Abstract
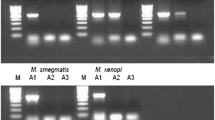
The polymerase chain reaction has facilitated the use of molecular approaches in microbiology including new strategies for the rapid identification of micro-organisms. Approaches based on the use of random primers and standard conditions, allows characteristic DNA fingerprints to be generated from any micro-organism even in the absence of information about its DNA sequence. Different primers can be used to produce genus-specific, species-specific, or even strain-specific DNA fingerprints. This article covers the background to this strategy, describes three different approaches to generating DNA fingerprints using random primers, and provides experimental detail for one method, RAPD.
Similar content being viewed by others
References
Howard, J. and Whitcombe, D. M. (1995)Methods in Molecular Biology, Vol. 46: Diagnostic Bacteriology Protocols. Humana, Totowa, NJ.
Welsh, J. and McClelland, M. (1990) Fingerprinting genomes using PCR with random primers.Nucleic Acids Res. 18, 7213–7218.
Williams, J. G. K., Kubelik, A. R., Livak, K. J., Rafalski, J. A., and Tingey, S. V. (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers,Nucleic Acids Res. 18, 6531–6535.
Caetano-Anolles, G., Bassam, B. J., and Gresshof, P. M. (1991) DNA amplification fingerprinting using very short arbitrary oligonucleotide primers.Biotechnology 9, 553–556.
Akopyanz, N., Bukanov, N. O., Westblom, T. U., and Berg, D. E. (1992) PCR-based RFLP analysis of DNA sequence diversity in the gastric pathogenHelicobacter pylori.Nucleic Acids Res. 20, 6221–6225.
Akopyanz, N., Bukanov, N. O., Westblom, T. U., Kresovich, S., and Berg, D. E. (1992) DNA diversity among clinical isolates ofHelicobacter pylori detected by PCR-based RAPD fingerprinting.Nucleic Acids Res. 20, 5137–5142.
Cancilla, M. R., Powell, I. B., Hillier, A. J., and Davidson, B. E. (1992) Rapid genomic fingerprinting ofLactococcus lactis strains by arbitrarily primed polymerase chain reaction with P32 and fluorescent labels.Appl. Environ. Microbiol. 58, 1772–1775.
Mazurier, S., Van de Giessen, A., Heuvelman, K., and Wernars, K. (1992) RAPD analysis ofCampylobacter isolates—DNA fingerprinting without the need to purify DNA.Lett. Appl. Microbiol. 14, 260–262.
Mazurier, S. I., Audurier, A., Marqet-Van der Mee, N., Notermans, S., and Wernars, K. A. (1992) A comparative study of randomly amplified polymorphic DNA analysis and conventional phage typing for epidemial studies ofListeria monocytogenes isolates.Res. Microbiol. 143, 507–512.
Mazurier, S. I. and Wernars, K. A. (1992) Typing of listeria strains by random amplification of polymorphic DNA.Res. Microbiol. 143, 499–505.
McMillin, D. E. and Muldrow, L. L. (1992) Typing of toxic strains ofClostridium difficile using DNA fingerprints generated with arbitrary polymerase chain reaction primers.FEMS Microbiol. Lett. 92, 5–10.
Menard, C., Brousseau, R., and Mouton, C. (1992) Application of polymerase chain reaction with arbitrary primer (AP-PCR) to strain identification ofPorphyromonas (Bacteroides) gingivalis.FEMS Microbiol. Lett. 95, 163–168.
Brousseau, R., Saintonge, A., Prefontaine, G., Masson, L., and Cabana, J. (1993) Arbitrary primer polymerase chain reaction, a powerful method to identifyBacillus thuringiensis serovars and strains.Appl. Environ. Microbiol. 59, 114–119.
Myers, L. E., Silva, S. V. P. S., Procunier, J. D., and Little, P. B. (1993) Genomic fingerprinting ofHaemophilus somnus isolates by using a random-amplified polymorphic DNA assay.J. Clin. Microbiol. 31, 512–517.
Kernodle, S. P., Cannon, R. E., and Scandalios, J. G. (1993) Concentration of primer and template quantitatively affects products in random-amplified polymorphic DNA PCR.BioFeedback 14, 362,363.
Ellsworth, D. L., Rittenhouse, K. D., and Honeycut, R. L. (1993) Artifactual variation in randomly amplified polymorphic DNA banding patterns.BioTechniques 14, 214–217.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Mileham, A.J. Identification of microorganisms using random primed PCR. Mol Biotechnol 8, 139–145 (1997). https://doi.org/10.1007/BF02752258
Issue Date:
DOI: https://doi.org/10.1007/BF02752258