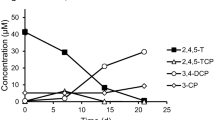
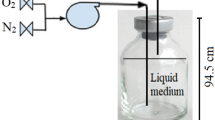
Abstract
From natural samples 11 isolates able to remove trichloroethene (CCl2CHl) from an aqueousenvironment were obtained which were capable of cometabolic degradation of CCl2CHCl by an enzyme system for phenol degradation. At an initial CCl2CHCl concentration of 1 mg/L, the resting cells of particular cultures degraded 33–94% CCl2CHCl during 1 d and their transformation capacity ranged from 0.3 to 3.1 mg CCl2CHCl per g organic fraction. An analysis of a mixed phenol-fed culture with an excellent trichloroethene-degrading ability found a markedly minority isolate represented in the consortium to be responsible for this property. This culture degraded CCl2CHCl even at a low inoculum concentration and attained a transformation capacity of 14.7 mg CCl2CHCl per g. The increase in chloride concentration after degradation was quantitative when compared with the decrease in organically bound chlorine. The degree of CCl2CHCl degradation was affected by Me2S2; this substance can significantly reduce the degrading ability of some tested cultures (>60%); however, it does not cause this inhibition with others.
Similar content being viewed by others
References
Arciero D., Vannelli T., Logan M., Hooper A.B.: Degradation of trichloroethylene by the ammonia-oxidizing bacteriumNitrosomonas europeaea.Biochem. Biophys. Res. Commun. 159, 640–643 (1989).
Chang H.L., Alvarez-Cohen L.: Transformation capacities of chlorinated organics by mixed cultures enriched on methane, progane, toluene or phenol.Biotechnol. Bioeng. 45 440–449 (1995).
Dabrock B., Riedel J., Bertram J., Gottschalk G.: Isopropylbenzene (cumene)—a new substrate for the isolation of trichloroethene-degrading bacteria.Arch. Microbiol. 158, 9–13 (1992).
Ensign S.A., Hyman M.R., Arp D.J.: Cometabolic degradation of chlorinated alkenes by alkene monooxygenase in a propylene-grownXanthobacter strain.Appl. Environ. Microbiol. 58, 3038–3046 (1992).
Folsom B.R., Chapman P.J., Pritchard P.H.: Phenol and trichloroethylene degradation byPseudomonas cepacia G4: kinetics and interaction between substrates.Appl. Environ. Microbiol.,56, 1279–1285 (1990).
Fries M.R., Forney L.J., Tiedje J.M.: Phenol- and toluene-degrading microbial populations from an aquifer in which successful trichloroethene cometabolism occurred.Appl. Environ. Microbiol..63, 1523–1530 (1997).
Futamata H., Watanabe K., Harayama S.: Relationships between the trichlorethylene-degrading activities and the amino acid sequences of phenol hydroxylases in phenol-degrading bacteria.Battelle 1st Internat. Conf. on Remediation of Chlorinated and Recalcitrant Compounds, Monterey (CA) 1998.
Ginzburg B., Chalifa I., Hadas O., Dor I., Lev O.: Formation of dimethyloligosulfides in lake Kinneret.Water Sci. Technol. 40, 73–78 (1999).
Ginzburg B., Chalifa I., Zohari T., Hadas O., Dor I., Lev O.: Identification of oligosulfide odorous compounds and their source in the sea of Galilee.Water Res. 32, 1789–1800 (1998).
Hopkins G.D., Munakata J., Semprini L., McCarty P.L.: Trichloroethylene concentration effects on pilot field-scalein situ groundwater bioremediation by phenol-oxidizing microorganisms.Environ. Sci. Technol. 27, 2542–2547 (1993).
Hopkins G.D., McCarty P.L.: Field evaluation ofin situ aerobic cometabolism of trichloroethylene and three dichloroethylene isomers using phenol and toluene as the primary substrates.Environ. Sci. Technol. 29, 1628–1637 (1995).
Ishida H., Nakamura K.: Trichloroethylene degradation byRalstonia sp. KN1-10A constitutively expressing phenol hydroxylase: transformation products, NADH limitation, and product toxicity.J. Biosci. Bioeng. 89, 438–445 (2000).
Iwasaki I., Utsumi S., Ozawa T.: New colorimetric determination of chloride using mercuric thiocyanate and ferric ions.Bull. Chem. Soc. Japan. 25, 226 (1952).
Kyung K.H., Fleming H.P.: Antimicrobial activity of sulphur compounds derived from cabbage.J. Food. Prot. 60, 67–71 (1997).
Saeki H., Akira M., Furuhashi K., Averhoff B., Gottschalk G.: Degradation of trichloroethylene by a linear-plasmid-encoded alkene monooxygenase inRhodococcus corallinus (Nocardia corallina) B-276.Microbiology 145, 1721–1730 (1999).
Schöller C., Molin S., Wilkins K.: Volatile metabolites from some gramnegative bacteria.Chemosphere 35, 1487–1495 (1997).
Shih C., Davey M.E., Zhou J., Tiedje J.M., Criddle C.S.: Effects of phenol feeding pattern on microbial community structure and cometabolism of trichloroethylene.Appl. Environ. Microbiol. 62, 2953–2960 (1996).
Shurtliff M.M., Parkin G.F., Weathers L.J., Gibson D.T.: Biotransformation of trichloroethylene by a phenol-induced mixed culture.J. Environ. Eng. 122, 581–589 (1996).
Steffan R.J., Sperry K.L., Walsh M.T., Vainberg S., Condee C.W.: Field-scale evaluation ofin situ bioaugmentation for remediation of chlorinated solvents in groudwater.Environ. Sci. Technol. 33, 2771–2781 (1999).
Sun A.K., Hong J., Wood T.K.: Modeling trichloroethylene degradation by a recombinant pseudomonad expressing tolueneortho-monooxygenase in a fixed-film bioreactor.Biotechnol. Bioeng. 59, 40–51 (1998).
Sun A.K., Wood T.K.: Trichloroethylene degradation and mineralization by pseudomonads andMethylosinus trichosporium OB3b.Appl. Microbiol. Biotechnol. 45, 248–256 (1996).
Takami W., Horinouchi M., Nojiri H., Yamane H., Omori T.: Evaluation of trichloroethylene degradation byE. coli transformed with dimethylsulphide monooxygenase genes and/or cumene dioxygenase genes.Biotechnol. Lett. 21, 259–264 (1999).
Tomita B., Inoue H., Chaya K., Nakamura A., Hamamura N., Ueno K., Watanabe K., Ose Y.: Identification of dimethyl disulfide-forming bacteria isolated from activated sludge.Appl. environ. Microbiol. 53, 1541–1547 (1987).
Vogel T.M., Criddle C.S., McCarty P.L.: Transformations of halogenated aliphatic compounds.Environ. Sci. Technol. 21, 722–736 (1987).
Wilson J.T., Wilson B.H.: Biotransformation of trichloroethylene in soil.Appl. Environ. Microbiol. 49 242–243 (1985).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Růžička, J., Müller, J., Vít, D. et al. Biotransformation of trichloroethene by pure bacterial cultures. Folia Microbiol 47, 467–472 (2002). https://doi.org/10.1007/BF02818782
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1007/BF02818782