Abstract
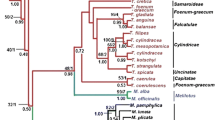
A phylogenetic reconstruction of eight species of the genusBrachypodium P. Beauv. (Poaceae) has been obtained combining sequence data from the chloroplastndhF gene and the nuclear ITS, and using five representatives of tribes Triticeae (Secale), Poeae (Lolium), Meliceae (Melica, Glyceria) and Oryzeae (Oryza) as out-groups. Similar numbers of informative substitutions for the ingroup species were provided by both the 3′ region of the chloroplastndhF gene and the nuclear ITS region. The Mediterranean annualBrachypodium distachyon appears to be the basal lineage, followed by the divergence of the New World non-rhizomatousB. mexicanum, which antedates the separation of a core of six European and Eurosiberian rhizomatous perennials (Brachypodium arbuscula, B. retusum, B. rupestre, B. phoenicoides, B. pinnatum, andB. sylvaticum). The evolutionary reconstruction based on sequences of the chloroplast and the nuclear genomes is congruent with topologies obtained from analysis of RAPD data.
Similar content being viewed by others
References
Asay K. H., Hsiao C., Chatterton N. J., Jensen K. B., Wang R. R. (1992) Nucleotide sequence of the internal transcribed spacer region of rDNA in mountain rye,Secale montanum Guss. (Gramineae). Plant Mol. Biol. 20: 161–162.
Ascherson P., Graebner P. (1901)Brachypodium. In: Synopsis der mitteleuropäischen Flora 2: 631–640. W. Engelmann, Leipzig.
Baldwin B. G., Sanderson M. J., Porter J. M., Wojciechowski M. F., Campbell C. S., Donoghue M. J. (1995) The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Ann. Missouri Bot. Gard. 82: 247–277.
Baum D. A., Sytsma K. J., Hoch P. C. (1994) A phylogenetic analysis ofEpilobium (Onagraceae) based on nuclear ribosomal DNA sequences. Syst. Bot. 19: 363–388.
Bluff M. J., Nees, Esenbeck C-G., Schauer J. C. (1836) Compendium Florae Germaniae 1(1). Schrag, J. L. Nürnberg.
Bremer K. (1988) The limits of amino acid sequence data in angiosperm phylogenetic reconstruction. Evolution 42: 795–803.
Catalán P., Shi Y., Amstrong L., Draper J., Stace C. A. (1995) Molecular phylogeny of the grass genus Brachypodium P. Beauv. based on RFLP and RAPD analysis. Bot. J. Linnean Soc. 117: 263–280.
Catalán P., Kellogg E. A., Olmstead R. G. (1997) Phylogeny of Poaceae subfamily Pooideae based on chloroplastndhF sequences. Mol. Phylog. Evol. 8: 1–17.
Clark L. G., Zhang W., Wendel J. F. (1995) A phylogeny of the grass family (Poaceae) based onndhF sequence data. Syst. Bot. 20: 436–460.
Clayton W. D., Renvoize S. A. (1986) Genera graminum. Kew Bulletin Additional Series XIII HMSO, London.
Davis J. I., Soreng R. J. (1993) Phylogenetic structure in the grass family (Poaceae) as inferred from chloroplast DNA restriction site variation. Amer. J. Bot. 80: 1444–1454.
Dumortier B. C. J. (1824) Observations sur les Graminées de la flore belgique. Castermann, Tournay.
Eriksson T. (1996) AutoDecay ver. 2.9.6. Botaniska Institutionen, Stockholm University, Stockholm.
Farris J. S., Källersjö M., Kluge A. G., Bult C. (1994) Testing significance of incongruence. Cladistics 10: 315–319.
Felsenstein J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791.
Furman B. J., Grattapaglia D., Dvorak W. S., O'Malley D. M. (1997) Analysis of genetic relationships of Central American and Mexican pines using RAPD markers that distinghish species. Mol. Ecol. 6: 321–331.
Harz C. O. (1880–1882) Beiträge zur Systematik der Gramineen. Linnaea 43: 1–30.
Hayek A. (1925) Zur Systematik der Gramineen. Österr. Bot. Z. 74: 249–255.
Higgins D. G., Bleasby A. J., Fuchs R. (1992) CLUSTAL V: Improved software for multiple sequence alignment. CABIOS 8: 189–191.
Hilu K. W. (1994) Evidence from RAPD markers in the evolution ofEchinochloa millets (Poaceae). Plant Syst. Evol. 189: 247–257.
Hilu K. W., Wright K. (1982) Systematics of Gramineae: A cluster analysis study. Taxon 31: 9–36.
Hsiao C., Chatterton N. J., Asay K. H., Jensen K. B. (1995) Molecular phylogeny of the Pooideae (Poaceae) based on nuclear rDNA (ITS) sequences. Theor. Appl. Genet. 90: 389–398.
Khan M-A. (1984) Biosystematic study inBrachypodium (Poaceae). Doctorate thesis, University of Leicester.
Khan M-A. (1992) Seed-protein electrophoretic pattern inBrachypodium P. Beauv. species. Annals of Botany, London 70: 61–68.
Kim K.-J., Jansen R. K. (1994) Comparisons of phylogenetic hypotheses among different data sets in dwarf dandelions (Krigia, Asteraceae): additional information from internal transcribed spacer sequences of nuclear ribosomal DNA. Plant Syst. Evol. 190: 157–185.
Link H. F. (1827) Hortus regius botanicus Berolinensis 1. G. Reimer, Berlin.
Nickrent D. L., Schuette K. P., Starr E. M. (1994) A molecular phylogeny ofArceuthobium (Viscaceae) based on nuclear ribosomal DNA internal transcribed spacer sequences. Amer. J. Bot. 81: 1149–1160.
Pillay M., Hilu K. W. (1995). Chloroplast-DNA restriction site analysis in the genusBromus (Poaceae). Amer. J. Bot. 82: 239–249.
Rieseberg L. H. (1996) Homology among RAPD fragments in interspecific comparisons. Mol. Ecol. 5: 99–105.
Shi Y. (1991) Molecular studies of the evolutionary relationships ofBrachypodium (Poaceae). Doctorate thesis, University of Leicester.
Shi Y., Draper J., Stace C. A. (1993) Ribosomal DNA variation and its phylogenetic implication in the genusBrachypodium (Poaceae). Plant Syst. Evol. 188: 125–138.
Schippmann U. (1991) Revision der europäischen Arten der GattungBrachypodium Palisot de Beauvois (Poaceae). Boissiera 45: 1–249.
Stammers M., Harris J., Evans G. M., Hayward M. D., Forster J. W. (1995) Use of random PCR (RAPD) technology to analyse phylogenetic relationships in theLolium/Festuca complex. Heredity 74: 19–27.
Swofford D. L. (1993) PAUP: Phylogenetic Analysis Using Parsimony, version 3.1. Illinois Natural History Survey. Champaign.
Takaiwa F., Oono K., Sugiura M. (1985) Nucleotide sequence of the 17S–25S spacer region from rice rDNA. Plant Mol. Biol. 4: 355–364.
Van de Zande L., Bijlsma R. (1995) Limitations of the RAPD technique in phylogeny reconstruction inDrosophila. J. Evol. Biol. 8: 645–656.
Watson L., M. J. Dallwitz R. (1992) The grass genera of the world. Oxon: Wallingford, CAB International.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Catalán, P., Olmstead, R.G. Phylogenetic reconstruction of the genusBrachypodium P. Beauv. (Poaceae) from combined sequences of chloroplastndhF gene and nuclear ITS. Pl Syst Evol 220, 1–19 (2000). https://doi.org/10.1007/BF00985367
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00985367