeicosatetraenoic acid (8S-HPETE)—products of murine 8-LOX [37, 40, 44, 45]. Yet, another important activator of PPARγ and PPARβ/δ is the 15-LOX product: 15-HETE [46, 47]. Among natural activators of PPARα and PPARγ are AA metabolites, products of many CYP isoforms with anti-inflammatory properties are also found. They include: 5,6-epoxyeicosatrienoic acid (EET), 8,9-EET, 11,12-EET, 14,15-EET, and 20-HETE (with its metabolite 20-COOH-AA) [13, 48,49,50,51].
Prostaglandins produced in the COX pathway have impact on activating PPARγ. Cyclopentenone prostaglandins activate PPARγ [37, 44, 52]. This group includes prostaglandin (PG)A2, PGC2, PGJ2, Δ12-PGJ2, and the most thoroughly studied 15-deoxy-Δ12,14-prostaglandin J2 (15d-PGJ2). All prostaglandins in this group arise from pro-inflammatory prostaglandins. PGA2 is formed as a result of non-enzymatic dehydration of PGE2. Subsequently, PGA2 may be isomerized to PGB2 and PGC2, whereas PGD2 may be subject to non-enzymatic dehydration and isomerisation to PGJ2, Δ12-PGJ2 and, finally, to 15d-PGJ2. All of the cyclopentenone prostaglandins contain an electrophilic carbon atom in their cyclopentenone ring [53, 54]. Owing to this, they can be subject to Michael addition to free thiol (–SH) groups in cysteine. If such residue is present in the catalytic center or in an important domain in the function of a given protein, such enzymes and transcription factors become inactive. IκB kinase β subunit (IKKβ) and nuclear factor κB (NF-κB) are examples of such proteins [52, 55]. Cysteine is also present in LBD PPARγ in position 285 (Cys285), which is specifically susceptible to reactions and covalent 15d-PGJ2 connection (Fig. 2) [40, 54, 56]. As a result of alkylation of this residue, a change in PPARγ conformation takes place and the protein is activated [37, 39, 40, 44]. The cyclopentenone prostaglandins constitute negative feedback to inflammatory responses. During the inflammatory responses, an increase in the production of PGE2 and PGD2 is observed [57]. With time, in a non-enzymatic way, further transformation of the prostaglandins into anti-inflammatory compounds takes place in PGA2 and 15d-PGJ2, respectively. As a consequence, the inflammatory response is reduced by transformed products of the responses [3, 58].
15d-PGJ2 as an agonist of PPARγ. Modeled (as purple) 15d-PGJ2 in LBD PPARγ. Amino acid residues interacting with this ligand and covalently bound Cys285 are shown. PDB:2zk1 [54]
In the course of inflammatory reactions, an increased synthesis occurs of nitric oxide (NO) and ROS generation, among others superoxide radical (O •−2 ), and hydrogen peroxide (H2O2) [59, 60]. NO may react with O •−2 creating peroxynitrite (ONOO−). Both compounds, NO and ONOO−, lead to nitration of double bonds in unsaturated fatty acids [42, 61]. As a result, nitrated derivatives of fatty acids are formed. These compounds have biological properties of an anti-inflammatory nature. One of their functions is binding-free thiol (–SH) groups in cysteine [62, 63]. Owing to this, they may cause nitroalkylation of Cys62 p50 NF-κB and Cys38 p65 NF-κB, which, as a consequence, inactivates these transcription factor subunits. A nitrated derivative of AA replaces heme in COX, which impacts the intensity of inflammatory responses. This results in irreversible inhibition of the enzymes Ki for COX-1, equal to 1.02 μM, and Ki for COX-2, equal to 1.76 μM [64]. Another anti-inflammatory characteristic of nitrated derivatives of fatty acids, in very low concentrations, is activation of PPARα and PPARγ [65, 66]. In PPARγ, meanwhile, there are residues of Arg288 and Glu343 which stabilize, respectively, the nitrated derivatives of fatty acids on 10 and 12 carbon [67]. The above-mentioned residues of Arg288 and Glu343 are not conservative and hence do not occur in other PPAR. Owing to such structure, PPARγ is activated by the nitrated derivatives of unsaturated fatty acids even at a concentration of 100 nM [65, 66]. Meanwhile, concentration of PPARα deprived of such residues occurs at a concentration of 300 nM [65]. For higher concentrations of about 4 μM, nitrated derivatives of fatty acids, and of AA in particular, unbalance the assembly of the NADPH oxidase complex [68]. This causes disturbance in ROS generation and, as a result, suppresses inflammatory reactions.
Activation of peroxisome proliferator-activated receptors
PPARγ is a transcription factor, whose activity is subject to regulation by SUMOylation and an assembly of complexes with various proteins, especially with nuclear receptor corepressor (NCoR) and silencing mediator of retinoid and thyroid hormone receptors (SMRT) [69,70,71,72,73]. Activation of PPARγ with ligand causes changes in SUMOylation of this protein. In particular, a decrease takes place in SUMOylation of Lys33 PPARγ1 in the domain of activation function 1 (AF1) [74]. This allows LBD to interact with the AF1 domain, whereas SUMOylation of the Lys365 in LBD PPARγ1 allows for full activation of PPARγ1 by ligand. PPARγ activation also causes suppression of proteolytic degradation of NCoR and dissociation of the complex of PPARγ with NCoR and SMRT [69, 72]. As a result, expression of genes is suppressed by these corepressors, especially of the genes dependent on NF-κB and AP-1, such as PTGS2 and NOS2 [75,76,77].
A similar mechanism is present in PPARα. The transcription factor in its inactive form is subject to SUMOylation and it is also in complex with NCoR [78, 79]. The process of SUMOylation is governed by enzymes Ubc9 and PIASy, which modify the Lys185 PPARα. Activation of PPARα by ligand leads to a decrease in SUMOylation and, as a consequence, to release of NCoR. NCoR is a corepressor which connects exclusively to SUMOylated receptors. Dissociation of the complex of PPARα with NCoR leads to activation of both proteins; PPARα increases expression of particular genes and NCoR is a corepressor suppressing the expression of other genes, including PTGS2 and NOS2 [77].
Changes in the stability of peroxisome proliferator-activated receptors as a result of activation by ligand
PPAR are transcription factors which are also subject to regulation by proteolytic degradation. Nonetheless, different PPAR are subject to different mechanisms of this regulation. Activating PPARα by ligand causes inhibition of ubiquitination, which leads to suppression of proteolytic degradation of this protein [80]. Ubiquitination of Lys292, Lys310, and Lys388 PPARα by muscle ring finger-(MuRF)1 results in export from the nucleus of this transcription factor and subsequently in proteolytic degradation of PPARα [81].
A similar mechanism occurs in regulation of the activity of PPARβ/δ by ligand. Activation by ligand causes inhibition of ubiquitination and inhibition of proteolytic degradation of PPARβ/δ [82].
Contrary to PPARα and PPARβ/δ, activation of PPARγ by ligand leads to ubiquitination and proteolytic degradation of this protein in proteasomes [83, 84]. In adipocytes, drosophila seven-in-absentia homolog 2 (Siah2) is responsible for this process [85]. Ubiquitination in this PPARγ mechanism causes changes in the structure of the activation function 2 (AF-2) domain, and as a consequence, the protein is directed to the proteolytic degradation pathway [86]. Nonetheless, PPARγ may also be subject to ubiquitination independently of ligand, which causes degradation or increase in the stability of the protein depending on the place of ubiquitination. Tripartite motif protein 23 (TRIM23) catalyzes polyubiquitination, owing to which PPARγ stability increases [87]. Meanwhile, F-box only protein 9 (FBXO9) [88] or makorin ring finger protein 1 (MKRN1) [89] has been identified as specific E3 ligase of PPARγ in adipocytes, which leads to ubiquitination and proteasome-dependent degradation of PPARγ. The regulation mechanism of PPARγ expression through proteolytic degradation is important in adipocyte differentiation, as this transcription factor impacts expression of genes responsible for lipogenesis of adipocytes. Meanwhile, MuRF2, which is present in cardiomiocytes, causes ubiquitination of PPARγ and thus leads to an increase in stability of the protein [90]. However, overactivity of MuRF2 causes polyubiquitination of PPARγ and, as a consequence, proteolytic degradation of the protein. It has been demonstrated that dysregulation of MuRF2 activity in diabetes has an impact on the development of cardiomyopathy [90].
Phosphorylation changing the activity of peroxisome proliferator-activated receptors
Activated PPAR may also be subject to phosphorylation, which modifies the activity of these nuclear receptors. PPARγ is subject to phosphorylation into the N-terminus A/B domain by such kinases as protein kinase A (PKA) [91], extracellular signal-regulated kinase (ERK) MAPK [92,93,94] or JNK MAPK [92, 95]. This results in a change of character of the PPARγ. As a result of phosphorylation at the N-terminus, PPARγ ceases to be a transcription factor and begins to physically bind the activated NF-κB. This mechanism may constitute regulation of inflammatory responses in which activation of the above-mentioned kinases takes place and ligands for PPARγ are present.
PPARα is also subject to phosphorylation by such kinases as PKA [96], p38 MAPK [97] or ERK MAPK [98]. This causes enhanced ligand-dependent activation. Phosphorylation by p38 MAPK can also inhibit the activity of PPARα [99].
Interaction between various peroxisome proliferator-activated receptors
All three PPAR have a similar LBD structure. This enables their simultaneous activation by the same ligand, for example, a given PUFA [33]. This results in interactions between different PPAR. As yet, unfortunately, not much research has been done into the interactions between various PPAR.
Activation of PPARγ leads to an increase in the expression of genes dependent on this transcription factor and on COX-2 in particular. This is associated with increased expression of PPARβ/δ [100]. However, sole activation of PPARβ/δ by ligand does not cause changes in COX-2 expression. It only cooperates with the activated PPARγ. Depending on the model, activation of PPARβ/δ inhibits [101] or increases [100] transcription activity of PPARγ. Activation of PPARβ/δ also increases expression of PPARα, whereas activation of PPARα decreases expression of PPARβ/δ [100]. Thus, simultaneous activation of PPARα and PPARγ leads to mutual abolishing of activity of both the PPAR through changes in expression of PPARβ/δ. However, the specific tissue expression of various PPAR should not be ignored, as it may modify the response to the activation of these nuclear receptors.
Activation of peroxisome proliferator-activated receptor-γ as an inducer of cyclooxygenase-2 expression
Activating PPAR reduces inflammatory reactions. However, when pro-inflammatory factors are absent, activation of PPARγ induces the expression of COX-2 [100]. In the promoter of gene PTGS2, a PPAR response (PPRE) element is found [102, 103]. Thus, when pro-inflammatory factors are absent, activation of PPARγ causes induction of COX-2 protein expression. Inducers of COX-2 expression, which are commonly known, as compounds decrease expression of COX-2 in inflammatory reactions, are PUFA [γ-linolenic acid (γ-LA), AA, EPA, and DHA] [102, 104,105,106] as well as 15d-PGJ2 [107,108,109]. Nonsteroidal anti-inflammatory drug (NSAID) is also agonists for PPARγ [110, 111]. This is why such compounds as diclofenac, flufenamic acid, flurbiprofen, indomethacin, or NS-398, despite inhibiting the activity of COX-2, induce expression of that enzyme [107,108,109, 112].
AA and EPA (both PUFA) are processed into 2- and 3-series prostanoids [113,114,115], respectively. Prostanoids serve a very important biological function, unrelated to inflammatory reactions; they are crucial for proper functioning of the kidneys [116] and blood vessels [117]. Therefore, PUFA must influence the expression and activity of COX-2 differently than in inflammatory reactions. In metabolism, very often, the substrate of a given enzymatic pathway stimulates an increase in the activity of enzymes participating in its processing. That is a likely explanation as to why AA and EPA, by activating PPARγ increase the expression of COX-2. That is, they increase the expression and activity of the enzyme which uses them as substrates. This effect is not related to inflammation, but more likely to the functions of AA and EPA as substrates for the production of prostanoids.
Peroxisome proliferator-activated receptor as a protein with anti-inflammatory properties: effect on NF-κB
Activation of all PPAR, PPARα [4, 118,119,120,121], PPARβ/δ [122, 123] and PPARγ [124,125,126,127] causes inhibition of NF-κB activation (Table 2). Nonetheless, the mechanism of anti-inflammatory properties is a very complex one and takes different forms.
PPAR are transcription factors, but they also have properties which are not associated with the expression of genes. They are capable of binding different proteins, by which means they inactivate them. It is predominantly the binding of PPARα [30, 128], PPARβ/δ [129], or PPARγ with p65 NF-κB that is responsible for anti-inflammatory properties [94, 130] with p65 NF-κB which reduces the pro-inflammatory response.
The direct impact of PPARγ on NF-κB may be associated with its enzymatic properties. PPARγ is E3 ubiquitin ligase, which cooperates with E2 UBCH3. PPARγ causes ubiquitination of the Lys48 p65 NF-κB, which leads to proteolytic degradation of this NF-κB subunit [131]. The intensity of NF-κB degradation is increased by PPARγ ligand activation. However, it is not only NF-κB that is subject to such regulation by PPARγ. It is possible to reduce the stability of the MUC1-C by PPARγ, which has anticancer properties [132].
Inactivated PPARβ/δ occurs in the complex with p65 NF-κB [133, 134]. During induction of inflammatory responses, the inactivated PPARβ/δ is involved in activation of NF-κB p65. PPARβ/δ, in particular, takes part in assembly of the complex from TAK1, TAB1, and HSP27 [135]. Activation by ligand PPARβ/δ results in lack of this cooperation, and consequently, activation of PPARβ/δ interferes with the function of NF-κB p65. As a result, inflammatory responses caused by a high concentration of glucose, activation of the receptor for TNFα, IL-1β, or activation of TLR4 are reduced [136, 137].
PPARα and PPARγ can also inhibit acetylation of p65 NF-κB, which inhibits activation of this pro-inflammatory transcription factor. After degradation of the IκB, p65 NF-κB is subject to acetylation of the Lys310 p65 NF-κB by p300. This modification is very important with respect to the proper functioning of p65 NF-κB [138, 139]. PPARα [30, 140] and PPARγ [141] bind p300. Assembly of these complexes leads to loss of enzymatic properties of p300 and, as a consequence, to inhibition of activation of p65 NF-κB through reduced of acetylation of this NF-κB subunit.
Besides this pathway, PPARα and PPARγ also cause deacetylation of p65 NF-κB. The process of deacetylation, which leads to inactivation of NF-κB, is catalyzed by sirtuin 1 (SIRT1) [142,143,144]. Activation of PPARα increases expression and activity of SIRT1, which inhibits the p65 NF-κB function [144,145,146,147]. The impact of PPARα on SIRT1 is dependent on AMP-activated protein kinase (AMPK) [145, 146]. Activation of AMPK leads to phosphorylation of p300, which decreases activity of the latter enzyme [123]. Nevertheless, SIRT1 and AMPK are enzymes which activate one another [148, 149]. Hence, no accurate data are available on whether PPARα activates SIRT1 directly or activation of SIRT1 is caused directly by activation of AMPK. Activation of PPARγ causes deacetylation of p65 NF-κB depending on SIRT1 [150]. Nonetheless, the SIRT1 protein itself forms a complex with NCoR, SMRT, and PPARγ [149, 151, 152]. The process inactivates PPARγ.
PPARα and PPARγ indirectly impact the pro-inflammatory transcription factor. In particular, the promoter of the gene-coding IκBα is controlled by PPAR [153]. Owing to this, PPARα [154, 155] and PPARγ [156] increase expression of IκBα—protein binding NF-κB. As a result, inactive NF-κB bound with its inhibitor, IκBα, occurs in the cell. During inflammatory reactions, phosphorylation and proteolytic degradation of IκBα takes place, which activates NF-κB. Increased expression of IκBα by PPARα and PPARγ prevents activation of NF-κB.
Activation of PPARγ [125, 157,158,159,160] or PPARβ/δ [161] causes an increase in expression and activity of phosphatase and tensin homolog (PTEN). This effect, however, may be dependent on the research model. In, A549 line lung carcinoma, H23 line adenocarcinoma, and squamous H157 cell line carcinoma activation of PPARβ/δ decrease expression of PTEN for a few hours [162]. He et al. [163] shows that after a day of being exposed to ligand, expression returns to its control level. PTEN is phosphatase what catalyzes the dephosphorylation of phosphate from position 3′ in phosphatidylinositol-3,4,5-trisphosphate. This enzyme catalyzes a reaction reverse to PI3K. In the transmission of pro-inflammatory factor signals, activation of the PI3K/PKB/IKK/NF-κB pathway takes place [164]. Thus, increased expression and activity of PTEN leads to inhibition of NF-κB activation by PI3K.
Peroxisome proliferator-activated receptor as protein with anti-inflammatory properties: effect on other signaling pathways
PPAR cause inhibition of inflammatory reactions not only by their effect on NF-κB. What is more, c-Jun is bound by PPARα or PPARγ. As a result of this reaction, inhibition of AP-1 activation and inhibition of AP-1 DNA-binding activity by activated PPARα occur [30, 50, 165], and PPARγ occurs [124, 141, 166]. AP-1-binding site occurs in promoters of many genes important in inflammatory reactions, including PTGS2 [167]. As a result of this mechanism, PPARα and PPARγ inhibit COX-2 protein expression.
PPAR disrupt activation of STATs. In particular, PPARγ causes an increase in the expression of the suppressor of cytokine signaling 3 (SOCS3) [168, 169]. This protein inhibits activation of JAK2/STAT3. The activity of STAT1 is disrupted by the activated PPARα [170]. STAT5b is also disrupted by the activated PPARα or PPARγ [171, 172]. Nonetheless, the exact mechanism of such activity of PPARα and PPARγ is not fully understood. PPARα and PPARγ probably compete with STATs for coactivators [171]. It is also possible that they have an effect on membrane receptors. The expression of β-defensin 1 is increased, especially by PPARα. This signaling element leads to decreased expression of TLR4 in J774 macrophages [173].
The effect on antioxidant enzymes
Besides the direct impact of the activated PPAR on NF-κB, an indirect effect on inflammatory reactions is also possible. PPAR reduce concentration of ROS by increasing the expression of antioxidant enzymes. Due to the fact that ROS fulfill a very important role as a second messenger in inflammatory reactions, the increase in the activity of antioxidant enzymes has an anti-inflammatory character [174, 175]. This leads to decreased activation of NF-κB, and thus to decreased expression of COX-2 [176, 177].
One of such ways is increased expression and activity of heme oxygenase-1 (HO-1) [2, 178]. In the HO-1 promoter, two PPAR responsive elements are present [179]. Owing to this, all the three isoforms of activated PPAR increase the expression of HO-1 [180,181,182,183,184,185]. An increase in HO-1 expression may also take place by other means. PPARγ forms a complex with Nrf2 and binds in antioxidant response element (ARE) on the HO-1 promoter, causing an increase in expression of this antioxidant enzyme [184]. PPARγ also leads to stabilization of mRNA HO-1, which prolongs the half-life of this transcript, thus increasing the expression of the HO-1 protein [186]. However, it must be remembered that activators of PPAR may also increase expression of HO-1 regardless of the transcription factor and depending on induction of oxidative stress [187].
HO-1 is an enzyme engaged in heme degradation to biliverdin and carbon monoxide (CO). These compounds have antioxidant properties. Biliverdin is converted to bilirubin, which is an antioxidant [188]. The second product of HO-1 activity, CO, decreases the activity of NADPH oxidase, which decreases the level of ROS in cells activated by pro-inflammatory factors [189, 190]. This also disrupts the activation of TLR4, which inhibits the pro-inflammatory effect of fatty acids and LPS [191]. In addition, CO, depending on ROS, causes S-glutathionylation of STAT3 and p65 NF-κB, which leads to deregulation of the function of these proteins [192, 193]. Owing to all of those properties, HO-1 is an antioxidant and anti-inflammatory enzyme [2, 194].
Activating PPAR causes induction of expression of other antioxidant enzymes, such as catalase, Mn-superoxide dismutase (SOD), and CuZn-SOD. In the promoter of gene Mn-SOD, a sequence of PPRE is present [195]. This allows PPARγ to increase the expression of this enzyme. The expression of CuZn-SOD is also increased after activation of PPARα [196,197,198,199] and PPARγ [185, 198, 200]. Consequently, an increase in SOD activity is followed by a decrease in O •−2 concentration, produced by NADPH oxidase.
In the promoter of the catalase gene, the PPRE is also present [201,202,203]. Owing to this, activation of PPARα or PPARγ causes an increase in expression of this antioxidant enzyme [176, 177, 199, 204,205,206]. As a consequence of the increase in the activity of catalase, the H2O2 level decreases and inflammatory reactions are reduced.
In this not fully understood mechanism, which is probably independent on ROS, an increase in catalase expression may lead to increased expression of COX-2 [207,208,209].
Mechanisms inhibiting inflammatory reactions as the aim of the therapy
Inflammatory reactions, and the expression of COX-2 as well as synthesis of PGE2 in particular, constitute an important element in pathogenesis of many illnesses, such as Parkinson’s disease [210, 211], type II diabetes and concomitant diseases [212] and cancer [213, 214]. Therefore, the use of specific COX-2 inhibitors has a preventive effect and may facilitate the process of curing the diseases. Nonetheless, COX-2 is only an enzyme, whose expression and activity depends on intracellular signal transduction pathways induced in the course of many diseases. Therefore, the best solution in therapy is interference with the signaling pathways of the inflammatory reactions. In particular, PPAR activators can be used, such as naturally occurring n-3 PUFA (EPA and DHA), or artificial pharmacological compounds (ciprofibrate). Activating PPAR reduces inflammatory reactions and, as a result, decreases the expression and activity of COX-2. This allows for cancer prevention or for combining PPAR activators with the current anticancer treatment [215,216,217,218,219,220,221,222,223,224]. In addition to the effect on inflammatory reactions, medications that activate PPAR also regulate metabolism, which has a therapeutic effect in diabetics [225].
It should be remembered, however, that the activation of PPARγ may induce the expression of COX-2 and an increase in synthesis of PGE2 [102, 105, 107,108,109,110,111,112]. This may lead to counterproductive side effects of preventive treatment.
Conclusion
Inflammatory reactions, such as all processes occur in living organisms, are strictly regulated. One group of such regulators are PPAR, receptors activated by nitrated fatty acid derivatives and cyclopentenone prostaglandins, products formed in the late stages of the inflammatory reaction. Activation of PPAR then results in the inhibition of core elements of the inflammatory reaction. Detailed knowledge of the regulatory mechanisms governing a given biological process facilitates interference with the functioning of cells and tissues, allowing for the development of therapeutic approaches for the treatment of diseases based on disorders in these processes.
Abbreviations
- 5(S)-HETE:
-
(S)-hydroxyeicosatetraenoic acid
- 15d-PGJ2 :
-
15-Deoxy-Δ12,14-prostaglandin J2
- AF1:
-
Activation function 1
- AMPK:
-
AMP-activated protein kinase
- ARE:
-
Antioxidant response element
- AA:
-
Arachidonic acid
- CO:
-
Carbon monoxide
- JNK:
-
c-Jun N-terminal kinase
- COX-2:
-
Cyclooxygenase-2
- CYP:
-
Cytochromes P450
- DHA:
-
Docosahexaenoic acid
- EP:
-
Prostaglandin E2 receptors
- EPA:
-
Eicosapentaenoic acid
- EET:
-
Epoxyeicosatrienoic acid
- ERK:
-
Extracellular signal-regulated kinase
- γ-LA:
-
γ-Linolenic acid
- HO-1:
-
Heme oxygenase-1
- H2O2 :
-
Hydrogen peroxide
- IKKβ:
-
IκB kinase β subunit
- LBD:
-
Ligand-binding domain
- LOX:
-
Lipoxygenases
- MAPK:
-
Mitogen-activated protein kinase
- NO:
-
Nitric oxide
- NSAID:
-
Nonsteroidal anti-inflammatory drug
- NF-κB:
-
Nuclear factor κB
- NCoR:
-
Nuclear receptor corepressor
- PPAR:
-
Peroxisome proliferator-activated receptors
- PPRE:
-
PPAR response element
- ONOO− :
-
Peroxynitrite
- PTEN:
-
Phosphatase and tensin homolog
- PI3K:
-
Phosphoinositide 3-kinase
- PUFA:
-
Polyunsaturated fatty acid
- PG:
-
Prostaglandin
- PGE2 :
-
Prostaglandin E2
- PKA:
-
Protein kinase A
- ROS:
-
Reactive oxygen species
- SMRT:
-
Silencing mediator of retinoid and thyroid hormone receptors
- SIRT1:
-
Sirtuin 1
- SOD:
-
Superoxide dismutase
- O •−2 :
-
Superoxide radical
References
Newton R, Kuitert LM, Bergmann M, Adcock IM, Barnes PJ. Evidence for involvement of NF-kappaB in the transcriptional control of COX-2 gene expression by IL-1beta. Biochem Biophys Res Commun. 1997;237:28–32.
Yang YC, Lii CK, Wei YL, Li CC, Lu CY, Liu KL, Chen HW. Docosahexaenoic acid inhibition of inflammation is partially via cross-talk between Nrf2/heme oxygenase 1 and IKK/NF-κB pathways. J Nutr Biochem. 2013;24:204–12.
Inoue H, Tanabe T, Umesono K. Feedback control of cyclooxygenase-2 expression through PPARgamma. J Biol Chem. 2000;275:28028–32.
Huang D, Zhao Q, Liu H, Guo Y, Xu H. PPAR-α agonist WY-14643 inhibits LPS-induced inflammation in synovial fibroblasts via NF-kB pathway. J Mol Neurosci. 2016;59:544–53.
Chang WC, Chang CC, Wang YS, Wang YS, Weng WT, Yoshioka T, Juo SH. Involvement of the epidermal growth factor receptor in Pb2+-induced activation of cPLA2/COX-2 genes and PGE2 production in vascular smooth muscle cells. Toxicology. 2011;279:45–53.
Korbecki J, Baranowska-Bosiacka I, Gutowska I, Piotrowska K, Chlubek D. Cyclooxygenase-1 as the main source of proinflammatory factors after sodium orthovanadate treatment. Biol Trace Elem Res. 2015;163:103–11.
Olszowski T, Gutowska I, Baranowska-Bosiacka I, Piotrowska K, Korbecki J, Kurzawski M, Chlubek D. The effect of cadmium on COX-1 and COX-2 gene, protein expression, and enzymatic activity in THP-1 macrophages. Biol Trace Elem Res. 2015;165:135–44.
Chen R, Zhao LD, Liu H, Li HH, Ren C, Zhang P, Guo KT, Zhang HX, Geng DQ, Zhang CY. Fluoride induces neuroinflammation and alters Wnt signaling pathway in BV2 microglial cells. Inflammation. 2017;40:1123–30.
Gutowska I, Baranowska-Bosiacka I, Safranow K, Jakubowska K, Olszewska M, Telesiński A, Siennicka A, Droździk M, Chlubek D, Stachowska E. Fluoride in low concentration modifies expression and activity of 15 lipoxygenase in human PBMC differentiated monocyte/macrophage. Toxicology. 2012;295:23–30.
Gutowska I, Baranowska-Bosiacka I, Goschorska M, Kolasa A, Łukomska A, Jakubczyk K, Dec K, Chlubek D. Fluoride as a factor initiating and potentiating inflammation in THP1 differentiated monocytes/macrophages. Toxicol In Vitro. 2015;29:1661–8.
Dennis EA, Cao J, Hsu YH, Magrioti V, Kokotos G. Phospholipase A2 enzymes: physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem Rev. 2011;111:6130–85.
Murakami M. Lipid mediators in life science. Exp Anim. 2011;60:7–20.
Jamieson KL, Endo T, Darwesh AM, Samokhvalov V, Seubert JM. Cytochrome P450-derived eicosanoids and heart function. Pharmacol Ther. 2017;179:47–83.
Stuhlmeier KM, Kao JJ, Bach FH. Arachidonic acid influences proinflammatory gene induction by stabilizing the inhibitor-kappaBalpha/nuclear factor-kappaB (NF-kappaB) complex, thus suppressing the nuclear translocation of NF-kappaB. J Biol Chem. 1997;272:24679–83.
Thommesen L, Sjursen W, Gåsvik K, Hanssen W, Brekke OL, Skattebøl L, Holmeide AK, Espevik T, Johansen B, Laegreid A. Selective inhibitors of cytosolic or secretory phospholipase A2 block TNF-induced activation of transcription factor nuclear factor-kappa B and expression of ICAM-1. J Immunol. 1998;161:3421–30.
Korbecki J, Baranowska-Bosiacka I, Gutowska I, Chlubek D. Cyclooxygenase pathways. Acta Biochim Pol. 2014;61:639–49.
Camandola S, Leonarduzzi G, Musso T, Varesio L, Carini R, Scavazza A, Chiarpotto E, Baeuerle PA, Poli G. Nuclear factor kB is activated by arachidonic acid but not by eicosapentaenoic acid. Biochem Biophys Res Commun. 1996;229:643–7.
Barry OP, Kazanietz MG, Praticò D, FitzGerald GA. Arachidonic acid in platelet microparticles up-regulates cyclooxygenase-2-dependent prostaglandin formation via a protein kinase C/mitogen-activated protein kinase-dependent pathway. J Biol Chem. 1999;274:7545–56.
Bradbury DA, Newton R, Zhu YM, El-Haroun H, Corbett L, Knox AJ. Cyclooxygenase-2 induction by bradykinin in human pulmonary artery smooth muscle cells is mediated by the cyclic AMP response element through a novel autocrine loop involving endogenous prostaglandin E2, E-prostanoid 2 (EP2), and EP4 receptors. J Biol Chem. 2003;278:49954–64.
Díaz-Muñoz MD, Osma-García IC, Fresno M, Iñiguez MA. Involvement of PGE2 and the cAMP signalling pathway in the up-regulation of COX-2 and mPGES-1 expression in LPS-activated macrophages. Biochem J. 2012;443:451–61.
Rösch S, Ramer R, Brune K, Hinz B. Prostaglandin E2 induces cyclooxygenase-2 expression in human non-pigmented ciliary epithelial cells through activation of p38 and p42/44 mitogen-activated protein kinases. Biochem Biophys Res Commun. 2005;338:1171–8.
Hughes-Fulford M, Li CF, Boonyaratanakornkit J, Sayyah S. Arachidonic acid activates phosphatidylinositol 3-kinase signaling and induces gene expression in prostate cancer. Cancer Res. 2006;66:1427–33.
van Puijenbroek AA, Wissink S, van der Saag PT, Peppelenbosch MP. Phospholipase A2 inhibitors and leukotriene synthesis inhibitors block TNF-induced NF-kappaB activation. Cytokine. 1999;11:104–10.
Cui XL, Douglas JG. Arachidonic acid activates c-jun N-terminal kinase through NADPH oxidase in rabbit proximal tubular epithelial cells. Proc Natl Acad Sci USA. 1997;94:3771–6.
Mazière C, Conte MA, Degonville J, Ali D, Mazière JC. Cellular enrichment with polyunsaturated fatty acids induces an oxidative stress and activates the transcription factors AP1 and NFkappaB. Biochem Biophys Res Commun. 1999;265:116–22.
Kapoor M, Kojima F, Qian M, Yang L, Crofford LJ. Microsomal prostaglandin E synthase-1 deficiency is associated with elevated peroxisome proliferator-activated receptor gamma: regulation by prostaglandin E2 via the phosphatidylinositol 3-kinase and Akt pathway. J Biol Chem. 2007;282:5356–66.
Necela BM, Su W, Thompson EA. Toll-like receptor 4 mediates cross-talk between peroxisome proliferator-activated receptor gamma and nuclear factor-kappaB in macrophages. Immunology. 2008;125:344–58.
Kono K, Kamijo Y, Hora K, Takahashi K, Higuchi M, Kiyosawa K, Shigematsu H, Gonzalez FJ, Aoyama T. PPAR{alpha} attenuates the proinflammatory response in activated mesangial cells. Am J Physiol Renal Physiol. 2009;296:F328–36.
García-Alonso V, López-Vicario C, Titos E, Morán-Salvador E, González-Périz A, Rius B, Párrizas M, Werz O, Arroyo V, Clària J. Coordinate functional regulation between microsomal prostaglandin E synthase-1 (mPGES-1) and peroxisome proliferator-activated receptor γ (PPARγ) in the conversion of white-to-brown adipocytes. J Biol Chem. 2013;288:28230–42.
Delerive P, De Bosscher K, Besnard S, Vanden Berghe W, Peters JM, Gonzalez FJ, Fruchart JC, Tedgui A, Haegeman G, Staels B. Peroxisome proliferator-activated receptor alpha negatively regulates the vascular inflammatory gene response by negative cross-talk with transcription factors NF-kappaB and AP-1. J Biol Chem. 1999;274:32048–54.
Contreras AV, Torres N, Tovar AR. PPAR-α as a key nutritional and environmental sensor for metabolic adaptation. Adv Nutr. 2013;4:439–52.
Monsalve FA, Pyarasani RD, Delgado-Lopez F, Moore-Carrasco R. Peroxisome proliferator-activated receptor targets for the treatment of metabolic diseases. Mediat Inflamm. 2013;2013:549627.
Xu HE, Lambert MH, Montana VG, Parks DJ, Blanchard SG, Brown PJ, Sternbach DD, Lehmann JM, Wisely GB, Willson TM, Kliewer SA, Milburn MV. Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Mol Cell. 1999;3:397–403.
Hihi AK, Michalik L, Wahli W. PPARs: transcriptional effectors of fatty acids and their derivatives. Cell Mol Life Sci. 2002;59:790–8.
Itoh T, Fairall L, Amin K, Inaba Y, Szanto A, Balint BL, Nagy L, Yamamoto K, Schwabe JW. Structural basis for the activation of PPARgamma by oxidized fatty acids. Nat Struct Mol Biol. 2008;15:924–31.
Muralikumar S, Vetrivel U, Narayanasamy A, Das UN. Probing the intermolecular interactions of PPARγ-LBD with polyunsaturated fatty acids and their anti-inflammatory metabolites to infer most potential binding moieties. Lipids Health Dis. 2017;16:17.
Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc Natl Acad Sci USA. 1997;94:4312–7.
Popeijus HE, van Otterdijk SD, van der Krieken SE, Konings M, Serbonij K, Plat J, Mensink RP. Fatty acid chain length and saturation influences PPARα transcriptional activation and repression in HepG2 cells. Mol Nutr Food Res. 2014;58:2342–9.
Kliewer SA, Sundseth SS, Jones SA, Brown PJ, Wisely GB, Koble CS, Devchand P, Wahli W, Willson TM, Lenhard JM, Lehmann JM. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha and gamma. Proc Natl Acad Sci USA. 1997;94:4318–23.
Krey G, Braissant O, L’Horset F, Kalkhoven E, Perroud M, Parker MG, Wahli W. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand assay. Mol Endocrinol. 1997;11:779–91.
Hostetler HA, Kier AB, Schroeder F. Very-long-chain and branched-chain fatty acyl-CoAs are high affinity ligands for the peroxisome proliferator-activated receptor alpha (PPARalpha). Biochemistry. 2006;45:7669–81.
Trostchansky A, Rubbo H. Nitrated fatty acids: mechanisms of formation, chemical characterization, and biological properties. Free Radic Biol Med. 2008;44:1887–96.
Narala VR, Adapala RK, Suresh MV, Brock TG, Peters-Golden M, Reddy RC. Leukotriene B4 is a physiologically relevant endogenous peroxisome proliferator-activated receptor-alpha agonist. J Biol Chem. 2010;285:22067–74.
Yu K, Bayona W, Kallen CB, Harding HP, Ravera CP, McMahon G, Brown M, Lazar MA. Differential activation of peroxisome proliferator-activated receptors by eicosanoids. J Biol Chem. 1995;270:23975–83.
Jisaka M, Iwanaga C, Takahashi N, Goto T, Kawada T, Yamamoto T, Ikeda I, Nishimura K, Nagaya T, Fushiki T, Yokota K. Double dioxygenation by mouse 8S-lipoxygenase: specific formation of a potent peroxisome proliferator-activated receptor alpha agonist. Biochem Biophys Res Commun. 2005;338:136–43.
Flores AM, Li L, McHugh NG, Aneskievich BJ. Enzyme association with PPARgamma: evidence of a new role for 15-lipoxygenase type 2. Chem Biol Interact. 2005;151:121–32.
Naruhn S, Meissner W, Adhikary T, Kaddatz K, Klein T, Watzer B, Müller-Brüsselbach S, Müller R. 15-Hydroxyeicosatetraenoic acid is a preferential peroxisome proliferator-activated receptor beta/delta agonist. Mol Pharmacol. 2010;77:171–84.
Cowart LA, Wei S, Hsu MH, Johnson EF, Krishna MU, Falck JR, Capdevila JH. The CYP4A isoforms hydroxylate epoxyeicosatrienoic acids to form high affinity peroxisome proliferator-activated receptor ligands. J Biol Chem. 2002;277:35105–12.
Fang X, Dillon JS, Hu S, Harmon SD, Yao J, Anjaiah S, Falck JR, Spector AA. 20-Carboxy-arachidonic acid is a dual activator of peroxisome proliferator-activated receptors alpha and gamma. Prostaglandins Other Lipid Mediat. 2007;82:175–84.
Liang CJ, Tseng CP, Yang CM, Ma YH. 20-Hydroxyeicosatetraenoic acid inhibits ATP-induced COX-2 expression via peroxisome proliferator activator receptor-α in vascular smooth muscle cells. Br J Pharmacol. 2011;163:815–25.
Chen W, Yang S, Ping W, Fu X, Xu Q, Wang J. CYP2J2 and EETs protect against lung ischemia/reperfusion injury via anti-inflammatory effects in vivo and in vitro. Cell Physiol Biochem. 2015;35:2043–54.
Straus DS, Glass CK. Cyclopentenone prostaglandins: new insights on biological activities and cellular targets. Med Res Rev. 2001;21:185–210.
Straus DS, Pascual G, Li M, Welch JS, Ricote M, Hsiang CH, Sengchanthalangsy LL, Ghosh G, Glass CK. 15-Deoxy-delta 12,14-prostaglandin J2 inhibits multiple steps in the NF-kappa B signaling pathway. Proc Natl Acad Sci USA. 2000;97:4844–9.
Waku T, Shiraki T, Oyama T, Fujimoto Y, Maebara K, Kamiya N, Jingami H, Morikawa K. Structural insight into PPARgamma activation through covalent modification with endogenous fatty acids. J Mol Biol. 2009;385:188–99.
Musiek ES, Gao L, Milne GL, Han W, Everhart MB, Wang D, Backlund MG, DuBois RN, Zanoni G, Vidari G, Blackwell TS, Morrow JD. Cyclopentenone isoprostanes inhibit the inflammatory response in macrophages. J Biol Chem. 2005;280:35562–70.
Shiraki T, Kodama TS, Shiki S, Nakagawa T, Jingami H. Spectroscopic analyses of the binding kinetics of 15d-PGJ2 to the PPARgamma ligand-binding domain by multi-wavelength global fitting. Biochem J. 2006;393:749–55.
Akundi RS, Candelario-Jalil E, Hess S, Hüll M, Lieb K, Gebicke-Haerter PJ, Fiebich BL. Signal transduction pathways regulating cyclooxygenase-2 in lipopolysaccharide-activated primary rat microglia. Glia. 2005;51:199–208.
Colville-Nash PR, Qureshi SS, Willis D, Willoughby DA. Inhibition of inducible nitric oxide synthase by peroxisome proliferator-activated receptor agonists: correlation with induction of heme oxygenase 1. J Immunol. 1998;161:978–84.
Lee CW, Lin CC, Lee IT, Lee HC, Yang CM. Activation and induction of cytosolic phospholipase A2 by TNF-α mediated through Nox2, MAPKs, NF-κB, and p300 in human tracheal smooth muscle cells. J Cell Physiol. 2011;226:2103–14.
Ko HM, Lee SH, Bang M, Kim KC, Jeon SJ, Park YM, Han SH, Kim HY, Lee J, Shin CY. Tyrosine kinase Fyn regulates iNOS expression in LPS-stimulated astrocytes via modulation of ERK phosphorylation. Biochem Biophys Res Commun. 2018;495:1214–20.
Rubbo H, Radi R, Trujillo M, Telleri R, Kalyanaraman B, Barnes S, Kirk M, Freeman BA. Nitric oxide regulation of superoxide and peroxynitrite-dependent lipid peroxidation. Formation of novel nitrogen-containing oxidized lipid derivatives. J Biol Chem. 1994;269:26066–75.
Batthyany C, Schopfer FJ, Baker PR, Durán R, Baker LM, Huang Y, Cerveñansky C, Branchaud BP, Freeman BA. Reversible post-translational modification of proteins by nitrated fatty acids in vivo. J Biol Chem. 2006;281:20450–63.
Cui T, Schopfer FJ, Zhang J, Chen K, Ichikawa T, Baker PR, Batthyany C, Chacko BK, Feng X, Patel RP, Agarwal A, Freeman BA, Chen YE. Nitrated fatty acids: endogenous anti-inflammatory signaling mediators. J Biol Chem. 2006;281:35686–98.
Trostchansky A, Bonilla L, Thomas CP, O’Donnell VB, Marnett LJ, Radi R, Rubbo H. Nitroarachidonic acid, a novel peroxidase inhibitor of prostaglandin endoperoxide H synthases 1 and 2. J Biol Chem. 2011;286:12891–900.
Baker PR, Lin Y, Schopfer FJ, Woodcock SR, Groeger AL, Batthyany C, Sweeney S, Long MH, Iles KE, Baker LM, Branchaud BP, Chen YE, Freeman BA. Fatty acid transduction of nitric oxide signaling: multiple nitrated unsaturated fatty acid derivatives exist in human blood and urine and serve as endogenous peroxisome proliferator-activated receptor ligands. J Biol Chem. 2005;280:42464–75.
Schopfer FJ, Lin Y, Baker PR, Cui T, Garcia-Barrio M, Zhang J, Chen K, Chen YE, Freeman BA. Nitrolinoleic acid: an endogenous peroxisome proliferator-activated receptor gamma ligand. Proc Natl Acad Sci USA. 2005;102:2340–5.
Li Y, Zhang J, Schopfer FJ, Martynowski D, Garcia-Barrio MT, Kovach A, Suino-Powell K, Baker PR, Freeman BA, Chen YE, Xu HE. Molecular recognition of nitrated fatty acids by PPAR gamma. Nat Struct Mol Biol. 2008;15:865–7.
González-Perilli L, Álvarez MN, Prolo C, Radi R, Rubbo H, Trostchansky A. Nitroarachidonic acid prevents NADPH oxidase assembly and superoxide radical production in activated macrophages. Free Radic Biol Med. 2013;58:126–33.
Yu C, Markan K, Temple KA, Deplewski D, Brady MJ, Cohen RN. The nuclear receptor corepressors NCoR and SMRT decrease peroxisome proliferator-activated receptor gamma transcriptional activity and repress 3T3-L1 adipogenesis. J Biol Chem. 2005;280:13600–5.
Jennewein C, Kuhn AM, Schmidt MV, Meilladec-Jullig V, von Knethen A, Gonzalez FJ, Brüne B. Sumoylation of peroxisome proliferator-activated receptor gamma by apoptotic cells prevents lipopolysaccharide-induced NCoR removal from kappaB binding sites mediating transrepression of proinflammatory cytokines. J Immunol. 2008;181:5646–52.
Liu MH, Li J, Shen P, Husna B, Tai ES, Yong EL. A natural polymorphism in peroxisome proliferator-activated receptor-alpha hinge region attenuates transcription due to defective release of nuclear receptor corepressor from chromatin. Mol Endocrinol. 2008;22:1078–92.
Lu Y, Zhou Q, Shi Y, Liu J, Zhong F, Hao X, Li C, Chen N, Wang W. SUMOylation of PPARγ by rosiglitazone prevents LPS-induced NCoR degradation mediating down regulation of chemokines expression in renal proximal tubular cells. PLoS ONE. 2013;8:e79815.
Guo C, Li Y, Gow CH, Wong M, Zha J, Yan C, Liu H, Wang Y, Burris TP, Zhang J. The optimal corepressor function of nuclear receptor corepressor (NCoR) for peroxisome proliferator-activated receptor γ requires G protein pathway suppressor 2. J Biol Chem. 2015;290:3666–79.
Diezko R, Suske G. Ligand binding reduces SUMOylation of the peroxisome proliferator-activated receptor γ (PPARγ) activation function 1 (AF1) domain. PLoS ONE. 2013;8:e66947.
Ogawa S, Lozach J, Benner C, Pascual G, Tangirala RK, Westin S, Hoffmann A, Subramaniam S, David M, Rosenfeld MG, Glass CK. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell. 2005;122:707–21.
Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG, Glass CK. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature. 2005;437:759–63.
Subbaramaiah K, Dannenberg AJ. Cyclooxygenase-2 transcription is regulated by human papillomavirus 16 E6 and E7 oncoproteins: evidence of a corepressor/coactivator exchange. Cancer Res. 2007;67:3976–85.
Dowell P, Ishmael JE, Avram D, Peterson VJ, Nevrivy DJ, Leid M. Identification of nuclear receptor corepressor as a peroxisome proliferator-activated receptor alpha interacting protein. J Biol Chem. 1999;274:15901–7.
Pourcet B, Pineda-Torra I, Derudas B, Staels B, Glineur C. SUMOylation of human peroxisome proliferator-activated receptor alpha inhibits its trans-activity through the recruitment of the nuclear corepressor NCoR. J Biol Chem. 2010;285:5983–92.
Blanquart C, Barbier O, Fruchart JC, Staels B, Glineur C. Peroxisome proliferator-activated receptor alpha (PPARalpha) turnover by the ubiquitin-proteasome system controls the ligand-induced expression level of its target genes. J Biol Chem. 2002;277:37254–9.
Rodríguez JE, Liao JY, He J, Schisler JC, Newgard CB, Drujan D, Glass DJ, Frederick CB, Yoder BC, Lalush DS, Patterson C, Willis MS. The ubiquitin ligase MuRF1 regulates PPARα activity in the heart by enhancing nuclear export via monoubiquitination. Mol Cell Endocrinol. 2015;413:36–48.
Genini D, Catapano CV. Block of nuclear receptor ubiquitination. A mechanism of ligand-dependent control of peroxisome proliferator-activated receptor delta activity. J Biol Chem. 2007;282:11776–85.
Hauser S, Adelmant G, Sarraf P, Wright HM, Mueller E, Spiegelman BM. Degradation of the peroxisome proliferator-activated receptor gamma is linked to ligand-dependent activation. J Biol Chem. 2000;275:18527–33.
Li JJ, Wang R, Lama R, Wang X, Floyd ZE, Park EA, Liao FF. Ubiquitin ligase NEDD4 regulates PPARγ stability and adipocyte differentiation in 3T3-L1 cells. Sci Rep. 2016;6:38550.
Kilroy G, Kirk-Ballard H, Carter LE, Floyd ZE. The ubiquitin ligase Siah2 regulates PPARγ activity in adipocytes. Endocrinology. 2012;153:1206–18.
Kilroy GE, Zhang X, Floyd ZE. PPAR-gamma AF-2 domain functions as a component of a ubiquitin-dependent degradation signal. Obesity. 2009;17:665–73.
Watanabe M, Takahashi H, Saeki Y, Ozaki T, Itoh S, Suzuki M, Mizushima W, Tanaka K, Hatakeyama S. The E3 ubiquitin ligase TRIM23 regulates adipocyte differentiation via stabilization of the adipogenic activator PPARγ. Elife. 2015;4:e05615.
Lee KW, Kwak SH, Koo YD, Cho YK, Lee HM, Jung HS, Cho YM, Park YJ, Chung SS, Park KS. F-box only protein 9 is an E3 ubiquitin ligase of PPARγ. Exp Mol Med. 2016;48:e234.
Kim JH, Park KW, Lee EW, Jang WS, Seo J, Shin S, Hwang KA, Song J. Suppression of PPARγ through MKRN1-mediated ubiquitination and degradation prevents adipocyte differentiation. Cell Death Differ. 2014;21:594–603.
He J, Quintana MT, Sullivan J, Parry TL, Grevengoed TJ, Schisler JC, Hill JA, Yates CC, Mapanga RF, Essop MF, Stansfield WE, Bain JR, Newgard CB, Muehlbauer MJ, Han Y, Clarke BA, Willis MS. MuRF2 regulates PPARγ1 activity to protect against diabetic cardiomyopathy and enhance weight gain induced by a high fat diet. Cardiovasc Diabetol. 2015;14:97.
Li HH, Hsu HH, Chang GJ, Chen IC, Ho WJ, Hsu PC, Chen WJ, Pang JS, Huang CC, Lai YJ. Prostanoid EP4 agonist L-902,688 activates PPARγ and attenuates pulmonary arterial hypertension. Am J Physiol Lung Cell Mol Physiol. 2018;314:L349–59.
Adams M, Reginato MJ, Shao D, Lazar MA, Chatterjee VK. Transcriptional activation by peroxisome proliferator-activated receptor gamma is inhibited by phosphorylation at a consensus mitogen-activated protein kinase site. J Biol Chem. 1997;272:5128–32.
Camp HS, Tafuri SR. Regulation of peroxisome proliferator-activated receptor gamma activity by mitogen-activated protein kinase. J Biol Chem. 1997;272:10811–6.
Chen F, Wang M, O’Connor JP, He M, Tripathi T, Harrison LE. Phosphorylation of PPARgamma via active ERK1/2 leads to its physical association with p65 and inhibition of NF-kappabeta. J Cell Biochem. 2003;90:732–44.
Camp HS, Tafuri SR, Leff T. c-Jun N-terminal kinase phosphorylates peroxisome proliferator-activated receptor-gamma1 and negatively regulates its transcriptional activity. Endocrinology. 1999;140:392–7.
Lazennec G, Canaple L, Saugy D, Wahli W. Activation of peroxisome proliferator-activated receptors (PPARs) by their ligands and protein kinase A activators. Mol Endocrinol. 2000;14:1962–75.
Barger PM, Browning AC, Garner AN, Kelly DP. p38 Mitogen-activated protein kinase activates peroxisome proliferator-activated receptor alpha: a potential role in the cardiac metabolic stress response. J Biol Chem. 2001;276:44495–501.
Juge-Aubry CE, Hammar E, Siegrist-Kaiser C, Pernin A, Takeshita A, Chin WW, Burger AG, Meier CA. Regulation of the transcriptional activity of the peroxisome proliferator-activated receptor alpha by phosphorylation of a ligand-independent trans-activating domain. J Biol Chem. 1999;274:10505–10.
Diradourian C, Le May C, Caüzac M, Girard J, Burnol AF, Pégorier JP. Involvement of ZIP/p62 in the regulation of PPARalpha transcriptional activity by p38-MAPK. Biochim Biophys Acta. 2008;1781:239–44.
Aleshin S, Grabeklis S, Hanck T, Sergeeva M, Reiser G. Peroxisome proliferator-activated receptor (PPAR)-gamma positively controls and PPARalpha negatively controls cyclooxygenase-2 expression in rat brain astrocytes through a convergence on PPARbeta/delta via mutual control of PPAR expression levels. Mol Pharmacol. 2009;76:414–24.
Zuo X, Wu Y, Morris JS, Stimmel JB, Leesnitzer LM, Fischer SM, Lippman SM, Shureiqi I. Oxidative metabolism of linoleic acid modulates PPAR-beta/delta suppression of PPAR-gamma activity. Oncogene. 2006;25:1225–41.
Meade EA, McIntyre TM, Zimmerman GA, Prescott SM. Peroxisome proliferators enhance cyclooxygenase-2 expression in epithelial cells. J Biol Chem. 1999;274:8328–34.
Pontsler AV, St Hilaire A, Marathe GK, Zimmerman GA, McIntyre TM. Cyclooxygenase-2 is induced in monocytes by peroxisome proliferator activated receptor gamma and oxidized alkyl phospholipids from oxidized low density lipoprotein. J Biol Chem. 2002;277:13029–36.
Lo CJ, Chiu KC, Fu M, Lo R, Helton S. Fish oil augments macrophage cyclooxygenase II (COX-2) gene expression induced by endotoxin. J Surg Res. 1999;86:103–7.
Chêne G, Dubourdeau M, Balard P, Escoubet-Lozach L, Orfila C, Berry A, Bernad J, Aries MF, Charveron M, Pipy B. n-3 and n-6 polyunsaturated fatty acids induce the expression of COX-2 via PPARgamma activation in human keratinocyte HaCaT cells. Biochim Biophys Acta. 2007;1771:576–89.
Sheldrick EL, Derecka K, Marshall E, Chin EC, Hodges L, Wathes DC, Abayasekara DR, Flint AP. Peroxisome-proliferator-activated receptors and the control of levels of prostaglandin-endoperoxide synthase 2 by arachidonic acid in the bovine uterus. Biochem J. 2007;406:175–83.
Paik JH, Ju JH, Lee JY, Boudreau MD, Hwang DH. Two opposing effects of non-steroidal anti-inflammatory drugs on the expression of the inducible cyclooxygenase Mediation through different signaling pathways. J Biol Chem. 2000;275:28173–9.
Kalajdzic T, Faour WH, He QW, Fahmi H, Martel-Pelletier J, Pelletier JP, Di Battista JA. Nimesulide, a preferential cyclooxygenase 2 inhibitor, suppresses peroxisome proliferator-activated receptor induction of cyclooxygenase 2 gene expression in human synovial fibroblasts: evidence for receptor antagonism. Arthritis Rheum. 2002;46:494–506.
Ayoub SS, Botting RM, Joshi AN, Seed MP, Colville-Nash PR. Activation of macrophage peroxisome proliferator-activated receptor-gamma by diclofenac results in the induction of cyclooxygenase-2 protein and the synthesis of anti-inflammatory cytokines. Mol Cell Biochem. 2009;327:101–10.
Nixon JB, Kamitani H, Baek SJ, Eling TE. Evaluation of eicosanoids and NSAIDs as PPARgamma ligands in colorectal carcinoma cells. Prostaglandins Leukot Essent Fatty Acids. 2003;68:323–30.
Puhl AC, Milton FA, Cvoro A, Sieglaff DH, Campos JC, Bernardes A, Filgueira CS, Lindemann JL, Deng T, Neves FA, Polikarpov I, Webb P. Mechanisms of peroxisome proliferator activated receptor γ regulation by non-steroidal anti-inflammatory drugs. Nucl Recept Signal. 2015;13:e004.
Pang L, Nie M, Corbett L, Knox AJ. Cyclooxygenase-2 expression by nonsteroidal anti-inflammatory drugs in human airway smooth muscle cells: role of peroxisome proliferator-activated receptors. J Immunol. 2003;170:1043–51.
Juan H, Sametz W. Uptake, stimulated release and metabolism of (1-14C)-eicosapentaenoic acid in a perfused organ of the rabbit. Naunyn Schmiedebergs Arch Pharmacol. 1983;324:207–11.
Nieves D, Moreno JJ. Effect of arachidonic and eicosapentaenoic acid metabolism on RAW 264.7 macrophage proliferation. J Cell Physiol. 2006;208:428–34.
Yang P, Chan D, Felix E, Cartwright C, Menter DG, Madden T, Klein RD, Fischer SM, Newman RA. Formation and antiproliferative effect of prostaglandin E(3) from eicosapentaenoic acid in human lung cancer cells. J Lipid Res. 2004;45:1030–9.
Jin J, Tang Q, Li Z, Zhao Z, Zhang Z, Lu L, Zhu T, Vanhoutte PM, Leung SW, Tu R, Shi Y. Prostaglandin E2 regulates renal function in C57/BL6 mouse with 5/6 nephrectomy. Life Sci. 2017;174:68–76.
Gryglewski RJ. Prostacyclin among prostanoids. Pharmacol Rep. 2008;60:3–11.
Staels B, Koenig W, Habib A, Merval R, Lebret M, Torra IP, Delerive P, Fadel A, Chinetti G, Fruchart JC, Najib J, Maclouf J, Tedgui A. Activation of human aortic smooth-muscle cells is inhibited by PPARalpha but not by PPARgamma activators. Nature. 1998;393:790–3.
Rival Y, Benéteau N, Taillandier T, Pezet M, Dupont-Passelaigue E, Patoiseau JF, Junquéro D, Colpaert FC, Delhon A. PPARalpha and PPARdelta activators inhibit cytokine-induced nuclear translocation of NF-kappaB and expression of VCAM-1 in EAhy926 endothelial cells. Eur J Pharmacol. 2002;435:143–51.
Dubrac S, Stoitzner P, Pirkebner D, Elentner A, Schoonjans K, Auwerx J, Saeland S, Hengster P, Fritsch P, Romani N, Schmuth M. Peroxisome proliferator-activated receptor-alpha activation inhibits Langerhans cell function. J Immunol. 2007;178:4362–72.
Ramanan S, Kooshki M, Zhao W, Hsu FC, Robbins ME. PPARalpha ligands inhibit radiation-induced microglial inflammatory responses by negatively regulating NF-kappaB and AP-1 pathways. Free Radic Biol Med. 2008;45:1695–704.
Zingarelli B, Piraino G, Hake PW, O’Connor M, Denenberg A, Fan H, Cook JA. Peroxisome proliferator-activated receptor delta regulates inflammation via NF-{kappa}B signaling in polymicrobial sepsis. Am J Pathol. 2010;177:1834–47.
Barroso E, Eyre E, Palomer X, Vázquez-Carrera M. The peroxisome proliferator-activated receptor β/δ (PPARβ/δ) agonist GW501516 prevents TNF-α-induced NF-κB activation in human HaCaT cells by reducing p65 acetylation through AMPK and SIRT1. Biochem Pharmacol. 2011;81:534–43.
Han S, Inoue H, Flowers LC, Sidell N. Control of COX-2 gene expression through peroxisome proliferator-activated receptor gamma in human cervical cancer cells. Clin Cancer Res. 2003;9:4627–35.
Bren-Mattison Y, Meyer AM, Van Putten V, Li H, Kuhn K, Stearman R, Weiser-Evans M, Winn RA, Heasley LE, Nemenoff RA. Antitumorigenic effects of peroxisome proliferator-activated receptor-gamma in non-small-cell lung cancer cells are mediated by suppression of cyclooxygenase-2 via inhibition of nuclear factor-kappaB. Mol Pharmacol. 2008;73:709–17.
Remels AH, Langen RC, Gosker HR, Russell AP, Spaapen F, Voncken JW, Schrauwen P, Schols AM. PPARgamma inhibits NF-kappaB-dependent transcriptional activation in skeletal muscle. Am J Physiol Endocrinol Metab. 2009;297:E174–83.
He X, Liu W, Shi M, Yang Z, Zhang X, Gong P. Docosahexaenoic acid attenuates LPS-stimulated inflammatory response by regulating the PPARγ/NF-κB pathways in primary bovine mammary epithelial cells. Res Vet Sci. 2017;112:7–12.
Zúñiga J, Cancino M, Medina F, Varela P, Vargas R, Tapia G, Videla LA, Fernández V. N-3 PUFA supplementation triggers PPAR-α activation and PPAR-α/NF-κB interaction: anti-inflammatory implications in liver ischemia-reperfusion injury. PLoS ONE. 2011;6:e28502.
Schnegg CI, Kooshki M, Hsu FC, Sui G, Robbins ME. PPARδ prevents radiation-induced proinflammatory responses in microglia via transrepression of NF-κB and inhibition of the PKCα/MEK1/2/ERK1/2/AP-1 pathway. Free Radic Biol Med. 2012;52:1734–43.
Chung SW, Kang BY, Kim SH, Pak YK, Cho D, Trinchieri G, Kim TS. Oxidized low density lipoprotein inhibits interleukin-12 production in lipopolysaccharide-activated mouse macrophages via direct interactions between peroxisome proliferator-activated receptor-gamma and nuclear factor-kappa B. J Biol Chem. 2000;275:32681–7.
Hou Y, Moreau F, Chadee K. PPARγ is an E3 ligase that induces the degradation of NFκB/p65. Nat Commun. 2012;3:1300.
Hou Y, Gao J, Xu H, Xu Y, Zhang Z, Xu Q, Zhang C. PPARγ E3 ubiquitin ligase regulates MUC1-C oncoprotein stability. Oncogene. 2014;33:5619–25.
Jové M, Laguna JC, Vázquez-Carrera M. Agonist-induced activation releases peroxisome proliferator-activated receptor beta/delta from its inhibition by palmitate-induced nuclear factor-kappaB in skeletal muscle cells. Biochim Biophys Acta. 2005;1734:52–61.
Aarenstrup L, Flindt EN, Otkjaer K, Kirkegaard M, Andersen JS, Kristiansen K. HDAC activity is required for p65/RelA-dependent repression of PPARdelta-mediated transactivation in human keratinocytes. J Invest Dermatol. 2008;128:1095–106.
Stockert J, Wolf A, Kaddatz K, Schnitzer E, Finkernagel F, Meissner W, Müller-Brüsselbach S, Kracht M, Müller R. Regulation of TAK1/TAB 1-mediated IL-1β signaling by cytoplasmic PPARβ/δ. PLoS ONE. 2013;8:e63011.
Yang X, Kume S, Tanaka Y, Isshiki K, Araki S, Chin-Kanasaki M, Sugimoto T, Koya D, Haneda M, Sugaya T, Li D, Han P, Nishio Y, Kashiwagi A, Maegawa H, Uzu T. GW501516, a PPARδ agonist, ameliorates tubulointerstitial inflammation in proteinuric kidney disease via inhibition of TAK1-NFκB pathway in mice. PLoS ONE. 2011;6:e25271.
Su X, Zhou G, Wang Y, Yang X, Li L, Yu R, Li D. The PPARβ/δ agonist GW501516 attenuates peritonitis in peritoneal fibrosis via inhibition of TAK1-NFκB pathway in rats. Inflammation. 2014;37:729–37.
Schmitz ML, Mattioli I, Buss H, Kracht M. NF-kappaB: a multifaceted transcription factor regulated at several levels. ChemBioChem. 2004;5:1348–58.
Diamant G, Dikstein R. Transcriptional control by NF-κB: elongation in focus. Biochim Biophys Acta. 2013;1829:937–45.
Mochizuki K, Suzuki T, Goda T. PPAR alpha and PPAR delta transactivity and p300 binding activity induced by arachidonic acid in colorectal cancer cell line Caco-2. J Nutr Sci Vitaminol. 2008;54:298–302.
Subbaramaiah K, Lin DT, Hart JC, Dannenberg AJ. Peroxisome proliferator-activated receptor gamma ligands suppress the transcriptional activation of cyclooxygenase-2. Evidence for involvement of activator protein-1 and CREB-binding protein/p300. J Biol Chem. 2001;276:12440–8.
Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–80.
Xue B, Yang Z, Wang X, Shi H. Omega-3 polyunsaturated fatty acids antagonize macrophage inflammation via activation of AMPK/SIRT1 pathway. PLoS ONE. 2012;7:e45990.
Wang W, Bai L, Qiao H, Lu Y, Yang L, Zhang J, Lin R, Ren F, Zhang J, Ji M. The protective effect of fenofibrate against TNF-α-induced CD40 expression through SIRT1-mediated deacetylation of NF-κB in endothelial cells. Inflammation. 2014;37:177–85.
Okayasu T, Tomizawa A, Suzuki K, Manaka K, Hattori Y. PPARalpha activators upregulate eNOS activity and inhibit cytokine-induced NF-kappaB activation through AMP-activated protein kinase activation. Life Sci. 2008;82:884–91.
Wang W, Lin Q, Lin R, Zhang J, Ren F, Zhang J, Ji M, Li Y. PPARα agonist fenofibrate attenuates TNF-α-induced CD40 expression in 3T3-L1 adipocytes via the SIRT1-dependent signaling pathway. Exp Cell Res. 2013;319:1523–33.
Pantazi E, Folch-Puy E, Bejaoui M, Panisello A, Varela AT, Rolo AP, Palmeira CM, Roselló-Catafau J. PPARα agonist WY-14643 induces SIRT1 activity in rat fatty liver ischemia–reperfusion injury. Biomed Res Int. 2015;2015:894679.
Yang Z, Kahn BB, Shi H, Xue BZ. Macrophage alpha1 AMP-activated protein kinase (alpha1AMPK) antagonizes fatty acid-induced inflammation through SIRT1. J Biol Chem. 2010;285:19051–9.
Jiang S, Wang W, Miner J, Fromm M. Cross regulation of sirtuin 1, AMPK, and PPARγ in conjugated linoleic acid treated adipocytes. PLoS ONE. 2012;7:e48874.
Zhang J, Zhang Y, Xiao F, Liu Y, Wang J, Gao H, Rong S, Yao Y, Li J, Xu G. The peroxisome proliferator-activated receptor γ agonist pioglitazone prevents NF-κB activation in cisplatin nephrotoxicity through the reduction of p65 acetylation via the AMPK-SIRT1/p300 pathway. Biochem Pharmacol. 2016;101:100–11.
Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–6.
Han L, Zhou R, Niu J, McNutt MA, Wang P, Tong T. SIRT1 is regulated by a PPAR{γ}-SIRT1 negative feedback loop associated with senescence. Nucleic Acids Res. 2010;38:7458–71.
Buroker NE, Barboza J, Huang JY. The IkappaBalpha gene is a peroxisome proliferator-activated receptor cardiac target gene. FEBS J. 2009;276:3247–55.
Delerive P, Gervois P, Fruchart JC, Staels B. Induction of IkappaBalpha expression as a mechanism contributing to the anti-inflammatory activities of peroxisome proliferator-activated receptor-alpha activators. J Biol Chem. 2000;275:36703–7.
Delerive P, De Bosscher K, Vanden Berghe W, Fruchart JC, Haegeman G, Staels B. DNA binding-independent induction of IkappaBalpha gene transcription by PPARalpha. Mol Endocrinol. 2002;16:1029–39.
Scirpo R, Fiorotto R, Villani A, Amenduni M, Spirli C, Strazzabosco M. Stimulation of nuclear receptor peroxisome proliferator-activated receptor-γ limits NF-κB-dependent inflammation in mouse cystic fibrosis biliary epithelium. Hepatology. 2015;62:1551–62.
Patel L, Pass I, Coxon P, Downes CP, Smith SA, Macphee CH. Tumor suppressor and anti-inflammatory actions of PPARgamma agonists are mediated via upregulation of PTEN. Curr Biol. 2001;11:764–8.
Teresi RE, Shaiu CW, Chen CS, Chatterjee VK, Waite KA, Eng C. Increased PTEN expression due to transcriptional activation of PPARgamma by Lovastatin and Rosiglitazone. Int J Cancer. 2006;118:2390–8.
Teresi RE, Waite KA. PPARgamma, PTEN, and the fight against cancer. PPAR Res. 2008;2008:932632.
Kim KY, Ahn JH, Cheon HG. Anti-angiogenic action of PPARγ ligand in human umbilical vein endothelial cells is mediated by PTEN upregulation and VEGFR-2 downregulation. Mol Cell Biochem. 2011;358:375–85.
Ham SA, Hwang JS, Yoo T, Lee H, Kang ES, Park C, Oh JW, Lee HT, Min G, Kim JH, Seo HG. Ligand-activated PPARδ inhibits UVB-induced senescence of human keratinocytes via PTEN-mediated inhibition of superoxide production. Biochem J. 2012;444:27–38.
Pedchenko TV, Gonzalez AL, Wang D, DuBois RN, Massion PP. Peroxisome proliferator-activated receptor beta/delta expression and activation in lung cancer. Am J Respir Cell Mol Biol. 2008;39:689–96.
He P, Borland MG, Zhu B, Sharma AK, Amin S, El-Bayoumy K, Gonzalez FJ, Peters JM. Effect of ligand activation of peroxisome proliferator-activated receptor-beta/delta (PPARbeta/delta) in human lung cancer cell lines. Toxicology. 2008;254:112–7.
Madrid LV, Mayo MW, Reuther JY, Baldwin AS Jr. Akt stimulates the transactivation potential of the RelA/p65 Subunit of NF-kappa B through utilization of the Ikappa B kinase and activation of the mitogen-activated protein kinase p38. J Biol Chem. 2001;276:18934–40.
Grau R, Punzón C, Fresno M, Iñiguez MA. Peroxisome-proliferator-activated receptor alpha agonists inhibit cyclo-oxygenase 2 and vascular endothelial growth factor transcriptional activation in human colorectal carcinoma cells via inhibition of activator protein-1. Biochem J. 2006;395:81–8.
Khandoudi N, Delerive P, Berrebi-Bertrand I, Buckingham RE, Staels B, Bril A. Rosiglitazone, a peroxisome proliferator-activated receptor-gamma, inhibits the Jun NH(2)-terminal kinase/activating protein 1 pathway and protects the heart from ischemia/reperfusion injury. Diabetes. 2002;51:1507–14.
Kang YJ, Mbonye UR, DeLong CJ, Wada M, Smith WL. Regulation of intracellular cyclooxygenase levels by gene transcription and protein degradation. Prog Lipid Res. 2007;46:108–25.
Yu JH, Kim KH, Kim H. SOCS 3 and PPAR-gamma ligands inhibit the expression of IL-6 and TGF-beta1 by regulating JAK2/STAT3 signaling in pancreas. Int J Biochem Cell Biol. 2008;40:677–88.
Ji HG, Piao JY, Kim SJ, Kim DH, Lee HN, Na HK, Surh YJ. Docosahexaenoic acid inhibits Helicobacter pylori-induced STAT3 phosphorylation through activation of PPARγ. Mol Nutr Food Res. 2016;60:1448–57.
Lee JH, Joe EH, Jou I. PPAR-alpha activators suppress STAT1 inflammatory signaling in lipopolysaccharide-activated rat glia. NeuroReport. 2005;16:829–33.
Shipley JM, Waxman DJ. Down-regulation of STAT5b transcriptional activity by ligand-activated peroxisome proliferator-activated receptor (PPAR) alpha and PPARgamma. Mol Pharmacol. 2003;64:355–64.
Shipley JM, Waxman DJ. Simultaneous, bidirectional inhibitory crosstalk between PPAR and STAT5b. Toxicol Appl Pharmacol. 2004;199:275–84.
Ann SJ, Chung JH, Park BH, Kim SH, Jang J, Park S, Kang SM, Lee SH. PPARα agonists inhibit inflammatory activation of macrophages through upregulation of β-defensin 1. Atherosclerosis. 2015;240:389–97.
Lei Y, Wang K, Deng L, Chen Y, Nice EC, Huang C. Redox regulation of inflammation: old elements, a new story. Med Res Rev. 2015;35:306–40.
Zhang J, Wang X, Vikash V, Ye Q, Wu D, Liu Y, Dong W. ROS and ROS-mediated cellular signaling. Oxid Med Cell Longev. 2016;2016:4350965.
Poynter ME, Daynes RA. Peroxisome proliferator-activated receptor alpha activation modulates cellular redox status, represses nuclear factor-kappaB signaling, and reduces inflammatory cytokine production in aging. J Biol Chem. 1998;273:32833–41.
Song EA, Lim JW, Kim H. Docosahexaenoic acid inhibits IL-6 expression via PPARγ-mediated expression of catalase in cerulein-stimulated pancreatic acinar cells. Int J Biochem Cell Biol. 2017;88:60–8.
Yang Y, Li X, Zhang L, Liu L, Jing G, Cai H. Ginsenoside Rg1 suppressed inflammation and neuron apoptosis by activating PPARγ/HO-1 in hippocampus in rat model of cerebral ischemia–reperfusion injury. Int J Clin Exp Pathol. 2015;8:2484–94.
Krönke G, Kadl A, Ikonomu E, Blüml S, Fürnkranz A, Sarembock IJ, Bochkov VN, Exner M, Binder BR, Leitinger N. Expression of heme oxygenase-1 in human vascular cells is regulated by peroxisome proliferator-activated receptors. Arterioscler Thromb Vasc Biol. 2007;27:1276–82.
Kitamura Y, Kakimura J, Matsuoka Y, Nomura Y, Gebicke-Haerter PJ, Taniguchi T. Activators of peroxisome proliferator-activated receptor-gamma (PPARgamma) inhibit inducible nitric oxide synthase expression but increase heme oxygenase-1 expression in rat glial cells. Neurosci Lett. 1999;262:129–32.
Bigo C, Kaeding J, El Husseini D, Rudkowska I, Verreault M, Vohl MC, Barbier O. PPARα: a master regulator of bilirubin homeostasis. PPAR Res. 2014;2014:747014.
Sodhi K, Puri N, Kim DH, Hinds TD, Stechschulte LA, Favero G, Rodella L, Shapiro JI, Jude D, Abraham NG. PPARδ binding to heme oxygenase 1 promoter prevents angiotensin II-induced adipocyte dysfunction in Goldblatt hypertensive rats. Int J Obes. 2014;38:456–65.
Wang Y, Yu M, Ma Y, Wang R, Liu W, Xia W, Guan A, Xing C, Lu F, Ji X. Fenofibrate increases heme oxygenase 1 expression and astrocyte proliferation while limits neuronal injury during intracerebral hemorrhage. Curr Neurovasc Res. 2017;14:11–8.
Lin CC, Yang CC, Chen YW, Hsiao LD, Yang CM. Arachidonic acid induces ARE/Nrf2-dependent heme oxygenase-1 transcription in rat brain astrocytes. Mol Neurobiol. 2018;55:3328–43.
Kim JS, Lee YH, Chang YU, Yi HK. PPARγ regulates inflammatory reaction by inhibiting the MAPK/NF-κB pathway in C2C12 skeletal muscle cells. J Physiol Biochem. 2017;73:49–57.
von Knethen A, Neb H, Morbitzer V, Schmidt MV, Kuhn AM, Kuchler L, Brüne B. PPARγ stabilizes HO-1 mRNA in monocytes/macrophages which affects IFN-β expression. Free Radic Biol Med. 2011;51:396–405.
Wang S, Hannafon BN, Zhou J, Ding WQ. Clofibrate induces heme oxygenase 1 expression through a PPARα-independent mechanism in human cancer cells. Cell Physiol Biochem. 2013;32:1255–64.
Jansen T, Daiber A. Direct antioxidant properties of bilirubin and biliverdin. Is there a role for biliverdin reductase? Front Pharmacol. 2012;3:30.
Nakahira K, Kim HP, Geng XH, Nakao A, Wang X, Murase N, Drain PF, Wang X, Sasidhar M, Nabel EG, Takahashi T, Lukacs NW, Ryter SW, Morita K, Choi AM. Carbon monoxide differentially inhibits TLR signaling pathways by regulating ROS-induced trafficking of TLRs to lipid rafts. J Exp Med. 2006;203:2377–89.
Chi PL, Liu CJ, Lee IT, Chen YW, Hsiao LD, Yang CM. HO-1 induction by CO-RM2 attenuates TNF-α-induced cytosolic phospholipase A2 expression via inhibition of PKCα-dependent NADPH oxidase/ROS and NF-κB. Mediat Inflamm. 2014;2014:279171.
Riquelme SA, Bueno SM, Kalergis AM. Carbon monoxide down-modulates toll-like receptor 4/MD2 expression on innate immune cells and reduces endotoxic shock susceptibility. Immunology. 2015;144:321–32.
Yang YC, Huang YT, Hsieh CW, Yang PM, Wung BS. Carbon monoxide induces heme oxygenase-1 to modulate STAT3 activation in endothelial cells via S-glutathionylation. PLoS ONE. 2014;9:e100677.
Yeh PY, Li CY, Hsieh CW, Yang YC, Yang PM, Wung BS. CO-releasing molecules and increased heme oxygenase-1 induce protein S-glutathionylation to modulate NF-κB activity in endothelial cells. Free Radic Biol Med. 2014;70:1–13.
Lee J, Kang U, Seo EK, Kim YS. Heme oxygenase-1-mediated anti-inflammatory effects of tussilagonone on macrophages and 12-O-tetradecanoylphorbol-13-acetate-induced skin inflammation in mice. Int Immunopharmacol. 2016;34:155–64.
Ding G, Fu M, Qin Q, Lewis W, Kim HW, Fukai T, Bacanamwo M, Chen YE, Schneider MD, Mangelsdorf DJ, Evans RM, Yang Q. Cardiac peroxisome proliferator-activated receptor gamma is essential in protecting cardiomyocytes from oxidative damage. Cardiovasc Res. 2007;76:269–79.
Liu X, Jang SS, An Z, Song H, Kim WD, Yu JR, Park WY. Fenofibrate decreases radiation sensitivity via peroxisome proliferator-activated receptor α-mediated superoxide dismutase induction in HeLa cells. Radiat Oncol J. 2012;30:88–95.
Inoue I, Noji S, Awata T, Takahashi K, Nakajima T, Sonoda M, Komoda T, Katayama S. Bezafibrate has an antioxidant effect: peroxisome proliferator-activated receptor alpha is associated with Cu2+, Zn2+-superoxide dismutase in the liver. Life Sci. 1998;63:135–44.
Inoue I, Goto S, Matsunaga T, Nakajima T, Awata T, Hokari S, Komoda T, Katayama S. The ligands/activators for peroxisome proliferator-activated receptor alpha (PPARalpha) and PPARgamma increase Cu2+, Zn2+-superoxide dismutase and decrease p22phox message expressions in primary endothelial cells. Metabolism. 2001;50:3–11.
Ibarra-Lara L, Hong E, Soria-Castro E, Torres-Narváez JC, Pérez-Severiano F, Del Valle-Mondragón L, Cervantes-Pérez LG, Ramírez-Ortega M, Pastelín-Hernández GS, Sánchez-Mendoza A. Clofibrate PPARα activation reduces oxidative stress and improves ultrastructure and ventricular hemodynamics in no-flow myocardial ischemia. J Cardiovasc Pharmacol. 2012;60:323–34.
Araújo TG, Oliveira AG, Vecina JF, Marin RM, Franco ES, Abdalla Saad MJ, de Sousa Maia MB. Treatment with Parkinsonia aculeata combats insulin resistance-induced oxidative stress through the increase in PPARγ/CuZn-SOD axis expression in diet-induced obesity mice. Mol Cell Biochem. 2016;419:93–101.
Girnun GD, Domann FE, Moore SA, Robbins ME. Identification of a functional peroxisome proliferator-activated receptor response element in the rat catalase promoter. Mol Endocrinol. 2002;16:2793–801.
Okuno Y, Matsuda M, Kobayashi H, Morita K, Suzuki E, Fukuhara A, Komuro R, Shimabukuro M, Shimomura I. Adipose expression of catalase is regulated via a novel remote PPARgamma-responsive region. Biochem Biophys Res Commun. 2008;366:698–704.
Okuno Y, Matsuda M, Miyata Y, Fukuhara A, Komuro R, Shimabukuro M, Shimomura I. Human catalase gene is regulated by peroxisome proliferator activated receptor-gamma through a response element distinct from that of mouse. Endocr J. 2010;57:303–9.
Toyama T, Nakamura H, Harano Y, Yamauchi N, Morita A, Kirishima T, Minami M, Itoh Y, Okanoue T. PPARalpha ligands activate antioxidant enzymes and suppress hepatic fibrosis in rats. Biochem Biophys Res Commun. 2004;324:697–704.
Khoo NK, Hebbar S, Zhao W, Moore SA, Domann FE, Robbins ME. Differential activation of catalase expression and activity by PPAR agonists: implications for astrocyte protection in anti-glioma therapy. Redox Biol. 2013;1:70–9.
Shin MH, Lee SR, Kim MK, Shin CY, Lee DH, Chung JH. Activation of peroxisome proliferator-activated receptor alpha improves aged and UV-irradiated skin by catalase induction. PLoS ONE. 2016;11:e0162628.
Fang X, Moore AS, Nwankwo JO, Weintraub LN, Oberley WL, Snyder DG, Spector AA. Induction of cyclooxygenase-2 by overexpression of the human catalase gene in cerebral microvascular endothelial cells. J Neurochem. 2000;75:614–23.
Jang BC, Kim DH, Park JW, Kwon TK, Kim SP, Song DK, Park JG, Bae JH, Mun KC, Baek WK, Suh MH, Hla T, Suh SI. Induction of cyclooxygenase-2 in macrophages by catalase: role of NF-kappaB and PI3K signaling pathways. Biochem Biophys Res Commun. 2004;316:398–406.
Jang BC, Paik JH, Kim SP, Shin DH, Song DK, Park JG, Suh MH, Park JW, Suh SI. Catalase induced expression of inflammatory mediators via activation of NF-kappaB, PI3K/AKT, p70S6K, and JNKs in BV2 microglia. Cell Signal. 2005;17:625–33.
Hunot S, Hirsch EC. Neuroinflammatory processes in Parkinson’s disease. Ann Neurol. 2003;53(Suppl 3):S49–58 (discussion S58–60).
Liang X, Wu L, Wang Q, Hand T, Bilak M, McCullough L, Andreasson K. Function of COX-2 and prostaglandins in neurological disease. J Mol Neurosci. 2007;33:94–9.
Agrawal NK, Kant S. Targeting inflammation in diabetes: newer therapeutic options. World J Diabetes. 2014;5:697–710.
Matsuyama M, Yoshimura R. The target of arachidonic acid pathway is a new anticancer strategy for human prostate cancer. Biologics. 2008;2:725–32.
Greenhough A, Smartt HJ, Moore AE, Roberts HR, Williams AC, Paraskeva C, Kaidi A. The COX-2/PGE2 pathway: key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis. 2009;30:377–86.
Borland MG, Kehres EM, Lee C, Wagner AL, Shannon BE, Albrecht PP, Zhu B, Gonzalez FJ, Peters JM. Inhibition of tumorigenesis by peroxisome proliferator-activated receptor (PPAR)-dependent cell cycle blocks in human skin carcinoma cells. Toxicology. 2018;404–405:25–32.
Chandran K, Goswami S, Sharma-Walia N. Implications of a peroxisome proliferator-activated receptor alpha (PPARα) ligand clofibrate in breast cancer. Oncotarget. 2016;7:15577–99.
Gutting T, Weber CA, Weidner P, Herweck F, Henn S, Friedrich T, Yin S, Kzhyshkowska J, Gaiser T, Janssen KP, Reindl W, Ebert MPA, Burgermeister E. PPARγ-activation increases intestinal M1 macrophages and mitigates formation of serrated adenomas in mutant KRAS mice. Oncoimmunology. 2018;7:e1423168.
Higuchi T, Takeuchi A, Munesue S, Yamamoto N, Hayashi K, Kimura H, Miwa S, Inatani H, Shimozaki S, Kato T, Aoki Y, Abe K, Taniguchi Y, Aiba H, Murakami H, Harashima A, Yamamoto Y, Tsuchiya H. Anti-tumor effects of a nonsteroidal anti-inflammatory drug zaltoprofen on chondrosarcoma via activating peroxisome proliferator-activated receptor gamma and suppressing matrix metalloproteinase-2 expression. Cancer Med. 2018;7:1944–54.
Huang G, Yin L, Lan J, Tong R, Li M, Na F, Mo X, Chen C, Xue J, Lu Y. Synergy between peroxisome proliferator-activated receptor γ agonist and radiotherapy in cancer. Cancer Sci. 2018;109:2243–55.
Tao T, Zhao F, Xuan Q, Shen Z, Xiao J, Shen Q. Fenofibrate inhibits the growth of prostate cancer through regulating autophagy and endoplasmic reticulum stress. Biochem Biophys Res Commun. 2018;503:2685–9.
Xiao YB, Cai SH, Liu LL, Yang X, Yun JP. Decreased expression of peroxisome proliferator-activated receptor alpha indicates unfavorable outcomes in hepatocellular carcinoma. Cancer Manag Res. 2018;10:1781–9.
Yao PL, Chen L, Dobrzański TP, Zhu B, Kang BH, Müller R, Gonzalez FJ, Peters JM. Peroxisome proliferator-activated receptor-β/δ inhibits human neuroblastoma cell tumorigenesis by inducing p53- and SOX2-mediated cell differentiation. Mol Carcinog. 2017;56:1472–83.
Zhang N, Chu ES, Zhang J, Li X, Liang Q, Chen J, Chen M, Teoh N, Farrell G, Sung JJ, Yu J. Peroxisome proliferator activated receptor alpha inhibits hepatocarcinogenesis through mediating NF-κB signaling pathway. Oncotarget. 2014;5:8330–40.
Zhong WB, Tsai YC, Chin LH, Tseng JH, Tang LW, Horng S, Fan YC, Hsu SP. A synergistic anti-cancer effect of troglitazone and lovastatin in a human anaplastic thyroid cancer cell line and in a mouse xenograft model. Int J Mol Sci. 2018;19:1834.
Rizos CV, Kei A, Elisaf MS. The current role of thiazolidinediones in diabetes management. Arch Toxicol. 2016;90:1861–81.
Funding
This study was supported by the statutory budget of the Faculty of Health Sciences, University of Bielsko-Biala.
Author information
Authors and Affiliations
Contributions
JK: manuscript concept, literature search and review, and writing the manuscript, MD: participated in writing the manuscript, RB: participated in writing the manuscript and translation the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible Editor: John Di Battista.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Korbecki, J., Bobiński, R. & Dutka, M. Self-regulation of the inflammatory response by peroxisome proliferator-activated receptors. Inflamm. Res. 68, 443–458 (2019). https://doi.org/10.1007/s00011-019-01231-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00011-019-01231-1