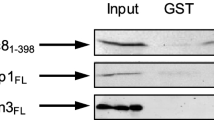
Summary
In the yeast Kluyveromyces lactis the β-galactosidase gene is induced by lactose or galactose. As shown here it can also be repressed by glucose but only in some strains. When the LAC9 gene of a repressible strain is substituted by an allele of a non-repressible strain, the β-galactosidase gene is no longer glucose repressed. LAC9 codes for a regulatory protein homologous to GAL4 which activates transcription in the presence of the inducer. Since the LAC9 product is also present in the repressed strain and binds to DNA in vitro, as shown by DNA footprinting, glucose repression cannot be caused by repression of LAC9 gene expression. Instead, our results demonstrate that glucose repression is mediated by the LAC9 gene product, and is separable from the ability of LAC9 to activate transcription.
Similar content being viewed by others
References
Bianchi MM, Falcone C, Chen XJ, Wésolowski-Louvel M, Frontali L, Fukuhara H (1987) Transformation of the yeast Kluyveromyces lactis by new vectors derived from the 1.6 μm circular plasmid pKD1. Curr Genet 12:185–192
Botstein D, Falco S, Stewart S, Brennan M, Scherer S, Stinchcomb D, Struhl K, Davis R (1979) Sterile host yeast (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene 8:17–24
Breunig KD (1989) Multicopy plasmids containing the gene for the transcriptional activator LAC9 are not tolerated by Kilactis cells. Curr Genet, in press
Breunig KD, Kuger P (1987) Functional homology between the yeast regulatory proteins GAL4 and LAC9: LAC9-mediated transcriptional activation in Kluyveromyces lactis involves protein binding to a regulatory sequence homologous to the GAL4 protein-binding site. Mol Cell Biol 7:4400–4406
Breunig KD, Dahlems U, Das S, Hollenberg CP (1984) Analysis of a eukaryotic β-galactosidase gene: the N-terminal end of the yeast Kluyveromyces lactis protein shows homology to the Escherichia coli lacZ gene product. Nucleic Acids Res 12:2327–2342
Chen XJ, Saliola M, Falcone C, Bianchi MM, Fukuhara H (1986) Sequence organization of the circular plasmid pKD1 from the yeast Kluyveromyces drosophilarum. Nucleic Acids Res 14:4471–4481
Das S, Hollenberg CP (1982) A high frequency transformation system for the yeast Kluyveromyces lactis. Curr Genet 6:123–128
Das S, Breunig KD, Hollenberg CP (1985) A positive regulatory element is involved in the induction of the β-galactosidase gene from Kluyveromyces lactis. EMBO J 4:793–798
Dickson RC, Markin JS (1980) Physiological studies of β-galactosidase induction in Kluyveromyces lactis. J Bacteriol 142:777–785
Ferrero I, Rossi C, Landini MP, Puglisi PP (1978) Role of the mitochondrial protein synthesis in the catabolite repression of the petite-negative yeast K. lactis. Biochem Biophys Res Commun 80:340–348
Gill G, Ptashne M (1988) Negative effect of the transcriptional activator GAL4. Nature 334:721–724
Giniger E, Varnum SM, Ptashne M (1985) Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell 40:767–774
Johnston M (1987) A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev 51:458–476
Khandjian EW (1987) Optimized hybridization of DNA blotted and fixed to nitrocellulose and nylon membranes. Biotechnology 5:165–167
Klebe LJ, Harris JV, Sharp ZD, Douglas MJ (1983) A general method for polyethylene-glycol-induced genetic transformation of bacteria and yeast. Gene 25:333–341
Lohr D, Hopper JE (1985) The relationship of regulatory proteins and DNase I hypersensitive sites in the yeast GAL1-10 genes. Nucleic Acids Res 13:8409–8423
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275
Mas J, Celis E, Pina E, Brunner A(1974) Effect of oxygen, ergosteron and sterol precursors on the mating ability of Kluyveromyces lactis. Biochem Biophys Res Commun 61:613–620
Matsumoto K, Yoshimatsu T, Oshima Y (1983) Recessive mutations conferring resistance to carbon catabolite repression of galactokinase synthesis in Saccharomyces cerevisiae. J Bacteriol 153:1405–1414
Miller J (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Orr-Weaver TL, Szostak JW (1983) Yeast recombination: The association between double-strand gap repair and crossing-over. Proc Natl Acad Sci USA 80:4417–4421
Post-Beittenmiller MA, Hamilton RW, Hopper JE (1984) Regulation of basal and induced levels of the MEL1 transcript in Saccharomyces cerevisiae. Mol Cell Biol 4:1238–1245
Riley MI, Hopper JE, Johnston SA, Dickson RC (1987a) GAL4 of Saccharomyces cerevisiae activates the lactose-galactose regulon of Kluyveromyces lactis and creates a new phenotype: glucose repression of the regulon. Mol Cell Biol 7:780–786
Riley MI, Sreekrishna K, Bhairi S, Dickson RC (1987b) Isolation and characterization of mutants of Kluyveromyces lactis defective in lactose transport. Mol Gen Genet 208:145–151
Ruzzi M, Breunig KD, Ficca AG, Hollenberg CP (1987) Positive regulation of the β-galactosidase gene from Kluyveromyces lactis is mediated by an upstream activation site that shows homology to the GAL upstream activation site of Saccharomyces cerevisiae. Mol Cell Biol 7:991–997
Salmeron JM, Johnston SA (1986) Analysis of the Kluyveromyces lactis positive regulatory gene LAC9 reveals functional homology to but sequence divergence from the Saccharomyces GAL4 gene. Nucleic Acids Res 14:7767–7781
Selleck SB, Majors JM (1987) In vivo DNA-binding properties of a yeast transcription activator protein. Mol Cell Biol 7:3260–3267
Sheetz RM, Dickson RC (1980) Mutations affecting synthesis of β-galactosidase in Kluyveromyces lactis. Genetics 95:877–890
Sherman F, Fink GR, Hicks JB (1986) Laboratory course manual for methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Sreekrishna K, Dickson RC (1985) Construction of strains of Saccharomyces cerevisiae that grow on lactose. Proc Natl Acad Sci USA 82:7909–7913
Struhl K (1985) Negative control at a distance mediates catabolite repression in yeast. Nature 317:822–824
Tschumper G, Carbon J (1980) Sequence of a yeast DNA fragment containing a chromosomal replicator and the TRP1 gene. Gene 10:157–166
Wesolowski M, Algeri A, Goffrini P, Fukuhara H (1982) Killer DNA plasmids of the yeast Kluyveromyces lactis. Curr Genet 5:191–197
Wickerham LJ, Burton KA (1952) Occurrence of yeast mating types in nature. J Bacteriol 63:449–451
Wray LV, Witte MM, Dickson RC, Riley MI (1987) Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol Cell Biol 7:1111–1121
Author information
Authors and Affiliations
Additional information
Communicated by D.Y. Thomas
Rights and permissions
About this article
Cite this article
Breunig, K.D. Glucose repression of LAC gene expression in yeast is mediated by the transcriptional activator LAC9. Mol Gen Genet 216, 422–427 (1989). https://doi.org/10.1007/BF00334386
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF00334386