Abstract
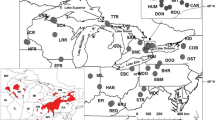
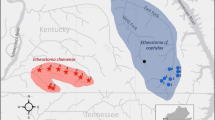
Riverine populations of shortnose sturgeon (Acipenser brevirostrum) once occurred in rivers and estuaries along the east coast of North America from the St. John River, New Brunswick, to the St. Johns River, Florida. Within this range, 19 population segments were identified by the U.S. Federal Shortnose Sturgeon Recovery Team; empirical data supporting this structure is limited. We obtained samples from 11 (12 including a small sample from the Cape Fear River, North Carolina) of these population segments and used PCR and direct sequence analysis of 440 base pairs of the mitochondrial DNA (mtDNA) control region to define the coast-wide genetic population structure of shortnose sturgeon. Collections from most population segments exhibited significant differences in haplotype frequencies with their nearest neighbors, including from the Ogeechee and Savannah Rivers, Georgia (despite the known movement of hatchery-reared offspring from the Savannah into the Ogeechee River). Collections from the Chesapeake Bay and Delaware River exhibited similar haplotype frequencies, suggesting that specimens collected in the Chesapeake Bay had dispersed from the Delaware River. Collections from the Kennebec River and Androscoggin River within a hypothesized single population segment did not exhibit significant differentiation of mtDNA haplotype frequencies. Haplotype frequencies were almost identical between collections from above and below the Holyoke Dam on the Connecticut River, indicating that these aggregations should be managed as a single unit. Our results support the population segment status afforded to shortnose sturgeon in at least the following 9 systems; St. John River, Kennebec-Androscoggin Rivers, upper-lower Connecticut River, Hudson River, Delaware River-Chesapeake Bay, Pee Dee River, Cooper River, Savannah River, and Ogeechee-Altamaha Rivers.
Similar content being viewed by others
Literature Cited
Brown, J. R., A. T. Beckenbach, andM. J. Smith. 1993. Intraspecific DNA sequence variation of the mitochondrial control region of white sturgeon (Acipenser transmontanus).Molecular Biology and Evolution 10:326–341.
Collins, M. R., D. Cooke, B. Post, J. Crane, J. Bulak, T. I. J. Smith, T. W. Greig, andJ. M. Quattro. 2003. Shortnose sturgeon in the Santee-Cooper Reservoir system, South Carolina.Transactions of the American Fisheries Society 132:1244–1250.
Dadswell, M. J. 1979. Biology and population characteristics of the shortnose sturgeon,Acipenser brevirostrum LeSueur 1818 (Osteichthyes: Acipenseridae), in the Saint John River estuary, New Brunswick, Canada.Canadian Journal of Zoology 57: 2186–2210.
Dadswell, M. J., B. D. Taubert, T. S. Squiers, D. Marchette, andJ. Buckley. 1984. Synopsis of biological data on shortnose sturgeon,Acipenser brevirostrum Le Sueur 1818.FAO Fisheries Synopsis 140:1–45.
Dizon, A. E., C. Lockyer, W. F. Perrin, D. P. Demaster, andJ. Sisson. 1992. Rethinking the stock concept: A phylogenetic approach.Conservation Biology 6:24–36.
Dovel, W. L., A. W. Pekovitch, andT. J. Berggren. 1992. Biology of the shortnose sturgeon (Acipenser brevirostrum Lesueur, 1818) in the Hudson River estuary, New York, p. 187–216.In C. L. Smith (ed.), Estuarine Research in the 1980s. State University of New York Press, Albany, New York.
Excoffier, L., P. E. Smouse, andJ. M. Quattro. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data.Genetics 131:479–491.
Felsenstein, J. 1985. Confidence limits on phylogenies: An approach using bootstrap.Evolution 39:783–791.
Gaggiotti, O. E. andR. D. Vetter. 1999. Effect of life history strategy, environmental variability, and overexploitation on the genetic diversity of pelagic fish populations.Canadian Journal of Fisheries and Aquatic Sciences 56:1376–1388.
Grunwald, C., J. R. Waldman, J. Stabile, andI. I. Wirgin. 2002. Population genetics of shortnose sturgeonAcipenser brevirostrum based on mitochondrial DNA control region sequences.Molecular Ecology 11:1885–1898.
Hillis, D. M. andJ. Bull. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis.Systematic Biology 42:182–192.
Hudson, R. R., M. Slatkin, andW. P. Maddison. 1992. Estimation of levels of gene flow from DNA sequence data.Genetics 132:583–589.
Kieffer, M. andB. Kynard. 1993. Annual movements of shortnose and Atlantic sturgeons in the Merrimack River, Massachusetts.Transactions of the American Fisheries Society 122:1088–1103.
Kretz, K. A. andJ. S. O’Brien. 1993. Direct sequencing of polymerase chain reaction products from low melting temperature agarose.Methods in Enzymology 218:72–79.
Kynard, B. 1997. Life history, latitudinal patterns, and status of shortnose sturgeon,Acipenser brevirostrum.Environmental Biology of Fishes 48:319–334.
Kynard, B. 1998. Twenty-two years of passing shortnose sturgeon in fish lifts on the Connecticut River: What has been learned?, p. 255–264In M. Jungwirth, S. Schmutz, and S. Weiss (eds.), Fish Migration and Fish Bypasses. Fishing News Books, London, U.K.
McElroy, D., P. Moran, E. Bermingham, andI. Kornfield. 1992. REAP: An integrated environment for the manipulation of and phylogenetic analysis of restriction data.Journal of Heredity 83:157–158.
Miller, M. P. 1997. Tools for population genetic analyses (TFPGA) 1.3; A Windows program for the analysis of allozyme and molecular population genetic data. Computer software distributed by author. http://bioweb.usu.edu/mpmbio/tfpga.asp.
Moser, M. L. andS. W. Ross. 1995. Habitat use and movements of shortnose and Atlantic sturgeons in the lower Cape Fear River, North Carolina.Transactions of the American Fisheries Society 124:225–234.
National Marine Fisheries Service (NMFS). 1996. Status review of the shortnose sturgeon in the Androscoggin and Kennebec Rivers. Northeast Regional Office, Gloucester, Massachusetts.
National Marine Fisheries Service (NMFS). 1998. Recovery Plan for the Shortnose Sturgeon (Acipenser brevirostrum), Prepared by the Shortnose Sturgeon Recovery Team for the National Marine Fisheries Service, Silver Spring, Maryland.
Nei, M. 1972. Genetic distances between populations.American Naturalist 106:283–292.
Nei, M. 1987. Molecular Evolutionary Genetics. Columbia University Press, New York.
Nielsen, R. andJ. Wakeley. 2001. Distinguishing migration from isolation: A Markov Chain Monte Carlo approach.Genetics 158:885–896.
Quattro, J. M., T. W. Greig, D. K. Coykendall, B. W. Bowen, andJ. D. Baldwin. 2002. Genetic issues in aquatic species management: The shortnose sturgeon (Acipenser brevirostrum) in the southeastern United States.Conservation Genetics 3:155–166.
Rivera, M. A., C. D. Kelley, andG. K. Roderick. 2004. Subtle population genetic structure in the Hawaiian grouper,Epinephelus quernus (Serranidae) as revealed by mtDNA analysis.Biological Journal of the Linnean Society 81:449–468.
Roff, D. A. andP. Bentzen. 1989. The statistical analysis of mitochondrial DNA polymorphism: χ2 and the problem of small samples.Molecular Biology and Evolution 6:539–545.
Schneider, S., D. Roessli, andL. Excoffier. 2000. Arlequin: A software for population genetic data analysis. Version 2.000. Genetics and Biometry Lab, Department of Anthropology, University of Geneva, Geneva, Switzerland.
Smith, H. M. andB. A. Bean. 1898. List of fishes known to inhabit the waters of The District of Columbia and vicinity.Bulletin of the United States Fish Commission 18:179–181.
Smith, T. I. J., M. C. Collins, W. C. Post, andJ. W. McCord. 2002a. Stock enhancement of shortnose sturgeon: A case study.American Fisheries Society Symposium 28:31–44.
Smith, T. I. J., J. W. McCord, M. R. Collins, andW. C. Post. 2002b. Occurrence of stocked shortnose sturgeonAcipenser brevirostrum in non-target rivers.Journal of Applied Ichthyology 18:470–474.
Sokal, R. R. andF. J. Rohlf. 1995. Biometry: The Principles and Practice of Statistics in Biological Research, 3rd edition. W. H. Freeman and Co., New York.
Squiers, T. S., M. Smith, and L. Flagg. 1982. American shad enhancement and status of sturgeon stocks in selected Maine waters. Final report to the National Marine Fisheries Service, Gloucester, Massachusetts.
Tallman, R. F. andM. C. Healy. 1994. Homing straying, and gene flow among seasonally separated populations of chum salmon (Oncorhynchus keta).Canadian Journal of Fisheries and Aquatic Sciences 51:577–588.
Tessier, N. andL. Bernatchez. 1999. Stability of population structure and genetic diversity across generations assessed by microsatellites among sympatric populations of landlocked Atlantic salmon (Salmo salar L.).Molecular Ecology 8:169–179.
Waldman, J. R., C. Grunwald, J. Stabile, andI. Wirgin. 2002. Impacts of life history and biogeography on genetic stock structure in Atlantic sturgeon,Acipenser oxyrinchus oxyrinchus, Gulf sturgeonA. oxyrinchus desotoi, and shortnose sturgeon,A. brevirostrum.Journal of Applied Ichthyology 18:509–518.
Walsh, M. G., M. B. Bain, T. Squiers, Jr.,J. R. Waldman, andI. Wirgin. 2001. Morphological and genetic variation among shortnose sturgeonAcipenser brevirostrum from adjacent and distant rivers.Estuaries 24:41–48.
Waples, R. S. 1991. Genetic interactions between hatchery and wild salmonids: Lessons from the Pacific Northwest.Canadian Journal of Fisheries and Aquatic Sciences 48:124–133.
Waters, J. M., J. M. Epifanio, T. Gunter, andB. L. Brown. 2000. Homing behaviour facilitates subtle genetic differentiation among river populations ofAlosa sapidissima: Microsatellites and mtDNA.Journal of Fish Biology 56:622–636.
Weber, W. 1996. Population size and habitat use of shortnose sturgeon,Acipenser brevirostrum, in the Ogeechee River system, Georgia. M.S. Thesis, University of Georgia, Athens Georgia.
Weir, B. S. andC. C. Cockerham. 1984. Estimating F-statistics for the analysis of population structure.Evolution 38:1358–1370.
Welsh, S. A., M. F. Mangold, J. E. Skjeveland, andA. J. Spells. 2002. Distribution and movement of shortnose sturgeonAcipenser brevirostrum in the Chesapeake Bay.Estuaries 25:101–104.
Wirgin, I. I., J. Stabile, andJ. R. Waldman. 1997a. Molecular analysis in the conservation of sturgeons and paddlefishes. Use of DNA techniques in the management and restoration of sturgeon and paddlefish populations.Environmental Biology of Fishes 48:385–398.
Wirgin, I. I., J. R. Waldman, L. Maceda, J. Stabile, andV. J. Vecchio. 1997b. Mixed-stock analysis of Atlantic coast striped bass using nuclear DNA and mitochondrial DNA markers.Canadian Journal of Fisheries and Aquatic Sciences 54: 2814–2826.
Wirgin, I. I., J. R. Waldman, J. Rosko, R. Gross, M. R. Collins, S. G. Rogers, andJ. Stabile. 2000. Genetic structure of Atlantic sturgeon populations based on mitochondrial DNA control region sequences.Transactions of the American Fisheries Society 129:476–486.
Wright, S. 1951. The genetical structure of populations.Annals of Eugenics 15:323–354.
Source of Unpublished Materials
Bryce, T. Personal Communication. Fish and Wildlife Branch Environmental and Natural Resources Division, 1113 Frank Cochoran Drive, Ft. Stewart, Georgia 31314.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wirgin, I., Grunwald, C., Carlson, E. et al. Range-wide population structure of shortnose sturgeonAcipenser brevirostrum based on sequence analysis of the mitochondrial DNA control region. Estuaries 28, 406–421 (2005). https://doi.org/10.1007/BF02693923
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF02693923