Abstract
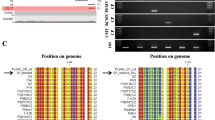
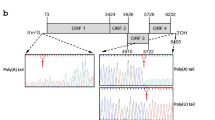
Peanut mottle virus (PeMoV) was identified for the first time in Israel in peanut plants expressing mottle symptoms. Particle morphology, biological properties and serology suggested that this virus belongs to the genusPotyvirus. The characteristics of the Israeli (IL) PeMoV were compared with those of previously reported isolates. Using RT-PCR, a 1393-bp fragment consisting of the helper component — proteinase (HC-Pro) and a 988-bp product containing the coat protein (CP) were amplified, cloned and sequenced. Comparison of the HC-Pro sequences for PeMoV-IL and PeMoV-M (reported previously), showed 98% homology at the amino acid (aa) level. The aa sequence homology of the entire CP of isolate IL and six other PeMoV isolates ranged from 92% to 98%. A phylogenetic analysis carried out using the CP nucleotide sequence data indicated close similarity between PeMoV-IL and an Australian isolate and between two M isolates. The conserved KITC and CCC motifs in the HC-Pro were replaced by KVSC and ASC, respectively, in PeMoV-IL as in strain M. The DAG motif in the CP was replaced by DAA in the PeMoV isolates including IL. These results prompted the examination of aphid transmissibility of PeMoV-IL which were low and variable among experiments. These results differ from a previous report showing high aphid transmission of PeMoV.
Similar content being viewed by others
References
Altschul, S.F., Gish, W., Miller, W., Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tools.J. Mol. Biol. 215:403–410.
Atreya, P.L., Lòpez-Moya, J.J., Chu, M., Atreya, C.D. and Pirone, T.P. (1995) Mutational analysis of the coat protein N-terminal amino acids involved in potyvirus transmission by aphids.J. Gen. Virol. 76:265–270.
Brunt, A.A., Crabtree, K., Dallwitz, M.J., Gibbs, A.J., Watson, L. and Zurcher, E.J. [Eds.] (1996) Plant Viruses Online: Descriptions and Lists from the VIDE Database. Version: 20th August 1996. URL.http://biology.anu.edu.au/groups/MES/vide/.
Desbiez, C. and Lecoq, H. (2004) The nucleotide sequence ofWatermelon mosaic virus (WMV, Potyvirus) reveals interspecific recombination between two related potyviruses in the 5′ part of the genome.Arch. Virol. 149:1619–1632.
Dietzgen, R.G., Callaghan, B., Higgins, C.M., Birch, R.G., Chen, K. and Xu, Z. (2001) Differentiation of peanut seedborne Potyviruses and Cucumoviruses by RT-PCR.Plant Dis. 85:989–992.
Flasinsky, S. and Cassidy, B.G. (1998) Potyvirus aphid transmission requires helper component and homologous coat protein for maximal efficiency.Arch. Virol. 143:2159–2172.
Gillaspie, A.G. Jr., Pittman, R.N., Pinnow, D.L. and Cassidy, B.G. (2000) Sensitive method for testing peanut seed lots for Peanut stripe and Peanut mottle viruss by immunocapture-reverse transcription-polymerase chain reaction.Plant Dis. 84:559–561.
Hall, T.A. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT.Nucl. Acids Symp. Ser. 41:95–98.
Hoffmann, K., Geske, S.M. and Moyer, J.W. (1998) Pathogenesis of tomato spotted wilt virus in peanut plants dually infected with peanut mottle virus.Plant Dis. 82:610–614.
Huet, H., Gal-On, A., Meir, E., Lecoq, H. and Raccah, B. (1994) Mutations in the helper component protease gene of Zucchini yellow mosaic virus affect its ability to mediate aphid transmissibility.J. Gen. Virol. 75:1404–1414.
Jeanmougin, F., Thompson, J.D., Gouy, M., Higgins, D.G. and Gibson, T.J. (1998) Multiple sequence alignment with Clustal X.Trends Biochem. Sci. 23:403–405.
Koenig, R. (1981) Indirect ELISA methods for the broad specificity detection of plant viruses.J. Gen. Virol. 55:53–62.
Kuhn, C.W. (1965) Symptomatology, host range and effect on yield of a seed-transmitted peanut virus.Phytopathology 55:880–884.
Lòpez-Moya, J.J., Wang, R.Y. and Pirone, T.P. (1999) Context of the coat protein DAG motif affects potyvirus transmissibility by aphids.J. Gen. Virol. 80:3281–3288.
Paguio, O.R. and Kuhn, C.W. (1973) Purification of a mild mottle strain of Peanut mottle virus.Phytopathology 63:720–724.
Pirone, T.P. and Blanc, S. (1996) Helper-dependent vector transmission of plant viruses.Annu. Rev. Phytopathol. 34:227–248.
Raccah, B., Huet, H. and Blanc, S. (2001) Potyviruses.in: Harris, K.F., Smith, O.P. and Duffus, J.E. [Eds.] Virus — Insect — Plant Interactions. Academic Press, New York, NY. pp. 181–206.
Saitou, N. and Nei, M. (1987) The neighbour-joining method: a new method for reconstructing phylogenetic trees.Mol. Biol. Evol. 4:406–425.
Singh, B. and Singh, U. (1991) Peanut as a source of protein for human foods.Plant Foods Hum. Nutr. 41:165–177.
Spiegel, S., Scott, S., Bowman-Vance, V., Tam, Y., Galiakparov, N.N. and Rosner, A. (1996) Improved detection of Prunus necrotic ringspot virus by the polymerase chain reaction.Eur. J. Plant Pathol. 102:681–685.
Teycheney, P.Y. and Dietzgen, R.G. (1994) Cloning and sequence analysis of the coat protein genes of an Australian strain of peanut mottle and an Indonesian ‘blotch’ strain of peanut stripe potyviruses.Virus Res. 31:235–244.
Thornbury, D.W., Patterson, C.A., Dessens, J.T. and Pirone, T.P. (1990) Comparative sequence of the helper component (HC) region of potato virus Y and a HC-defective strain, potato virus C.Virology 178:573–578.
Towbin, H., Staehelin, T. and Gordon, J. (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications.Proc. Natl. Acad. Sci. USA 76:4350–4354.
Urcuqui-Inchima, S., Haenni, A.L. and Bernardi, F. (2001) Potyvirus proteins: a wealth of functions.Virus Res. 74:157–175.
Author information
Authors and Affiliations
Corresponding author
Additional information
The nucleotide sequences reported in this manuscript have been deposited in GenBank under the accession numbers DQ868539 and DQ868540.
http://www.phytoparasitica.org posting March 11, 2008.
Rights and permissions
About this article
Cite this article
Spiegel, S., Sobolev, I., Dombrovsky, A. et al. Note: Characterization of aPeanut mottle virus isolate infecting peanut in Israel. Phytoparasitica 36, 168–174 (2008). https://doi.org/10.1007/BF02981329
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF02981329