Abstract
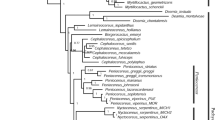
The Papilionoideae subfamily comprises more than 14,000 species, 501 genera and 32 tribes, representing two-thirds of all genera and species in the Fabaceae family. Papillonoids are recognized for their food and forage importance, wide distribution in different biomes and variation in floral architecture as well as plastome structure. Due to the high-level conservation of chloroplast genomes, when compared to nuclear and mitochondrial genomes, phylogenetic analysis based on chloroplast DNA (cpDNA) have been elucidating the relationships among the main Papilionoideae’s taxon. However, the phylogeny of some clades of the subfamily remains unresolved. Aiming at the phylogenetic reconstruction of the deep branching species of Papilionoideae, concatenated sequences of six loci (matK, psaA, psbA, psbD, rbcL and rpoC2) of cpDNA from 117 species of Papilionoideae were analyzed using the maximum likelihood methodology. The plastomes of Papilionoideae showed low conservation and similarity. Phylogenetic analysis resulted in a monophyletic tree, confirming the division of the subfamily into four main clades (NPAAA, ADA, Genistoids and Dalbergioids). The sibling group relationship of the ADA clade with the Genistoids clade was demonstrated, with high support. The paraphyly of the Phaseoleae and Millettiae tribes was evidenced within the NPAAA clade with unresolved phylogeny of the Genistoids clade. As well, was observed that only species of the ADA clade have no rhizobium nodules, which may be a possible synapomorphy to support the relationships of this group. The analysis also suggest that the main Papilionoideae clades diverged from the Paleocene onwards.



Similar content being viewed by others
References
Anderson SJ, Kramer MC, Gosai SJ, Yu X, Vandivier LE, Nelson ADL, Anderson ZD, Beilstein MA, Fray RG, Lyons E, Gregory BD (2018) N6-Methyladenosine Inhibits Local Ribonucleolytic Cleavage to Stabilize mRNAs in Arabidopsis. Cell Rep 25:1146–1157. https://doi.org/10.1016/j.celrep.2018.10.020
Antunes AM, Soares TN, Targueta CP, Novaes E, Coelho ASG, Telles MPC (2020) The chloroplast genome sequence of Dipteryx alata Vog. (Fabaceae: Papilionoideae): genomic features and comparative analysis with other legume genomes. Braz J Biol 43:271–282. https://doi.org/10.1007/s40415-020-00599-3
Asaf S, Khan AL, Khan MA, Imran QM, Kang SM, Al-Hosni K, Jeong EJ, Lee KE, Lee I-J (2017) Comparative analysis of complete plastid genomes from wild soybean (Glycine soja) and nine other Glycine species. PLoS ONE 8:e0182281. https://doi.org/10.1371/journal.pone.0182281
Baudoin JP (1988) Genetic resources, domestication and evolution of lima bean, Phaseolus lunatus. J Emerg Med 39:253–260
Bell EA (1981) Non-protein amino acids in the Leguminosae. In: Polhill RM, Raven PH (eds) Advances in legume systematics, royal botanic gardens. Kew, England, pp 489−499
Benton MJ (2010) The origins of modern biodiversity on land. Philos Trans R Soc Lond B Biol Sci 365:3667–3679. https://doi.org/10.1098/rstb.2010.0269
Boatwright JS, Le Roux MM, Wink M, Morozova T, Van Wyk BE (2008) Phylogenetic relationships of tribe Crotalarieae (Fabaceae) inferred from DNA sequences and morphology. Syst Bot 33:752–761. https://doi.org/10.1600/036364408786500271
Brudno M, Cooper GM, Kim MF, Davydov E (2003) LAGAN and Multi LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res 13:721–731. https://doi.org/10.1101/gr.926603
Bunsupa S, Saito K, Yamazaki M (2013) Molecular biology and biotechnology of quinolizidine alkaloid biosynthesis in leguminosae plants. In: Chandra S, Lata H, Varma A (eds) Biotechnology for Medicinal Plants. Springer, Berlin, pp 263–273
Cannon SB, McKain MR, Harkess A, Nelson MN, Dash S, Deyholos MK, Peng Y, Joyce B, Stewart CN Jr, Rolf M Jr, Kutchan T, Tan X, Chen C, Zhang Y, Carpenter E, Wong GK-S, Doyle JJ, Leebens-Mack J (2015) Multiple polyploidy events in the early radiation of nodulating and nonnodulating legumes. Mol Biol Evol 32:193–210. https://doi.org/10.1093/molbev/msu296
Cardoso D, Queiroz LP, Pennington RT, Lima HC, Fonty E, Wojciechowski MF, Lavin M (2012) Revisiting the phylogeny of papilionoid legumes: New insights from comprehensively sampled early-branching lineages. Am J Bot 99:1991–2013. https://doi.org/10.3732/ajb.1200380
Cardoso D, Pennington RT, Queiroz LP, Boatwright JS, Van Wyk BE, Wojciechowski MF, Lavin M (2013) Reconstructing the deep-branching relationships of the papilionoid legumes. S African J Bot 89:58–75. https://doi.org/10.1016/j.sajb.2013.05.001
Choi I-S, Choi B-H (2017) The distinct plastid genome structure of Maackia fauriei (Fabaceae: Papilionoideae) and its systematic implications for genistoids and tribe Sophoreae. PLoS ONE 4:e0173766. https://doi.org/10.1371/journal.pone.0173766
Choi I-S, Cardoso D, Queiroz LP, Lima HC, Lee C, Ruhlman TA, Jansen RK, Wojciechowski MF (2022) Highly resolved papilionoid legume phylogeny based on plastid phylogenomics. Front Plant Sci 13:823190. https://doi.org/10.3389/fpls.2022.823190
Compton JA, Schrire BD, Könyves K, Forest F, Malakasi P, Mattapha S, Sirichamorn Y (2019) The callerya group redefined and tribe wisterieae (Fabaceae) emended based on morphology and data from nuclear and chloroplast DNA sequences. PhytoKeys 124:1–112. https://doi.org/10.3897/phytokeys.125.34877
Condamine FL, Rolland J, Morlon H (2013) Macroevolutionary perspectives to environmental change. Ecol Lett 16:72–85. https://doi.org/10.1111/ele.12062
Davis JI, Soreng RJ (2010) Migration of endpoints of two genes relative to boundaries among regions of the plastid genome in the grass family (Poaceae). Am J Bot 97:874–892. https://doi.org/10.3732/ajb.0900228
Dodd ME, Silvertown J, Chase MW (1999) Phylogenetic analysis of trait evolution and species diversity variation among angiosperm families. Evolution 53:732–744. https://doi.org/10.2307/2640713
Doyle JJ (2016) Chasing unicorns: nodulation origins and the paradox of novelty. Am J Bot 103:1865–1868. https://doi.org/10.3732/ajb.1600260
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Egan AN, Vatanparast M, Cagle W (2016) Parsing polyphyletic Pueraria: delimiting distinct evolutionary lineages through phylogeny. Mol Phylogenet Evol 104:44–59. https://doi.org/10.1016/j.ympev.2016.08.001
Eriksson O, Bremer B (1992) Pollinationsystems dispersalmodes lifeforms and diversification rates in angiosperm families. Evolution 46:258–266. https://doi.org/10.2307/2409820
Erwin DH (2009) Climate as a driver of evolutionary change. Curr Biol 19:575–583. https://doi.org/10.1016/j.cub.2009.05.047
Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I (2004) VISTA: computational tools for comparative genomics. Nucleic Acids Res 32:273–279. https://doi.org/10.1093/nar/gkh458
Hallauer A (2011) Evolution of plant breeding. CBAB 11:197–206. https://doi.org/10.1590/S1984-70332011000300001
Hu JM (2000a) Phylogenetic relationships of the tribe Millettieae and allies - the current status. In: Herendeen PS, Bruneau A (eds) Advances in Legume Systematics part 9. Royal Botanic Gardens, Kew, pp 299–310
Hu JM, Lavin M, Wojciechowski MF, Sanderson J (2000b) Phylogenetic Systematics of the Tribe Millettieae (Leguminosae) Based on Chloroplast trnK/matK Sequences and its Implications for Evolutionary Patterns in Papilionoideae. Am J Bot 87:418–430. https://doi.org/10.2307/2656638
Hyland EG, Sheldon ND, Cotton JM (2015) Terrestrial evidence for a two-stage mid-Paleocene biotic event. Palaeogeogr Palaeoclimatol Palaeoecol 417:371–378. https://doi.org/10.1016/j.palaeo.2014.09.031
Jansen RK, Wojciechowski MF, Sanniyasi E, Lee SB, Daniell H (2008) Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Leguminosae). Mol Phylogenet Evol 48:1204–1217. https://doi.org/10.1016/j.ympev.2008.06.013
Jin DP, Choi IS, Choi BH (2019) Plastid genome evolution in tribe Desmodieae (Fabaceae: Papilionoideae). PLoS ONE 6:e0218743. https://doi.org/10.1371/journal.pone.0218743
Kajita T, Ohashi H, Tateishi Y, Bailey CD, Doyle JJ (2001) rbcL and legume phylogeny, with particular reference to Phaseoleae, Millettieae, and allies. Syst Bot 26:515–536
Keller J, Rousseau-Gueutin M, Martin GE, Morice J, Boutte J, Coissac E, Ourari M, Aïnouche M, Salmon A, Cabello-Hurtado F, Aïnouche A (2017) The evolutionary fate of the chloroplast and nuclear rps16 genes as revealed through the sequencing and comparative analyses off our novel legume chloroplast genomes from Lupinus. DNA Res 24:343–358. https://doi.org/10.1093/dnares/dsx006
Kite GC, Cardoso D, Veitch NC, Lewis GP (2013) Quinolizidine alkaloid status of Acosmium s.s., Guianodendron and Leptolobium, the segregate genera of Acosmium s.l. South African J Botany 89:176–180. https://doi.org/10.1016/j.sajb.2013.05.009
Klitgaard BB, Lavin M (2005) Dalbergieae sensu lato. In: Lewis GP, Schrire B, Mackinder B, Lock M (eds) Legumes of the world (mansc.), Royal Botanic Gardens, Kew, UK, p 557
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Lavin M, Herendeen PS, Wojciechowski MF (2005) Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the Tertiary. Syst Biol 54:575–594. https://doi.org/10.1080/10635150590947131
Lewis G, Schrire B, Mackinder B, Lock M (2005) Legumes of the World. Royal Botanic Gardens, Kew, UK
Li H, Wang W, Lin L, Zhu X, Li J, Zhu X, Chen Z (2013) Diversification of the phaseoloid legumes: effects of climate change, range expansion and habit shift. Front Plant Sci 4:386. https://doi.org/10.3389/fpls.2013.00386
Lohse M, Drechsel O, Kahlau S, Bock R (2013) OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res 41:W575–W581. https://doi.org/10.1093/nar/gkt289
LPWG (Legume Phylogeny Working Group) (2017) A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny. Taxon 66:44–77. https://doi.org/10.5061/dryad.61pd6
LPWG (Legume Phylogeny Working Group) (2013) Legume phylogeny and classification in the 21st century: progress, prospects and lessons for other species-rich clades. Taxon 62:217–248. https://doi.org/10.12705/622.8
LPWG (Legume Phylogeny Working Group) (2021) The World Checklist of Vascular Plants (WCVP): Fabaceae, vers. June 2021. Govaerts, R. ed. Available online at: http://sftp.kew.org/pub/data_collaborations/Fabaceae/DwCA/. Accessed in 13 December 2021
Machado LO, Vieira LN, Stefenon VM, Pedrosa FO, Souza EM, Guerra MP, Nodari RO (2017) Phylogenomic relationship of feijoa (Acca sellowiana (O.Berg) Burret) with other Myrtaceae based on complete chloroplast genome sequences. Genetica 145:163–174. https://doi.org/10.1007/s10709-017-9954-1
Martínez-Pérez M, Aparicio F, López-Gresa MP, Bellés JM, Sánchez-Navarro JÁ, Pallás V (2017) Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. PNAS 114:10755–10760. https://doi.org/10.1073/pnas.1703139114
Moghaddam M, Ohta A, Shimizu M, Terauchi R, Kazempour-Osaloo S (2022) The complete chloroplast genome of Onobrychis gaubae (Fabaceae-Papilionoideae): comparative analysis with related IR-lacking clade species. BMC Plant Biol 22:75. https://doi.org/10.1186/s12870-022-03465-4
Oyebanji O, Zhang R, Chen SY, Yi T-S (2020) New insights into the plastome evolution of the millettioid/phaseoloid clade (Papilionoideae, Leguminosae). Front Plant Sci 11:151. https://doi.org/10.3389/fpls.2020.00151
Pälike H, Norris RD, Herrle JO, Wilson PA, Coxall HK, Lear CH (2006) The heartbeat of the Oligocene climate system. Science 314:1894–1898. https://doi.org/10.1126/science.113382
Pennington RT, Stirton CH, Schrire BD (2005) Tribe Sophoreae. In: Lewis G, Schrire B, Mackinder B, Lock M (eds) Legumes of theWorld. Royal Botanic Gardens, Kew, UK, pp 227–249
Polhill RM, Raven PH (1981) Advances in Legume Systematics, Part 1. Royal Botanic Gardens, Kew, UK
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256. https://doi.org/10.1093/molbev/msn083
Queiroz LP, Pastore JFB, Cardoso D, Snak C, Lima ALC, Gagnon L, Vatanparast M, Holland AE, Egand AN (2015) A multilocus phylogenetic analysis reveals the monophyly of a recircumscribed papilionoid legume tribe Diocleae with well-supported generic relationships. Mol Phylogenet Evol 90:1–19. https://doi.org/10.1016/j.ympev.2015.04.016
Sabir J, Schwarz E, Ellison N, Zhang J, Baeshen NZ, Mutwakil M, Jansen R, Ruhlman T (2014) Evolutionary and biotechnology implications of plastid genome variation in the inverted-repeat-lacking clade of legumes. Plant Biotechnol J 12:743–754. https://doi.org/10.1111/pbi.12179
Santos V, Almeida C (2019) The complete chloroplast genome sequences of three Spondias species reveal close relationship among the species. Genet Mol Biol 42:132–138. https://doi.org/10.1590/1678-4685-GMB-2017-0265
Schrire BD (2005) Phaseoleae. In: Lewis G, Schrire B, Mackinder B, Lock M (eds) Legumes of the world. Royal Botanic Gardens, Kew, pp 393–431
Schwarz EM, Ruhlman TA, Sabir JSM, Hajrah NH, Alharbi NS, Al-Malki AL, Bailey CD, Jansen RK (2015) Plastid genome sequences of legumes reveal parallel inversions and multiple losses of rps16 in papilionoids. J Syst Evol 53:458–468. https://doi.org/10.1111/jse.12179
Soltis PS, Soltis DE (2020) Plant genomes: markers of evolutionary history and drivers of evolutionary change. Plants People Planet 3:74–82. https://doi.org/10.1002/ppp3.10159
Song Y, Chen Y, Lv J, Zhu S, Li M (2019) Comparative chloroplast genomes of Sorghum species: sequence divergence and phylogenetic relationships. Biomed Res Int 1:11. https://doi.org/10.1155/2019/5046958
Sprent JI (2008) Evolution and diversity of legumes symbiosis. In: Dilworth MJ, James EK, Sprent JI, Newton WE (eds) Nitrogen fixing leguminous symbioses. Springer, The Netherlands, pp 1–21
Sprent JI (2009) Legume nodulation: a global perspective. Wiley-Blackwell, Oxford
Stefanovic S, Pfeil BE, Palmer JD, Doyle JJ (2009) Relationships among phaseoloid legumes based on sequences from eight chloroplast regions. Syst Bot 34:115–128. https://doi.org/10.1600/036364409787602221
Tian S, Lu P, Zhang Z, Wu JQ, Zhang H, Shen H (2021) Chloroplast genome sequence of Chongming lima bean (Phaseolus lunatus L.) and comparative analyses with Other legume chloroplast genomes. BMC Genom 22:194. https://doi.org/10.1186/s12864-021-07467-8
Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R and Greiner S (2017) GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res 45:W6–W11. https://doi.org/10.1093/nar/gkx391
Van Wyk B-E (2003) The value of chemosystematics in clarifying relationships in the Genistoid tribes of papilionoid legumes. Biochem Syst Ecol 31:875–884. https://doi.org/10.1016/S0305-1978(03)00083-8
Vatanparast M, Powell A, Doyle JJ, Egan AN (2018) Targeting legume loci: a comparison of three methods for target enrichment bait design in Leguminosae phylogenomics. APPS 6:e1036. https://doi.org/10.1002/aps3.1036
Verdú M (2002) Age at maturity and diversification in woody angiosperms. Evol 56:1352–1361. https://doi.org/10.1111/j.0014-3820.2002.tb01449.x
Wade BS, Pälike H (2004) Oligocene climate dynamics. Paleoceanography. https://doi.org/10.1029/2004PA001042
Wicke S, Schneeweiss GM, Pamphilis CW, Muller KF, Quandt D (2011) The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol 76:273–297. https://doi.org/10.1007/s11103-011-9762-4
Wink M, Mohamed GIA (2003) Evolution of chemical defense traits in the Leguminosae: mapping of distribution patterns of secondary metabolites on a molecular phylogeny inferred from nucleotide sequences of the rbcL gene. Biochem Syst Ecol 31:897–917. https://doi.org/10.1016/S0305-1978(03)00085-1
Wojciechowski MF (2003) Reconstructing the phylogeny of legumes (Leguminosae): an early 21st century perspective. In: Klitgaard BB, Bruneau A (eds) Advances in legume systematics, part 10, Royal Botanic Gardens, Kew, England, pp 5–35
Wojciechowski MF (2013) Towards a new classification of Leguminosae: naming clades using non-Linnaean phylogenetic nomenclature. S Afr J Bot 89:85–93. https://doi.org/10.1016/j.sajb.2013.06.017
Wojciechowski MF, Lavin M, Sanderson MJ (2004) A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am J Bot 11:1846–1862. https://doi.org/10.3732/ajb.91.11.1846
Yang Z, Wang G, Ma Q, Liang L, Zhao T (2019) The complete chloroplast genomes of three Betulaceae species: implications for molecular phylogeny and historical biogeography. PeerJ 7:e6320. https://doi.org/10.7717/peerj.6320
Zachos J, Pagani M, Sloan L, Thomas E, Billups K (2001) Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 292:686–693. https://doi.org/10.1126/science.10594
Zha X, Wang X, Li J, Gao F, Zhou Y (2020) Complete chloroplast genome of Sophora alopecuroides (Papilionoideae): molecular structures, comparative genome analysis and phylogenetic analysis. J Genet 99:13. https://doi.org/10.1007/s12041-019-1173-3
Zhang R, Wang YH, Jin JJ, Stull GW, Bruneau A, Cardoso D, Queiroz LP, Moore MJ, Zhang SD, Chen SY, Wang J, Li DZ, Yi TS (2020) Exploration of plastid phylogenomic conflict yields new insights into the deep relationships of leguminosae. Syst Biol 4:613–622. https://doi.org/10.1093/sysbio/syaa013
Zhao Y, Zhang R, Jiang K-W, Qi J, Hu Y, Guo J, Zhu R, Zhang T, Egan AN, Yi TS, Huang CH, Ma H (2021) Nuclear phylotranscriptomics and phylogenomics support numerous polyploidization events and hypotheses for the evolution of rhizobial nitrogen fixing symbiosis in Fabaceae. Mol Plant 14:1–26. https://doi.org/10.1016/j.molp.2021.02.006w
Funding
This work was supported by the Coordination for the Improvement of Higher Education Personnel—Brazil (CAPES)—Financing Code 001. Author FG Silva was supported by a CAPES Doctoral Scholarship, RM Moraes Filho was supported by a CAPES Postdoctoral Scholarship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
da Silva, F.G., de Moraes Filho, R.M., Martins, L.S.S. et al. Plastid marker-based phylogeny reveals insights into relationships among Papilionoideae species. Genet Resour Crop Evol 71, 439–454 (2024). https://doi.org/10.1007/s10722-023-01637-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-023-01637-1