Abstract
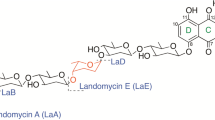
LanK is a TetR type regulatory protein that coordinates the late steps of the biosynthesis of the landomycin family of antitumor angucyclic polyketides and their export from the cells of Streptomyces cyanogenus S136. We recently described the structure of LanK and showed that it is the carbohydrate portion of the landomycins that is responsible for abrogating the repressing effect of LanK on landomycin production and export. The effect has been established in a series of in vitro tests using synthetic analogs of the landomycin carbohydrate chains. Whether such synthetic compounds would function as effector molecules for LanK under in vivo conditions remained unknown. Furthermore, the location and identity of LanK operator sites within the lanK-lanJ intergenic region (lanKJp) was unknown. Here we report that methoxyphenyl analogs of tri- and hexasaccharide chains of landomycins cannot function as LanK ligands when applied externally to the reporter strain. The lability of these compounds to cellular media and/or their poor penetration into the cells could explain our observations. The LanK operator site has been mapped to a 14-bp region of lanKJp that includes a plausible -35 site upstream of the lanK start codon in a series of electrophoretic DNA mobility shift assays. This opens the door to studies of the DNA-LanK interaction at a single nucleotide resolution level.



Similar content being viewed by others
Data Availability
All the data are present in the manuscript and Supplementary Materials. Original versions of all images will be provided by the corresponding author upon reasonable request.
Code Availability
Not applicable.
References
Ostash B, Korynevska A, Stoika R, Fedorenko V (2009) Chemistry and biology of landomycins, an expanding family of polyketide natural products. Mini Rev Med Chem 9:1040–1051. https://doi.org/10.2174/138955709788922593
Yushchuk O, Kharel M, Ostash I, Ostash B (2019) Landomycin biosynthesis and its regulation in Streptomyces. Appl Microbiol Biotechnol 103:1659–1665. https://doi.org/10.1007/s00253-018-09601-1
Panchuk RR, Lehka LV, Terenzi A, Matselyukh BP, Rohr J, Jha AK, Downey T, Kril IJ, Herbacek I, van Schoonhoven S, Heffeter P, Stoika RS, Berger W (2017) Rapid generation of hydrogen peroxide contributes to the complex cell death induction by the angucycline antibiotic landomycin E. Free Radic Biol Med 106:134–147. https://doi.org/10.1016/j.freeradbiomed.2017.02.024
Terenzi A, La Franca M, van Schoonhoven S, Panchuk R, Martinez A, Heffeter P, Gober R, Pirker C, Vician P, Kowol CR, Stoika R, Salassa L, Rohr J, Berger W (2021) Landomycins as glutathione-depleting agents and natural fluorescent probes for cellular Michael adduct-dependent quinone metabolism. Commun Chem 4:162. https://doi.org/10.1038/s42004-021-00600-4
Rebets Y, Dutko L, Ostash B, Luzhetskyy A, Kulachkovskyy O, Yamaguchi T, Nakamura T, Bechthold A, Fedorenko V (2008) Function of lanI in regulation of landomycin A biosynthesis in Streptomyces cyanogenus S136 and cross-complementation studies with Streptomyces antibiotic regulatory proteins encoding genes. Arch Microbiol 189:111–120. https://doi.org/10.1007/s00203-007-0299-5
Gessner A, Heitzler T, Zhang S, Klaus C, Murillo R, Zhao H, Vanner S, Zechel DL, Bechthold A (2015) Changing biosynthetic profiles by expressing bldA in Streptomyces strains. ChemBioChem 16(15):2244–2252. https://doi.org/10.1002/cbic.201500297
Yushchuk O, Ostash I, Vlasiuk I, Gren T, Luzhetskyy A, Kalinowski J, Fedorenko V, Ostash B (2018) Heterologous AdpA transcription factors enhance landomycin production in Streptomyces cyanogenus S136 under a broad range of growth conditions. Appl Microbiol Biotechnol 102:8419–8428. https://doi.org/10.1007/s00253-018-9249-1
Ostash I, Ostash B, Luzhetskyy A, Bechthold A, Walker S, Fedorenko V (2008) Coordination of export and glycosylation of landomycins in Streptomyces cyanogenus S136. FEMS Microbiol Lett 285(2):195–202. https://doi.org/10.1111/j.1574-6968.2008.01225.x
Ostash B, Ostash I, Zhu L, Kharel MK, Luzhetskiĭ A, Bechthold A, Walker S, Rohr J, Fedorenko V (2010) Properties of lanK-based regulatory circuit involved in landomycin biosynthesis in Streptomyces cyanogenus S136. Genetika 46(5):604–609. https://doi.org/10.1134/S1022795410050030
Tsugita A, Uehara S, Matsui T, Yokoyama T, Ostash I, Deneka M, Yalamanchili S, Bennett CS, Tanaka Y, Ostash B (2022) The carbohydrate tail of landomycin A is responsible for its interaction with the repressor protein LanK. FEBS J. https://doi.org/10.1111/febs.16460
Yalamanchili S, Lloyd D, Bennett CS (2019) Synthesis of the Hexasaccharide fragment of Landomycin A using a mild, reagent-controlled approach. Org Lett 21:3674–3677. https://doi.org/10.1021/acs.orglett.9b01118
Gromyko O, Rebets Y, Ostash B, Luzhetskyy A, Fukuhara M, Bechthold A, Nakamura T, Fedorenko V (2004) Generation of Streptomyces globisporus SMY622 strain with increased landomycin E production and it’s initial characterization. J Antibiot 57(6):383–389. https://doi.org/10.7164/antibiotics.57.383
Henkel T, Rohr J, Beale JM, Schwenen L (1990) Landomycins, new angucycline antibiotics from streptomyces sp. I. structural studies on landomycins A–D. J Antibiot 43(5):492–503. https://doi.org/10.7164/antibiotics.43.492
Overend WG, Rees CW, Sequeira JS (1962) Reactions at position 1 of carbohydrates. Part III. The acid-catalyzed hydrolysis of glycosides. J Chem Soc. https://doi.org/10.1039/JR9620003429
Kirschning A, Becthold AEW, Rohr J (1998) Chemical and biochemical aspects of deoxysugars and deoxysugar oligosaccharides Top. Curr Chem 188:1–84. https://doi.org/10.1007/BFb0119234
Yu Z, Reichheld SE, Savchenko A, Parkinson J, Davidson AR (2010) A comprehensive analysis of structural and sequence conservation in the TetR family transcriptional regulators. J Mol Biol 400:847–864. https://doi.org/10.1016/j.jmb.2010.05.062
Hemmerling F, Piel J (2022) Strategies to access biosynthetic novelty in bacterial genomes for drug discovery. Nat Rev Drug Discov 21:359–378. https://doi.org/10.1038/s41573-022-00414-6
Cimermancic P, Medema MH, Claesen J, Kurita K, Wieland Brown LC, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J, Birren BW, Takano E, Sali A, Linington RG, Fischbach MA (2014) Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158:412–421. https://doi.org/10.1016/j.cell.2014.06.034
Olano C, García I, González A, Rodriguez M, Rozas D, Rubio J, Sánchez-Hidalgo M, Braña AF, Méndez C, Salas JA (2014) Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb Biotechnol 7:242–256. https://doi.org/10.1111/1751-7915.12116
Grkovic S, Brown MH, Skurray RA (2002) Regulation of bacterial drug export systems. Microbiol Mol Biol Rev 66:671–701. https://doi.org/10.1128/MMBR.66.4.671-701.2002
Funding
The work was supported by grant BG-21F from the Ministry of Education and Science of Ukraine (to B.O.).
Author information
Authors and Affiliations
Contributions
Conceptualization: BO, Methodology: IO, SY, Formal analysis and investigation: MD, BO, Writing—original draft preparation: BO, MD, Writing—review and editing: BO, CB, Funding acquisition: BO, Resources: SY, IO, CB, Supervision: BO.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Deposition into repositories
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Deneka, M., Ostash, I., Yalamanchili, S. et al. Insights into the Biological Properties of Ligands and Identity of Operator Site for LanK Protein Involved in Landomycin Production. Curr Microbiol 81, 5 (2024). https://doi.org/10.1007/s00284-023-03528-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03528-1