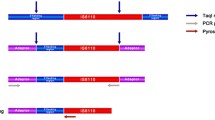
Abstract
Several species of mycobacteria cause infections in humans. Species identification of clinical isolates of mycobacteria is very important for the decision of treatment and in choosing the appropriate treatment regimen. We have developed a multiplex PCR method that can identify practically all known species of mycobacteria, by determination of single-nucleotide differences at a total of 13 different polymorphic regions in the genes of rRNA and hsp65, in four PCR mixes. To achieve this goal, single-nucleotide differences in these polymorphic regions were used to divide mycobacterial species into two groups, than four, eight, etc., in an algorithmic manner. It was sufficient to reach single species level by evaluating 13 polymorphic regions. Evaluation of the multiplex PCR patterns by observable real-time electrophoresis (ORTE) simplified species identification. This new method may enable easy, rapid, and cost-effective identification of all species of mycobacteria.


Similar content being viewed by others
References
Bloom BR (1994) Tuberculosis Pathogenesis, Protection, and Control, ASM Press, Washington
Sezgin E, Van Natta M, Thorne J, Puhan M, Jabs D (2018) Secular trends in opportunistic infections, cancers and mortality in patients with AIDS during the era of modern combination antiretroviral therapy. HIV Med 19:411–419
Gagneux S (2018) Ecology and evolution of Mycobacterium tuberculosis. Nat Rev Microbiol 16:202
Driesen M, Kondo Y, de Jong BC, Torrea G, Asnong S, Desmaretz C, Mostofa KSM, Tahseen S, Whitfield MG, Cirillo DM, Miotto P, Cabibbe AM, ve Rigouts L (2018) Evaluation of a novel line probe assay to detect resistance to pyrazinamide, a key drug used for tuberculosis treatment. Clin Microbiol Infect 24:60–64
REPORT ET (2018) Handbook on tuberculosis laboratory diagnostic methods in the European Union
Betty A, Forbes GSH, Melissa B, Miller SM, Novak M-C, Rowlinson M, Salfinger A, Somoskövi DM, Warshauer ML, Wilson (2018) Practice Guidelines for Clinical Microbiology Laboratories: Mycobacteria, ASM, Washington
PrasadMyneedua AKK (2018) Amplification of Hsp 65 gene and usage of restriction endonuclease for identification of non tuberculous rapid grower mycobacterium, Indian J Tuberc 65(1):57–62
Kim J-U, Ryu D-S, Cha C-H, ve Park S-H (2018) Paradigm for diagnosing mycobacterial disease: direct detection and differentiation of Mycobacterium tuberculosis complex and non-tuberculous mycobacteria in clinical specimens using multiplex real-time PCR. J Clin Pathol 71(9):774–780
Ryan S, Sadda S, Schachat A, Wilkinson C, Schachat A, Hinton D, Wiedemann P, Wiedemann P (2013) Mycobacterial infections, retina. Elsevier, Amsterdam. ISBN: 9781455707379
Kocagöz T, Özkara EY,Ş, Kocagöz S, Hayran M, Sachedeva M, Chambers HF (1993) Detection of Mycobacterium tuberculosis in sputum samples by polymerase chain reaction using a simplified procedure, J Clin Microbiol 31(6):1435–1438
Mozioğlu E, Tamerler AM, Kocagöz C, ZT (2014) A simple guanidinium isothiocyanate method for bacterial genomic DNA isolation, Turk J Biol 38(1):125–129
Kocagöz T (2013) Real-time observable electrophoresis system, which does not require any kind of light filter, Patent, US2013112562 (A1)
Gilpin C, Korobitsyn A, Migliori GB, Raviglione MC, ve Weyer K (2018) The World Health Organization standards for tuberculosis care and management. Eur Respir J 51:1800098
Fariz Nurwidya DH, Burhan E, Yunus F (2018) Molecular Diagnosis of Tuberculosis, Chonnam Medical Journa
Haworth CS, ve Floto RA (2017) Introducing the new BTS guideline: management of non-tuberculous mycobacterial pulmonary disease (NTM-PD). Thorax 72:969–970
Forbes BA, Hall GS, Miller MB, Novak SM, Rowlinson M-C, Salfinger M, Somoskövi A, Warshauer DM ve Wilson ML (2018) Practice guidelines for clinical microbiology laboratories: mycobacteria. Clin Microbiol Rev 31:e00038–e00017
Balows A, Hausler WJ, Herrmann JR, Kenneth L, Isenberg HD, Shadomy HJ,, (1991). Manual of clinical microbiology, ASM Press, Washington
Butler WR, Cage G, Desmond E, Duffey PS, Floyd MM, Gross WM, Guthertz LS, Jost WM, Ramos LS, Silcox V, Thibert L, Warren N,, (1996). Standardized Method for HPLC Identification of Mycobacteria, U.S Department of Health and Human Services
Frazier KS, Hines ME,, (1993). Differentiation of mycobacteria on the basis of chemotype profiles by using matrix solid-phase dispersion and thin-layer chromatography Clin Microbiol Infect 31(3):610–614
Novakova MNVKS (2017) Identification of Mycobacterium Species by MALDI-TOF Mass Spectrometry, ed. Pulm Care Clin Med 37–42
Acosta FL, Ruiz Serrano MJ, Marín M, Kohl TA, Lozano N, Niemann S, Valerio M, Olmedo M, Pérez-Granda MJ, Pérez MP, Bouza E, Muñoz P, de Viedma DG (2018) Fast update of undetected Mycobacterium chimaera infections to reveal unsuspected cases, J Hosp Infect 100(4):451–455
Olaru ID, Kranzer K, Perera N (2018) Turnaround time of whole genome sequencing for mycobacterial identification and drug susceptibility testing in routine practice, Clin Microbiol Infect 24(6):659–e5
Broda A, Nikolayevskyy V, Casali N, Khan H, Bowker R, Blackwell G, Patel B, Hume J, Hussain W, ve Drobniewski F (2018) Experimental platform utilising melting curve technology for detection of mutations in Mycobacterium tuberculosis isolates. Eur J Clin Microbiol Infect Dis 37:1273–1279
Tortone C, Zumárraga M, Gioffr A, ve Oriani D (2018) Utilization of molecular and conventional methods for the identification of nontuberculous mycobacteria isolated from different water sources. Int J Mycobacteriol 7:53–60
Germer S, Higuchi R, Holland MJ (2000) High-Throughput SNP Allele-Frequency Determination in Pooled DNA Samples by Kinetic PCR, Genome Res 10(2):258–266
Dae-Hoon L, Kyoung I, In-Taek HL, Jong-Kee K, Jong-Yoon C, Kyoung-Joong K, Yun-Jee K, (2007) Dual priming oligonucleotide system for the multiplex detection of respiratory viruses and SNP genotyping of CYP2C19 gene, Nucleic Acids Res 35(6):e40
Acknowledgements
This work was supported by TUBİTAK 107S014 (SBAG-3541) Identification of Mycobacterial Species by Sequence-Specific Polymerase Chain Reaction; for financial aid, I wish to thank TÜBİTAK.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Unubol, N., Kizilkaya, I.T., Okullu, S.O. et al. Simple Identification of Mycobacterial Species by Sequence-Specific Multiple Polymerase Chain Reactions. Curr Microbiol 76, 791–798 (2019). https://doi.org/10.1007/s00284-019-01661-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-019-01661-4